Spectral Signatures of Macroalgae on Hawaiian Reefs
Abstract
1. Introduction
2. Materials and Methods
2.1. Spectral Reflectance
2.2. Data Processing and Analysis
3. Results
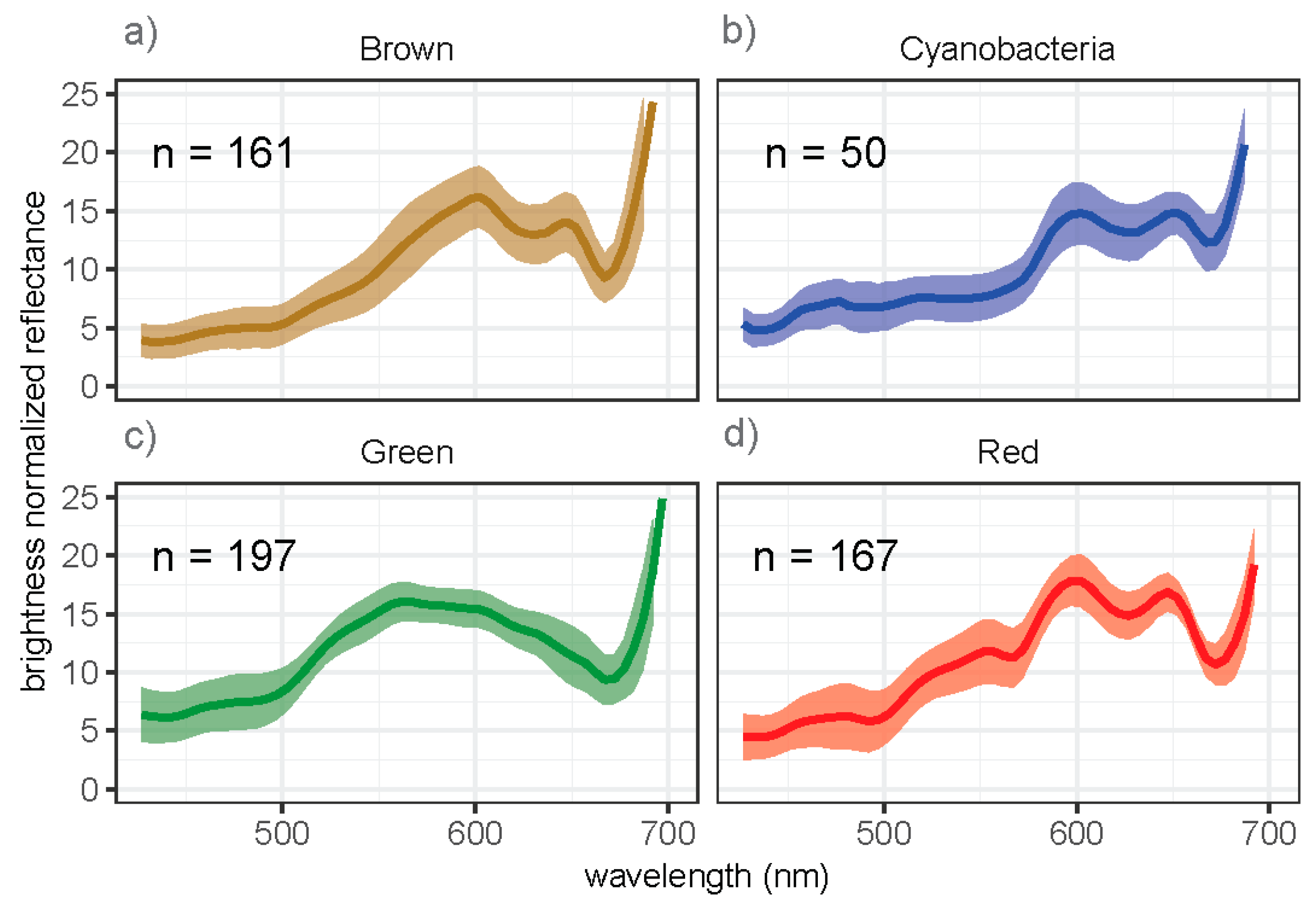
3.1. Spectral Signatures of Algae
3.2. Discriminant Analyses
3.3. Spectral Separability of Invasive and Native Algae
4. Discussion
4.1. Spectral Separability of Algae by Taxonomic Division
4.2. Spectral Separability of Algal Species
4.3. Spectral Separability of Native and Invasive Algae
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Division | Status | Island | Site | Genus | Species | Method | Number of Individuals |
|---|---|---|---|---|---|---|---|
| Brown | Native | Hawaiʻi | Miloliʻi | Sargassum | echinocarpum | ex situ | 23 |
| Brown | Native | Hawaiʻi | Miloliʻi | Sargassum | obtusifolium | ex situ | 24 |
| Brown | Native | Hawaiʻi | Miloliʻi | Turbinaria | ornata | ex situ | 24 |
| Brown | Native | Hawaiʻi | Papa | Chrysocystis | fragilis | in situ | 24 |
| Brown | Native | Hawaiʻi | Papa | Lobophora | variegata | in situ | 27 |
| Brown | Native | Hawaiʻi | Papa | Lobophora | variegata | ex situ | 3 |
| Brown | Native | Hawaiʻi | Papa | Padina | australis | ex situ | 1 |
| Brown | Native | Hawaiʻi | Papa | Sargassum | echinocarpum | ex situ | 1 |
| Brown | Native | Hawaiʻi | Papa | Sargassum | obtusifolium | ex situ | 1 |
| Brown | Native | Hawaiʻi | Papa | Turbinaria | ornata | ex situ | 1 |
| Brown | Native | Oʻahu | Maunalua | Dictyota | acutiloba | ex situ | 29 |
| Brown | Native | Oʻahu | Maunalua | Padina | australis | ex situ | 3 |
| Complex | Unknown | Hawaiʻi | Papa | Turf | spp. | in situ | 23 |
| Complex | Unknown | Hawaiʻi | Papa | Turf | spp. | ex situ | 6 |
| Cyanobacteria | Native | Hawaiʻi | Papa | Leptolyngbya | crosbyana | ex situ | 25 |
| Cyanobacteria | Native | Hawaiʻi | Papa | Symploca | hydnoides | in situ | 16 |
| Cyanobacteria | Native | Hawaiʻi | Papa | Symploca | hydnoides | ex situ | 9 |
| Green | Invasive | Oʻahu | Maunalua | Avrainvillea | lacerata | ex situ | 32 |
| Green | Native | Hawaiʻi | Miloliʻi | Chaetomorpha | antennina | ex situ | 5 |
| Green | Native | Hawaiʻi | Papa | Caulerpa | taxifolia | ex situ | 4 |
| Green | Native | Hawaiʻi | Papa | Halimeda | opuntina | in situ | 22 |
| Green | Native | Hawaiʻi | Papa | Halimeda | opuntina | ex situ | 2 |
| Green | Native | Oʻahu | Kāneʻohe | Dictyosphaeria | cavernosa | in situ | 25 |
| Green | Native | Oʻahu | Kāneʻohe | Dictyosphaeria | versluysii | in situ | 24 |
