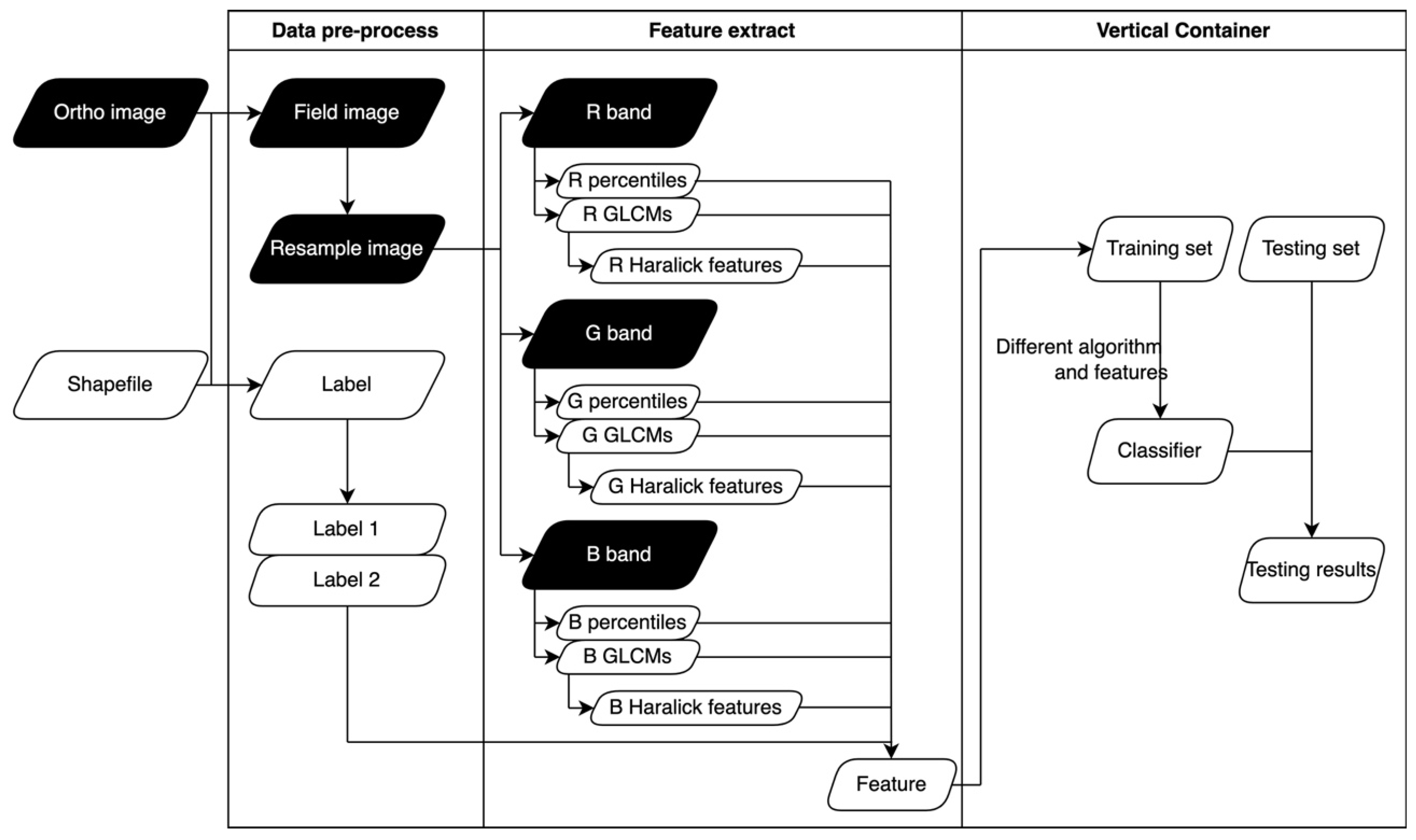
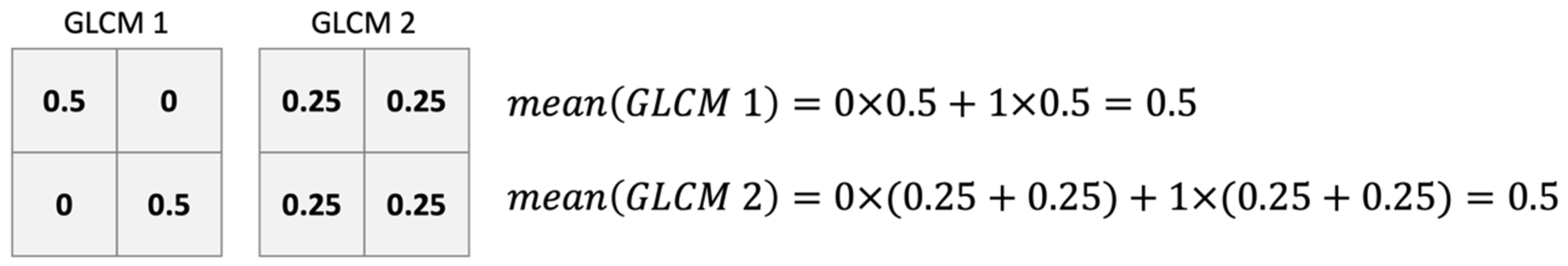1. Introduction
Using information about the types of crops cultivated in a region and the specific growing conditions to generate “crop maps” has long been a crucial tool for policy-making in agricultural organizations and governmental agencies. In Taiwan, like many other Southeast Asian countries, agricultural land typically comprises small fields, each less than 1 hectare in size, where various crops are grown simultaneously. At the same time, since each field may belong to a different farmer, even if the same crop type is grown, there may be differences in growth stages and appearance due to differences in planting time, cultivar, or management. Conducting detailed crop surveys in such a landscape poses significant challenges for agricultural agencies. Currently, Taiwan’s agricultural authorities rely heavily on manual surveys conducted by trained personnel, including local farmers and local agricultural officers [
1]. However, this approach has notable drawbacks, including high training costs, a high turnover rate, prolonged survey durations, and the potential for inaccurate record-keeping. Recognizing the limitations of conventional crop surveys, researchers are increasingly turning to remote sensing technology, specifically drone imagery, to generate crop maps. Previous research shows that this alternative method offers advantages such as reduced costs, higher resolution, and faster survey speeds [
2,
3,
4]. In addition to the traditional single-time image, time-series image collection and analysis are also important due to the change in appearance of crops as they grow over time [
5].
