Airborne Hyperspectral Images and Machine Learning Algorithms for the Identification of Lupine Invasive Species in Natura 2000 Meadows
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Areas and Objects
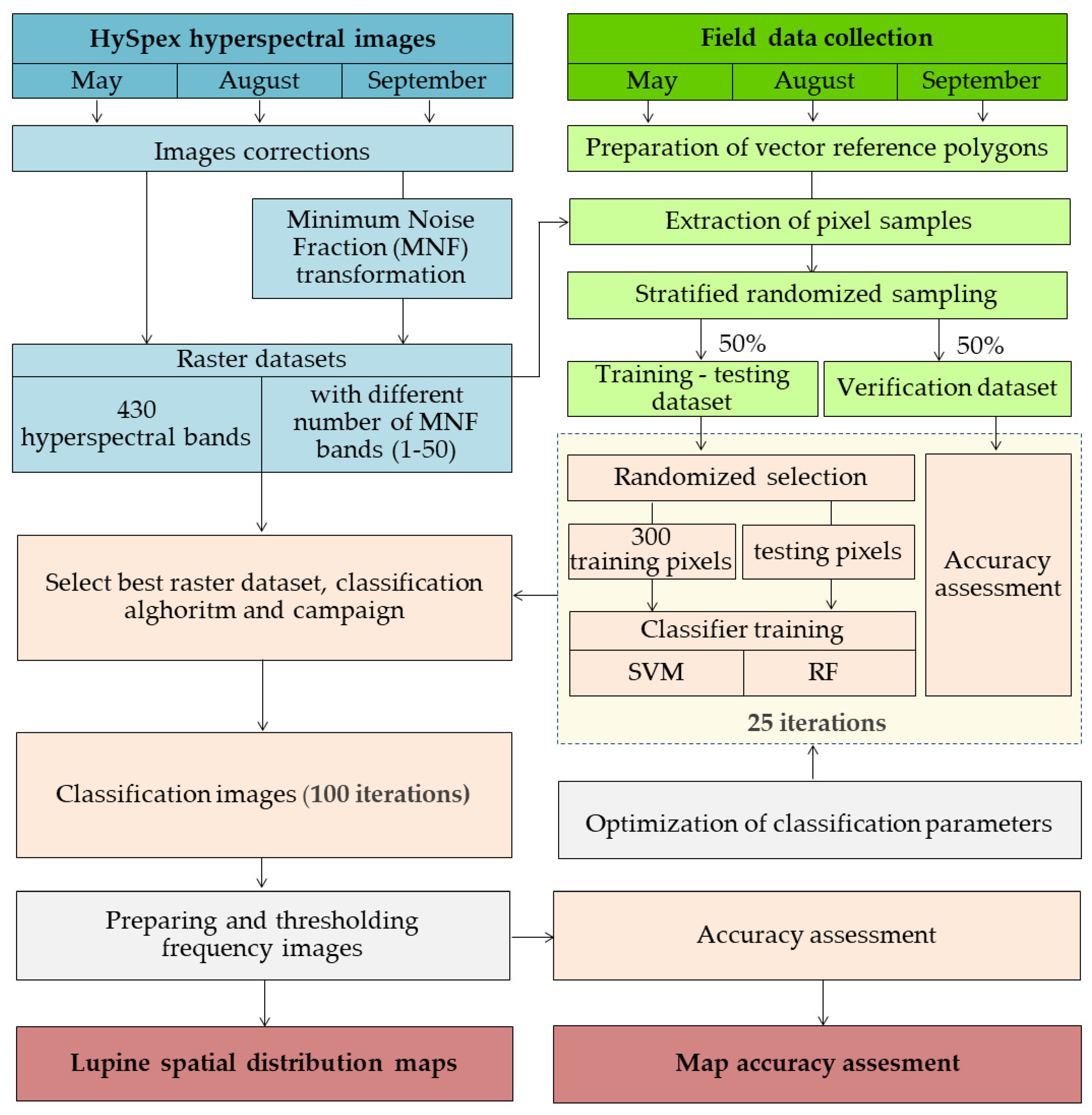
2.2. Research Methodology Overview
- The acquisition and processing of airborne hyperspectral images.