| Green | Native | Oʻahu | Kāneʻohe | Halimeda | discoidea | in situ | 25 |
| Green | Native | Oʻahu | Kāneʻohe | Dictyosphaeria | versluysii | ex situ | 6 |
| Green | Native | Oʻahu | Maunalua | Caulerpa | serularioides | ex situ | 43 |
| Green | Native | Oʻahu | Maunalua | Dictyosphaeria | cavernosa | ex situ | 3 |
| Green | Native | Oʻahu | Maunalua | Enteromorpha | prolifera | ex situ | 3 |
| Green | Native | Oʻahu | Maunalua | Halimeda | discoidea | ex situ | 3 |
| Red | Invasive | Oʻahu | Kāneʻohe | Acanthophora | spicifera | in situ | 25 |
| Red | Invasive | Oʻahu | Kāneʻohe | Eucheuma | spp. | in situ | 25 |
| Red | Invasive | Oʻahu | Kāneʻohe | Eucheuma | spp. | ex situ | 3 |
| Red | Invasive | Oʻahu | Kāneʻohe | Gracillaria | salicornia | ex situ | 3 |
| Red | Invasive | Oʻahu | Kāneʻohe | Kappaphycus | spp. | ex situ | 5 |
| Red | Invasive | Oʻahu | Maunalua | Acanthophora | spicifera | ex situ | 2 |
| Red | Invasive | Oʻahu | Maunalua | Gracillaria | salicornia | ex situ | 27 |
| Red | Native | Hawaiʻi | Miloliʻi | Anfeltiopsis | concinna | ex situ | 20 |
| Red | Native | Hawaiʻi | Papa | Anfeltiopsis | concinna | ex situ | 1 |
| Red | Native | Hawaiʻi | Papa | Dichotomaria | marginata | in situ | 19 |
| Red | Native | Hawaiʻi | Papa | Dichotomaria | marginata | ex situ | 3 |
| Red | Native | Hawaiʻi | Papa | Ramicrusta | hawaiiensis | in situ | 26 |
| Red | Native | Oʻahu | Kāneʻohe | Green CCA | spp_007 | ex situ | 1 |
| Red | Native | Oʻahu | Kāneʻohe | Red CCA | spp_006 | ex situ | 1 |
| Red | Unknown | Oʻahu | Kāneʻohe | Red Branching | spp_005 | ex situ | 3 |
| Red | Unknown | Oʻahu | Kāneʻohe | Red Filamentous | spp_004 | ex situ | 3 |


References
- Friedlander, A.; Aeby, G.; Brown, E.; Clark, A.; Coles, S.; Dollar, S.; Hunter, C.; Jokiel, P.; Smith, J.; Walsh, B.; et al. The State of Coral Reef Ecosystems of the Main Hawaiian Islands; University of Hawaii Press: Honolulu, HI, USA, 2005. [Google Scholar]
- Huisman, J.M.; Abbott, I.A.; Smith, C.M. Hawaiian Reef Plants; University of Hawaii Sea Grant College Program: Honolulu, HI, USA, 2007. [Google Scholar]
- Hart, G.M.; Ticktin, T.; Kelman, D.; Wright, A.D.; Tabandera, N. Contemporary Gathering Practice and Antioxidant Benefit of Wild Seaweeds in Hawai’i. Econ. Bot. 2014, 68, 30–43. [Google Scholar] [CrossRef]
- McDermid, K.J.; Martin, K.J.; Haws, M.C. Seaweed Resources of the Hawaiian Islands. Bot. Mar. 2019, 62, 443–462. [Google Scholar] [CrossRef]
- Smith, J.E.; Hunter, C.L.; Smith, C.M. Distribution and Reproductive Characteristics of Nonindigenous and Invasive Marine Algae in the Hawaiian Islands. Pac. Sci. 2002, 56, 299–315. [Google Scholar] [CrossRef]