Following the capture of drone imagery, a systematic image analysis protocol is essential to extract information for policy-making purposes. Typically, the initial steps involve filtering, orthorectifying, and stitching together the images. Subsequently, key data, such as specific vegetative indices (e.g., normalized difference vegetative index, NDVI), are extracted and modeled alongside ground truth data collected in the field. In the context of image analysis, the conversion of images into representative “features” is a common practice. This not only simplifies the complexity of image data but also eliminates noise and enhances the information. Predominantly, color and texture features are utilized in agricultural image analyses [
3,
6]. Color features describe the light intensity collected by sensors, often represented by band statistics or combinations, such as vegetation indices (VIs), commonly used in agricultural tasks [
7,
8]. For instance, a previous study applied various RGB-based VIs to high-throughput phenotyping of forage grasses, achieving an R
2 of 0.73 for predicting the breeder score [
9]. Another research effort utilized the percentile of infrared images to detect tea diseases and count lesions on tea leaves, achieving an impressive R
2 of 0.97 compared to lesion counting through human observation [
10].
Texture features act as descriptors for pixel positions and spatial relationships within an image, offering insights into the surface texture of the subject. Among all of the texture features, the Haralick texture feature stands out, relying on the gray-level co-occurrence matrix (GLCM) [
11]. Specifically, GLCM is a
K ×
K matrix that records the combinations of neighboring pixels with different brightness levels, also called gray levels, at a given distance and angle in the grayscale image with
K level of brightness [
12,
13]. Typically, one or a few summary statistics, also called the Haralick feature, summarize the GLCM for ease of interpretation [
11]. The Haralick features would then be combined with machine learning algorithms to build classifiers or prediction models.