- Obtaining and preparing reference field data.
- Classifier training and iterative accuracy assessment.
- The preparation of final maps using thresholding frequency images and statistical accuracy reports.
2.3. Airborne HySpex Hyperspectral Images
2.4. Field Research
2.5. Classification Process and Accuracy Assessment
- the random selection of 300 training pixels for each class from the training–testing dataset (number of pixels recommended according to a previous study [57]);
- RF and SVM classifiers trained on a variable number of MNF bands (from 1 to 50) and a set of 430 HySpex hyperspectral bands for each campaign;
- accuracy assessment based on the spatially unchanging validation dataset (spatially separated from the training set).
2.6. Image Post-Classification Analysis
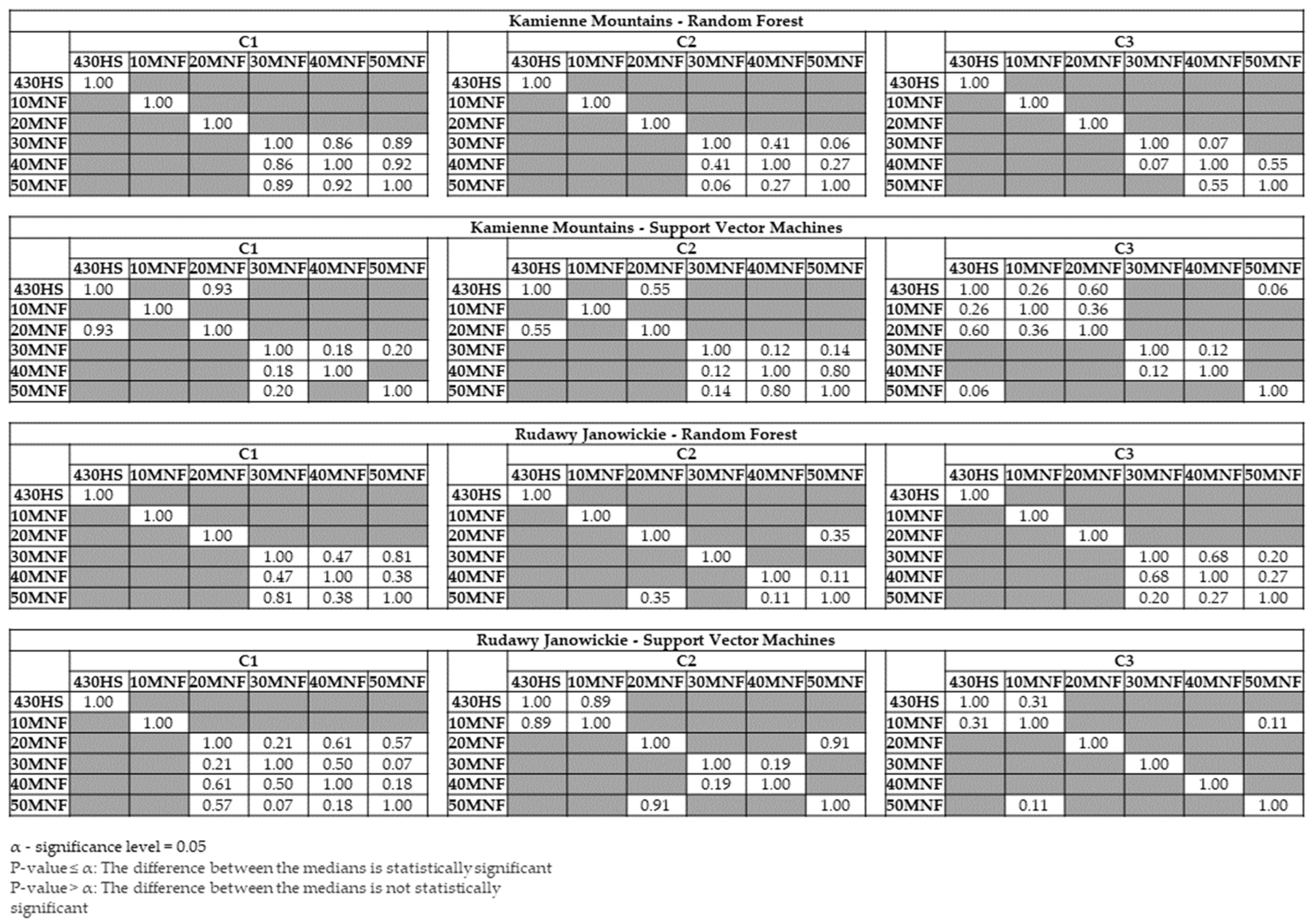
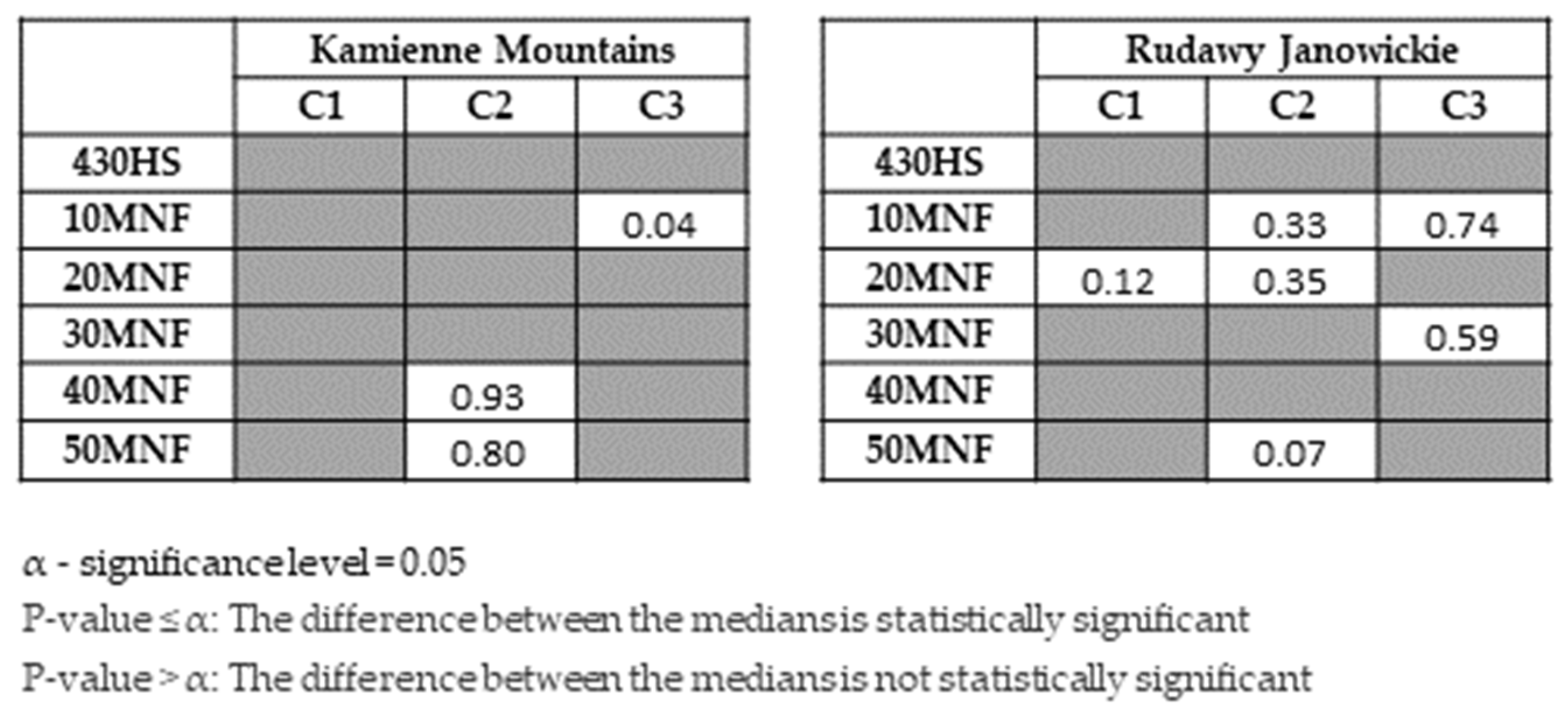
3. Results
4. Discussion
5. Conclusions
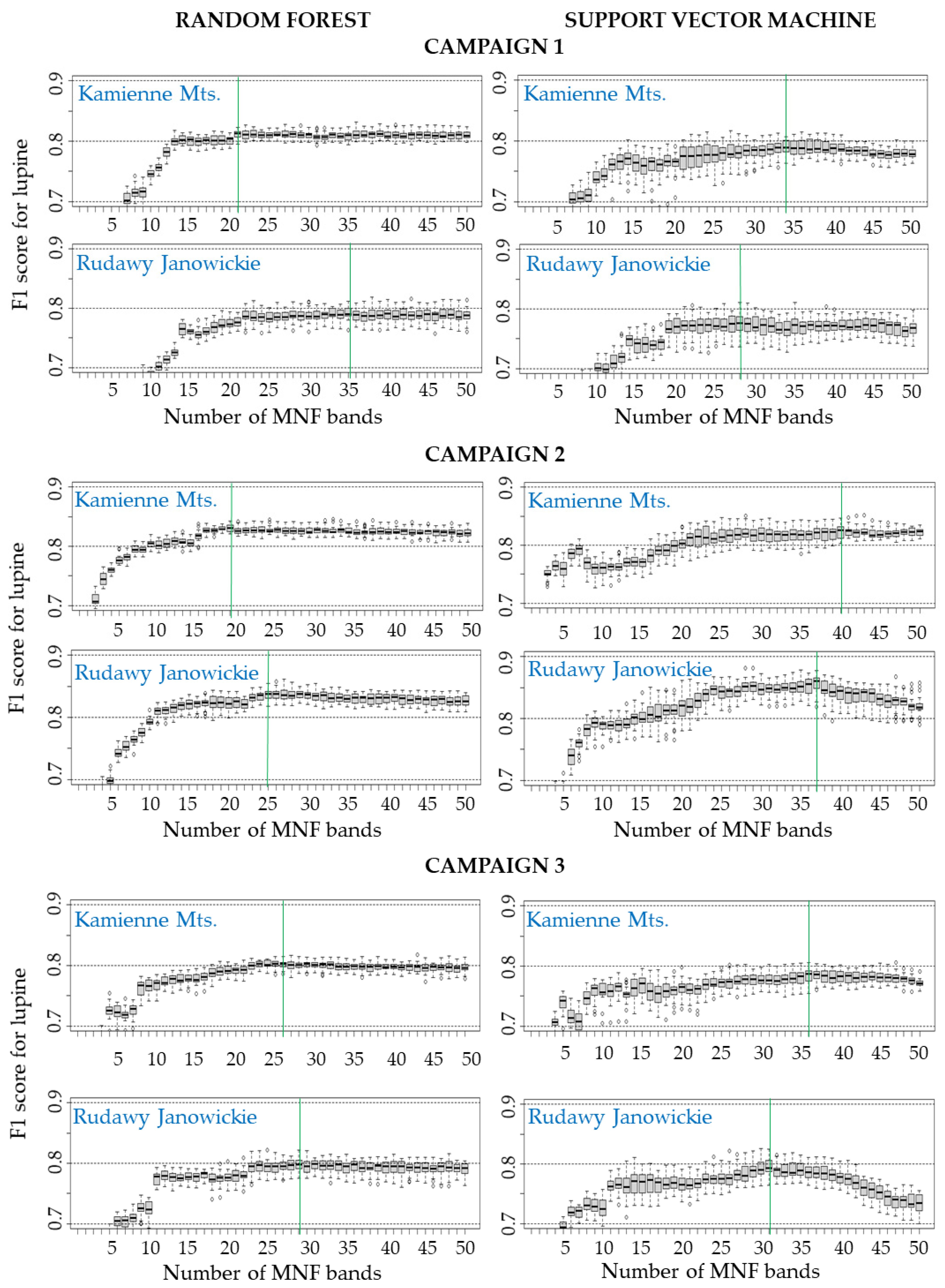
- The results show that aerial and field data should be collected at the peak of flowering of the identified plant to obtain the most accurate maps. The highest accuracy of the lupine class in both research areas was obtained during the summer campaign (August, median F1 score ranging from 0.82 to 0.85). Statistically significantly lower accuracies were obtained for the spring (F1 score: 0.77–0.81) and autumn (F1 score: 0.78–0.80) campaigns.