- Vroom, P.; Page, K.; Kenyon, J.; Brainard, R. Algae-Dominated Reefs: Numerous Reports Suggest That Reefs Must Be Dominated by Coral to Be Healthy, but Many Thriving Reefs Depent More on Algae. Am. Sci. 2006, 94, 430–437. [Google Scholar] [CrossRef]
- Jouffray, J.-B.; Nyström, M.; Norström, A.V.; Williams, I.D.; Wedding, L.M.; Kittinger, J.N.; Williams, G.J. Identifying Multiple Coral Reef Regimes and Their Drivers across the Hawaiian Archipelago. Phil. Trans. R. Soc. B 2015, 370, 20130268. [Google Scholar] [CrossRef]
- Littler, M.M.; Littler, D.S. Models of Tropical Reef Biogenesis: The Contribution of Algae. Prog. Phycol. Res. 1984, 3, 323–364. [Google Scholar]
- Rasher, D.B.; Engel, S.; Bonito, V.; Fraser, G.J.; Montoya, J.P.; Hay, M.E. Effects of Herbivory, Nutrients, and Reef Protection on Algal Proliferation and Coral Growth on a Tropical Reef. Oecologia 2012, 169, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.E.; Hunter, C.L.; Smith, C.M. The Effects of Top–down versus Bottom–up Control on Benthic Coral Reef Community Structure. Oecologia 2010, 163, 497–507. [Google Scholar] [CrossRef]
- Conklin, E.J.; Smith, J.E. Abundance and Spread of the Invasive Red Algae, Kappaphycus Spp., in Kane’ohe Bay, Hawai’i and an Experimental Assessment of Management Options. Biol Invasions 2005, 7, 1029–1039. [Google Scholar] [CrossRef]
- Neilson, B.J.; Wall, C.B.; Mancini, F.T.; Gewecke, C.A. Herbivore Biocontrol and Manual Removal Successfully Reduce Invasive Macroalgae on Coral Reefs. PeerJ 2018, 6, e5332. [Google Scholar] [CrossRef]
- Foster, A.D.; Spalding, H.L.; Cox, T.E.; La Valle, F.F.; Philippoff, J. The Invasive Green Alga Avrainvillea Sp. Transforms Native Epifauna and Algal Communities on a Tropical Hard Substrate Reef. Phycol. Res. 2019, 67, 164–169. [Google Scholar] [CrossRef]
- Chao Rodríguez, Y.; Domínguez Gómez, J.A.; Sánchez-Carnero, N.; Rodríguez-Pérez, D. A Comparison of Spectral Macroalgae Taxa Separability Methods Using an Extensive Spectral Library. Algal Res. 2017, 26, 463–473. [Google Scholar] [CrossRef]
- Olmedo-Masat, O.M.; Raffo, M.P.; Rodríguez-Pérez, D.; Arijón, M.; Sánchez-Carnero, N. How Far Can We Classify Macroalgae Remotely? An Example Using a New Spectral Library of Species from the South West Atlantic (Argentine Patagonia). Remote Sens. 2020, 12, 3870. [Google Scholar] [CrossRef]