In a previous study, the Haralick feature, combined with color features, was utilized to classify land use in multispectral images, resulting in improved accuracy of the support vector machine (SVM) classifier by up to 7.72%. Notably, the Haralick feature demonstrated increased relevance when spectral information was limited [
3]. Similarly, in another previous study about crop classification by high-resolution drone RGB images, the authors compared the use of the Haralick feature or grayscale image with four different machine learning algorithms, namely, Random Forest (RF), Naive Bayes (NB), Neural Network (NN), and Support Vector Machine (SVM), to build a classification model. The results showed that the RF-based classifier with a GLCM image can achieve an overall accuracy of 90.9% [
14]. Furthermore, a study focusing on wheat Fusarium head blight detection achieved an impressive 90% accuracy using the Haralick texture feature, underscoring the importance of an appropriate GLCM kernel size [
15]. These investigations collectively underscore the unique information offered by texture features, showing their potential to enhance classification and prediction tasks related to plant images.
Even though most studies utilized the summary statistics of GLCMs (usually referred to as Haralick features) for subsequent classification applications, we argue that GLCMs with identical summary statistics may possibly possess distinct structures upon revisiting the structure of the GLCM. This implies that selecting representative summary statistics for the GLCM could be pivotal for successful classification. While it is possible to address this by selecting multiple summary statistics simultaneously by incorporating the standard deviation of the GLCM, a broader perspective suggests the direct utilization of the GLCM itself. Considering the inherent risk of information loss associated with any summary statistics, relying solely on the complete probability distribution might best represent all facets of data characteristics. Several methods exist for calculating the empirical probability distribution.
On the other hand, the selection of machine learning models was also a critical issue, which may greatly affect the accuracy of the final prediction result. Previous work includes a comprehensive review about the machine learning or deep learning techniques applied to predict field images obtained from satellites or UAVs. Deep learning models based on Convolution Neural Network (CNN) technology have achieved good accuracy and become the most widely used algorithms [
16]. However, other algorithms might be utilized with different considerations. For example, SVM might have higher performance on some occasions [
3,
16,
17]. Classification and regression tree (CART) and other conventional decision tree-based models were easy to interpret with tree-diagram structures [
16]. The models based on ensemble learning, such as Extreme Gradient Boosting (XGBoost), have started to attract more attention due to their exceptional performance in complex systems [
16,
18].
Continuing the above argument, this study proposes to use the entire GLCM empirical distribution as the characteristics of the field to classify the fields. Our study used multitemporal visible-light RGB images taken in agricultural areas of Taiwan and captured by UAV. Since the spectral information that RGB images can provide is less than that of multispectral images, we wish to combine the texture and spectral features to extract more information from the images. A GLCM-based feature extraction process is developed in this study, and the effects of using a GLCM versus Haralick features as additional features were examined for the task of feature recognition. This study also evaluates the performance of three algorithms, CART, SVM, and XGBoost, in field classification with the selected features.
3. Results
3.1. Feature Extraction
Table 4 shows the percentiles of the R, G, and B bands for each category, and
Table 5 shows the time cost of extracting features in each image date and category. Due to the many percentiles and combinations of bands and angles, only the 0%, 50%, and 100% percentiles for each category and band are shown.
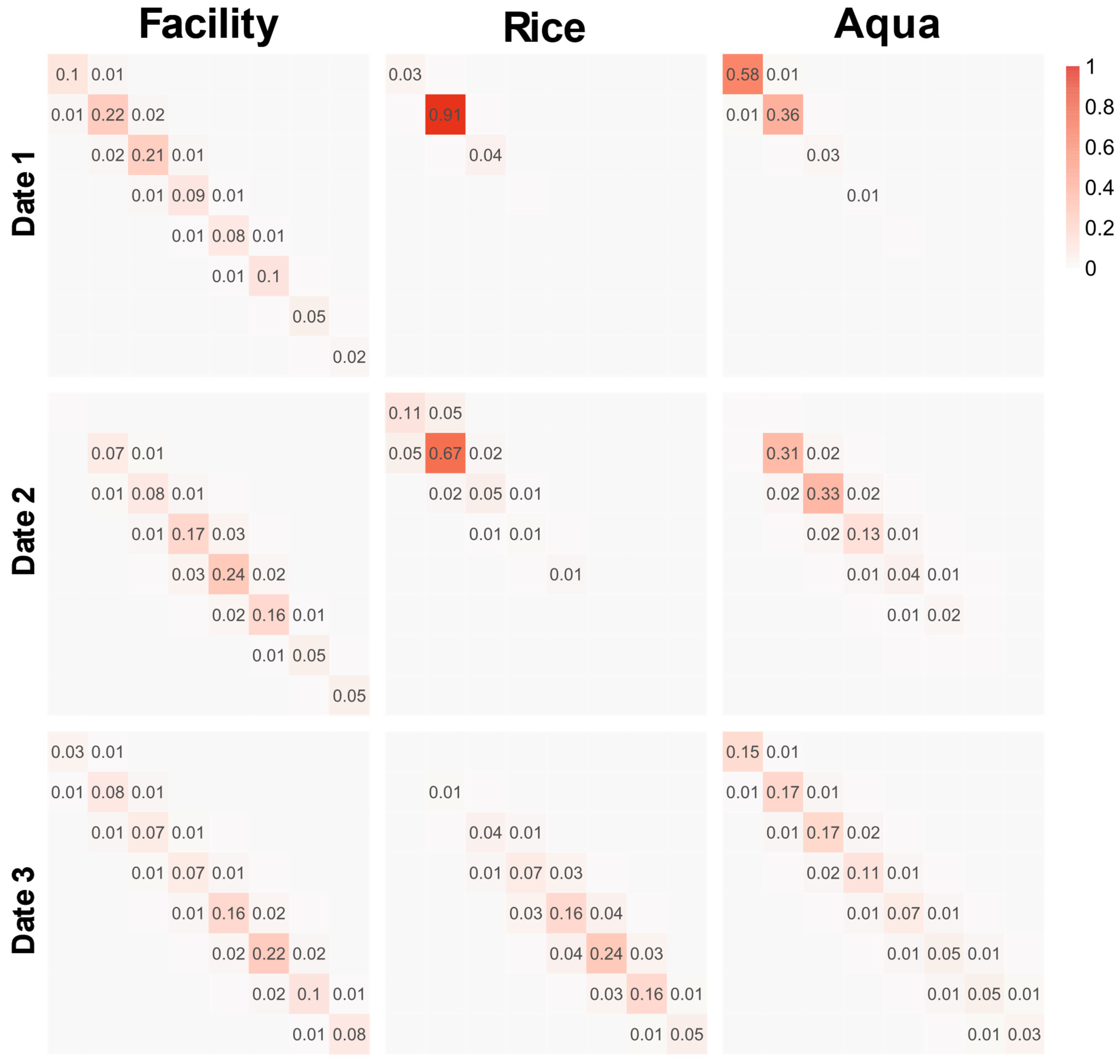
Figure 8 shows the GLCM for Facility, Rice, and Aqua on 2019/12, 2020/03, and 2020/06. Due to the vast number of possible combinations of angles, dates, bands, and categories, the plot only displays the average GLCM for all four angles and three bands. Additionally, only three categories deemed significant for discussion have been selected for display.