- Douay, F.; Verpoorter, C.; Duong, G.; Spilmont, N.; Gevaert, F. New Hyperspectral Procedure to Discriminate Intertidal Macroalgae. Remote Sens. 2022, 14, 346. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Pattaro, M.; Herrig Furlanetto, R.; Nanni, M.R.; Camargos Antunes, W. High Resolution Leaf Spectral Signature as a Tool for Foliar Pigment Estimation Displaying Potential for Species Differentiation. J. Plant Physiol. 2020, 249, 153161. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Jones, M.O.; Martin, R.E.; Knapp, D.E.; Hughes, R.F. Remote Sensing of Native and Invasive Species in Hawaiian Forests. Remote Sens. Environ. 2008, 112, 1912–1926. [Google Scholar] [CrossRef]
- Tsuda, R.T. Endemism of Marine Algae in the Hawaiian Islands. Bish. Mus. Occassional Pap. 2014, 115, 23–27. [Google Scholar]
- Carlton, J.T.; Eldredge, L.G. Marine Bioinvasions of Hawaiʻi: The Introduced and Cryptogenic Marine and Estuarine Animals and Plants of the Hawaiian Archipelago; Bishop Museum Bulletin in Cultural and Environmental Studies; Bernice P. Bishop Museum, Bishop Museum Press: Honolulu, HI, USA, 2009; ISBN 978-1-58178-096-3. [Google Scholar]
- Carlton, J.T.; Eldredge, L.G. Update and Revisions of The Marine Bioinvasions of Hawai‘i: The Introduced and Cryptogenic Marine and Estuarine Animals and Plants of the Hawaiian Archipelago; Bishop Museum Bulletin in Zoology; Bernice P. Bishop Museum, Bishop Museum Press: Honolulu, HI, USA, 2015; Volume 9, pp. 25–47. [Google Scholar]
- Department of Land and Natural Resources, (DLNR). State of Hawaii Aquatic Invasive Species Management Plan; Department of Land and Natural Resources: Honolulu, HI, USA, 2003; 205p. [Google Scholar]
- Li, J.; Schill, S.R.; Knapp, D.E.; Asner, G.P. Object-Based Mapping of Coral Reef Habitats Using Planet Dove Satellites. Remote Sens. 2019, 11, 1445. [Google Scholar] [CrossRef]
- Asner, G.P.; Vaughn, N.R.; Heckler, J.; Knapp, D.E.; Balzotti, C.; Shafron, E.; Martin, R.E.; Neilson, B.J.; Gove, J.M. Large-Scale Mapping of Live Corals to Guide Reef Conservation. Proc. Natl. Acad. Sci. USA 2020, 117, 33711–33718. [Google Scholar] [CrossRef]
- Asner, G.P.; Vaughn, N.; Foo, S.; Heckler, J.; Martin, R. Abiotic and Human Drivers of Reef Habitat Complexity Throughout the Main Hawaiian Islands. Front. Mar. Sci. 2021, 8, 631842. [Google Scholar] [CrossRef]
- Thompson, D.R.; Hochberg, E.J.; Asner, G.P.; Green, R.O.; Knapp, D.E.; Gao, B.-C.; Garcia, R.; Gierach, M.; Lee, Z.; Maritorena, S.; et al. Airborne Mapping of Benthic Reflectance Spectra with Bayesian Linear Mixtures. Remote Sens. Environ. 2017, 200, 18–30. [Google Scholar] [CrossRef]