The values of the R and G bands shown in
Table 4 were similar, with the differences at the three percentiles being 2.47, 2.18, and 0.05. However, the values of the B band were relatively lower, and the maximum differences to the other two bands at each percentile were 8, 14.1, and 15.32. Nevertheless, such a relationship does not necessarily exist when observing each category. For example, the 0% percentile values of Aqua were 23.2, 27.58, and 25.01, where the G band had the highest value. However, the 100% percentile values of Facility were 167.64, 110.22, and 170.42, where the R band had the highest value. In addition, certain relationships existed among the percentile values of each category. For example, the 100% percentile values of Aqua were 56.92, 60.96, and 57.39, which were all lower than the 100% percentile values of the other categories, which were all higher than 100. Another example was the 100% percentile values of Facility, which were 167.64, 169.58, and 170.42, which were the highest. The percentile value of different categories shows different patterns, and these patterns can be used for classification in a machine learning model.
The GLCM shown in
Figure 8 was concentrated on the diagonal line of the matrix irrespective of the category. The main difference among the categories was the concentration trend and the location of the maximum probability obtained. For example, on 2019/12, the GLCM of Rice showed a high concentration at (2, 2), while the other two categories displayed a more scattered pattern. In addition, the GLCM of the same category may vary considerably at different times. Again, in the case of Rice, for example, there is an obvious difference in the GLCMs for the three dates. In contrast, there is not much difference in the GLCMs for Facility on the three dates.
3.2. Overall Classification Results
Table 6 shows the OA and the time spent on model training and testing of six classifiers constructed by the combination of three algorithms and two labeling methods. Because the XGBoost algorithm allows training of the models by parallel computing, the CPU time spent on training, which means the sum of computing resources consumed by each CPU core, was also recorded.
The relation among the overall classification accuracies of the six classifiers was XGBoost with label 2 > XGBoost with label 1 > SVM with label 2 > SVM with label 1 > CART with label 2 > CART with label 1. The classifier with the XGBoost algorithm and label 2 had the highest OA of 0.82, followed by the classifier with the XGBoost algorithm and label 1. The worst-performing classifier used CART and label 1 and had an OA of only 0.527.
In terms of time spent on model training, the classifier with the CART algorithm and using label 1 took 19.22 s, which was the shortest time. In comparison, the classifier with the SVM algorithm and using label 1 took 590.67 s, which was the longest time. For the two tree-based algorithms CART and XGBoost, when label 2 was used, because the number of categories increased, the training time also increased by about 50–80%. However, for the SVM classifier, when using label 2, the training time was reduced by 30%, which showed a different trend from the other two classifiers. In terms of CPU time spent on training, the classifier with XGBoost and label 2 consumed the most computing resources, taking 3109.56 s.
In terms of time spent on model testing, the difference between the classifiers with the same algorithm but using different labels was relatively minor. The SVM classifiers took 37.28 and 36.77 s for the two labels. For the CART and XGBoost classifiers, the testing time consumed was relatively lower and was about 0.1 s, which was about 0.2% of what the SVM classifiers took.
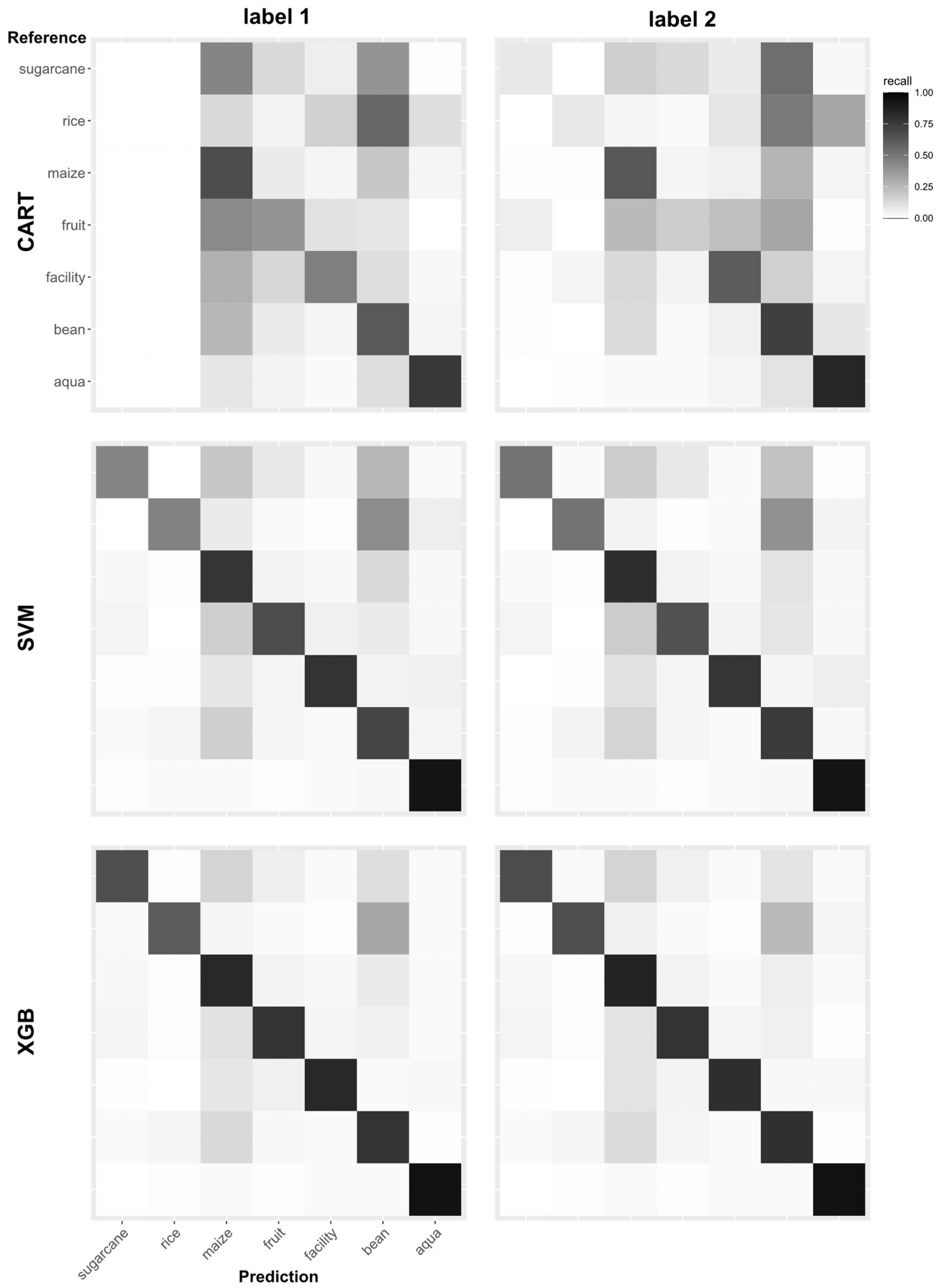
3.3. Classification Results for Each Category
Figure 9 and
Table 7 show the testing results of the six classifiers by confusion matrix and accuracy, respectively. Due to mismatch in the size of testing data across categories, balance accuracy was used in
Table 7 for more accurate evaluation.
The confusion matrixes show that none of the data were predicted in the categories Rice and Sugarcane of the classifier with CART and label 1, which means that the classifier ignores these two categories during model training and misclassifies those data into other categories. Although those two categories were predicted by the classifier with CART and label 2, it does not achieve comparable accuracy to the other categories. In the classification results of the classifiers with SVM and XGBoost, the problem that some categories were ignored was not observed. The confusion matrixes of XGBoost were highly concentrated on the diagonal line, which indicates that the classification was performed well. The results of each category for the same classifier were different in the confusion matrixes. For example, Aqua showed high accuracies for all classifiers while Rice and Maize performed poorly.
Table 7 also shows that the balance accuracy of Aqua for the classifier with XGBoost and label 2 reaches the highest value of 0.97, and the classifier with XGBoost and label 1 reaches the second-highest value of 0.96. The categories Rice and Sugarcane with CART and label 1 both had the lowest balance accuracy of 0.5, which means that all of the data belonging to Rice or Sugarcane are predicted as other categories, as in the confusion matrix. When observing results within a category, the classifiers with the XGBoost algorithm obtained the most accurate prediction results, followed by those with SVM and CART. For each classifier, model building using the XGBoost algorithm obtained the closest balance accuracy among the categories, where the maximum difference between the best and the worst performance was only 0.15. In contrast, the classifier with the CART algorithm had the largest accuracy difference among the categories, which reached 0.36.