- Kutser, T.; Hedley, J.; Giardino, C.; Roelfsema, C.; Brando, V.E. Remote Sensing of Shallow Waters–A 50 Year Retrospective and Future Directions. Remote Sens. Environ. 2020, 240, 111619. [Google Scholar] [CrossRef]
- Drury, C.; Martin, R.E.; Knapp, D.E.; Heckler, J.; Levy, J.; Gates, R.D.; Asner, G.P. Ecosystem-scale Mapping of Coral Species and Thermal Tolerance. Front. Ecol. Environ. 2022, 20, 285–291. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E.; Schmidtlein, S. Brightness-Normalized Partial Least Squares Regression for Hyperspectral Data. J. Quant. Spectrosc. Radiat. Transf. 2010, 111, 1947–1957. [Google Scholar] [CrossRef]
- SAS Institute Inc. JMP®, Version 16; SAS Institute Inc.: Cary, NC, USA, 2022. [Google Scholar]


| Division | Invasive | Native | Unknown | Total |
|---|---|---|---|---|
| Cyanobacteria | - | 2 (50) | - | 2 (50) |
| Red | 4 (90) | 5 (71) | 2 (6) | 11 (167) |
| Green | 1 (32) | 8 (165) | - | 9 (197) |
| Brown | - | 7 (161) | - | 7 (161) |
| Complex | - | - | 1(29) | 1(29) |
| Total | 5 (122) | 22 (447) | 3 (35) | 30 (604) |
| Source | Count | Number Misclassified | Percent Misclassified | Entropy R2 |
|---|---|---|---|---|
| Training | 345 | 1 | 0.3 | 0.95 |
| Validation | 115 | 4 | 3.5 | 0.23 |
| Test | 115 | 4 | 3.5 | 0.29 |
| Source | Count | Number Misclassified | Percent Misclassified | Entropy R2 |
|---|---|---|---|---|
| Training | 348 | 6 | 1.7 | 0.98 |
| Validation | 113 | 19 | 16.8 | −1.81 |
| Test | 114 | 19 | 16.7 | −4.40 |
| Division | Source | Count | Number Misclassified | Percent Misclassified | Entropy R2 |
|---|---|---|---|---|---|
| Green | Training | 118 | 0 | 0 | 1 |
| Green | Validation | 38 | 1 | 2.63 | 0.65 |
| Green | Test | 41 | 0 | 0 | 1 |
| Red | Training | 97 | 0 | 0 | 1 |
| Red | Validation | 33 | 0 | 0 | 1 |
| Red | Test | 31 | 0 | 0 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuller, K.; Martin, R.E.; Asner, G.P. Spectral Signatures of Macroalgae on Hawaiian Reefs. Remote Sens. 2024, 16, 1140. https://doi.org/10.3390/rs16071140
Fuller K, Martin RE, Asner GP. Spectral Signatures of Macroalgae on Hawaiian Reefs. Remote Sensing. 2024; 16(7):1140. https://doi.org/10.3390/rs16071140
Chicago/Turabian StyleFuller, Kimberly, Roberta E. Martin, and Gregory P. Asner. 2024. "Spectral Signatures of Macroalgae on Hawaiian Reefs" Remote Sensing 16, no. 7: 1140. https://doi.org/10.3390/rs16071140
APA StyleFuller, K., Martin, R. E., & Asner, G. P. (2024). Spectral Signatures of Macroalgae on Hawaiian Reefs. Remote Sensing, 16(7), 1140. https://doi.org/10.3390/rs16071140