Irrespective of the algorithm applied, the OA of the classifiers trained with label 2 was better than that of classifiers trained with label 1, and the testing OA was improved by 1–5%. For the classifiers with the XGBoost and SVM algorithms, label 2 improved the balance accuracy by 1–5% for each category. However, for the classifiers with the CART algorithm, label 2 reduced the accuracy of Fruit by 9% but improved the accuracies of the other categories by 3–7%.
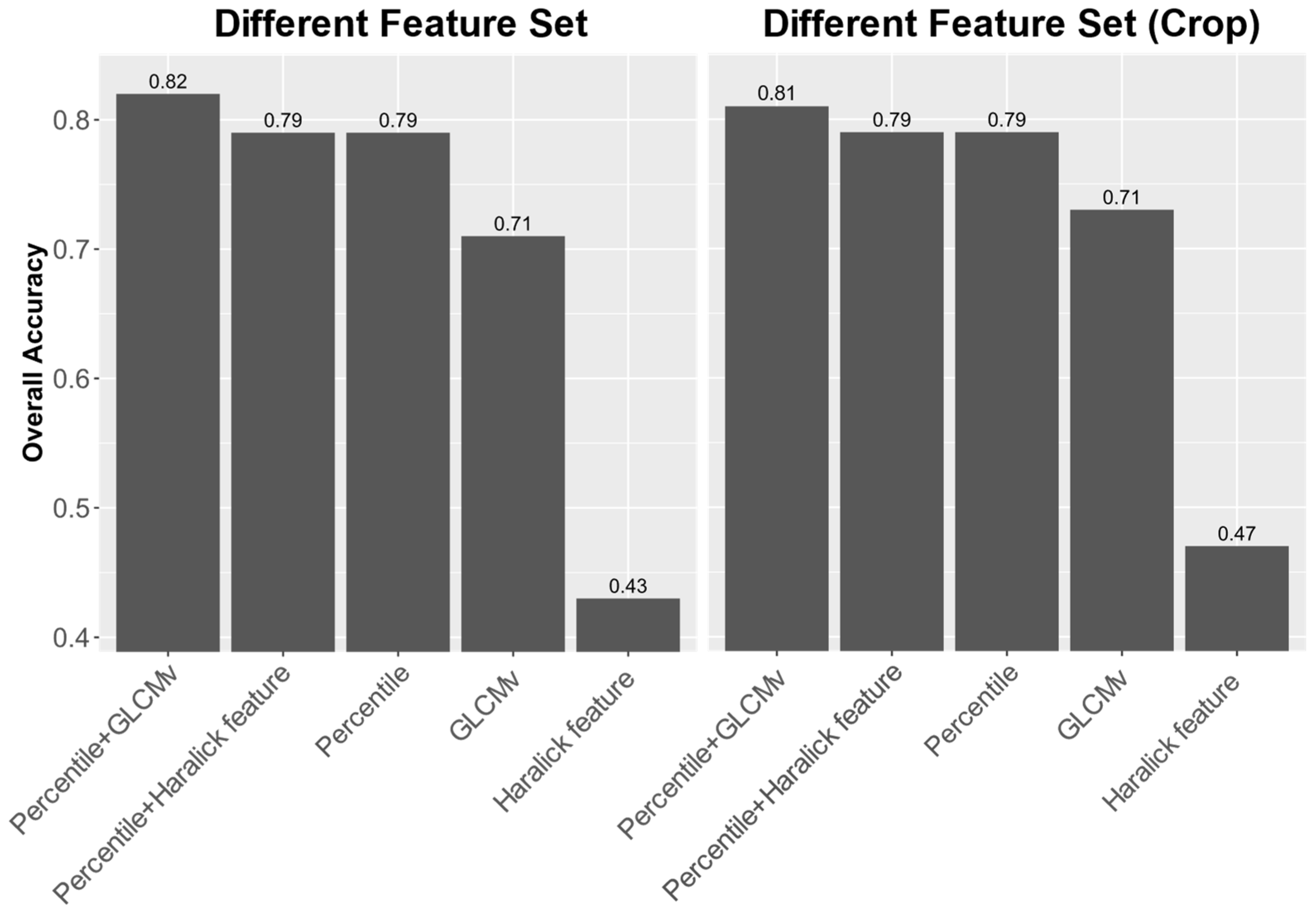
3.4. Classification Results for GLCMv, Haralick Feature, and Percentile
The left part of
Figure 10 shows the testing accuracy when different feature combinations were used. As discussed in
Section 2.3.2, the percentiles, GLCMv, and Haralick features (mean and standard deviation of the GLCM) of each band were extracted and combined differently. Five feature sets, namely Haralick feature, GLCMv, percentiles, percentiles + Haralick feature, and percentiles + GLCMv, were used to test their contribution to the classification problem. The algorithm and label used in the test were XGBoost and label 2, respectively, to ensure the best performance. The highest accuracy was obtained using percentiles + GLCMv, the same as the result in
Section 3.2, with a total accuracy of 0.82. The classifiers using only percentiles and percentiles + Haralick feature showed the second-highest accuracy (0.79), while the accuracies of using only GLCMv and Haralick feature were 0.71 and 0.43, respectively. The result shows an improvement of about 3.6% on overall accuracy when adding GLCMv to the classifier. We used McNemar’s test [
29] to compare the prediction results of percentiles, and percentiles + GLCMv. The
p-value was less than 10
−15, indicating a significant difference between the two results.
To understand the effect of features on the classification of different crops, the categories Aqua and Facility, which did not belong to crops, were removed to observe the classification accuracy of different crop types only. The results are shown in the right part of
Figure 10. The accuracy trend among the feature sets was the same as in the left part of the figure, which means that the classification results were not highly affected by Aqua and Facility, and this system can perform well on different types of crops. The OA of using the feature set percentiles + GLCMv dropped slightly to 0.81, while the accuracies of using only the GLCMv and Haralick feature increased a little to 0.73 and 0.47, respectively. The percentage of improvement and the result of McNemar’s test were the same as above.
5. Conclusions
In this investigation, we employ high-resolution and multitemporal visible-light RGB UAV imagery for analysis. By integrating the texture and color features and utilizing machine learning classification algorithms, we develop an effective multiclass classification model to discern Taiwan’s fragmented and diverse agricultural lands. The most successful model attains an overall accuracy of up to 82%. Exploiting the repetitive patterns within the field images allows us to adapt the feature extraction process through resampling, addressing the unequal number of samples, and facilitating the use of GLCMv as a texture feature.
Advancements in machine learning algorithms have enabled us to achieve enhanced classification accuracy using the same features. The XGBoost algorithm, rooted in ensemble learning, surpasses its traditional CART or SVM counterparts in accuracy while exhibiting shorter training times and rapid recognition capabilities. Regarding image features, we utilize the percentiles of the three RGB bands as color features for distinguishing various land-use types. Examining texture differences among land use types reveals that employing GLCMv as the texture feature yields more informative results, leading to improved classification accuracy compared to using the Haralick feature. Leveraging more powerful machine learning models and features containing richer information, such as GLCMv, while entrusting the task of information extraction to the machine learning model, is a preferable choice for enhancing model accuracy.
Concerning the temporal effect, our observations indicate that temporal disparities in crop types were crucial in influencing classification outcomes. This underscores the need for a more nuanced consideration of temporal differences in future analyses, where temporal variations in crop imagery will be a focal point in our ongoing research. Moving forward, we aim to delve into temporal and spatial variations more comprehensively. Simultaneously, we aspire to leverage the features of GLCMs to furnish analyzable information on texture differences that can serve as indices for tracking such variations.