- The use of approximately 30 MNF bands must be considered for classification purposes when hyperspectral data are used. Input datasets consisting of 30 MNF bands produced the highest accuracies for the lupine class (median F1 score ranging from 0.77 to 0.85), and the use of a higher number of MNF bands did not significantly increase the identification accuracy. The classification accuracies obtained on the original 430 spectral bands were lower (median F1 score from 0.62 to 0.81) in both study areas.
- The classifiers gave similar results for lupine identification in both research areas (F1 score: 0.80–0.86), which confirms that both RF and SVMs can be successfully used to identify IAS.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Seebens, H.; Essl, F.; Dawson, W.; Fuentes, N.; Moser, D.; Pergl, J.; Pyšek, P.; van Kleunen, M.; Weber, E.; Winter, M.; et al. Global Trade Will Accelerate Plant Invasions in Emerging Economies under Climate Change. Glob. Chang. Biol. 2015, 21, 4128–4140. [Google Scholar] [CrossRef]
- Sittaro, F.; Hutengs, C.; Vohland, M. Which Factors Determine the Invasion of Plant Species? Machine Learning Based Habitat Modelling Integrating Environmental Factors and Climate Scenarios. Int. J. Appl. Earth Obs. Geoinf. 2023, 116, 103158. [Google Scholar] [CrossRef]
- Kumar Rai, P.; Singh, J.S. Invasive Alien Plant Species: Their Impact on Environment, Ecosystem Services and Human Health. Ecol. Indic. 2020, 111, 106020. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, B.; Clavero, M.; Sánchez, M.I.; Vilà, M. Global Ecological Impacts of Invasive Species in Aquatic Ecosystems. Glob. Chang. Biol. 2016, 22, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Haubrock, P.J.; Turbelin, A.J.; Cuthbert, R.N.; Novoa, A.; Taylor, N.G.; Angulo, E.; Ballesteros-Mejia, L.; Bodey, T.W.; Capinha, C.; Diagne, C.; et al. Economic Costs of Invasive Alien Species across Europe. NeoBiota 2021, 67, 153–190. [Google Scholar] [CrossRef]
- Ludewig, K.; Klinger, Y.P.; Donath, T.W.; Bärmann, L.; Eichberg, C.; Thomsen, J.G.; Görzen, E.; Hansen, W.; Hasselquist, E.M.; Helminger, T.; et al. Phenology and Morphology of the Invasive Legume Lupinus polyphyllus along a Latitudinal Gradient in Europe. NeoBiota 2022, 78, 185–206. [Google Scholar] [CrossRef]
- Lambdon, P.W.; Pyšek, P.; Basnou, C.; Hejda, M.; Arianoutsou, M.; Essl, F.; Jarošík, V.; Pergl, J.; Winter, M.; Anastasiu, P.; et al. Alien Flora of Europe: Species Diversity, Temporal Trends, Geographical Patterns and Research Needs. Preslia 2008, 80, 101–149. [Google Scholar]
- Walsh, S.J. Multi-Scale Remote Sensing of Introduced and Invasive Species: An Overview of Approaches and Perspectives; Springer: Cham, Switzerland, 2018; pp. 143–154. [Google Scholar]
- Kopeć, D.; Zakrzewska, A.; Halladin-Dąbrowska, A.; Wylazłowska, J.; Sławik, Ł. The Essence of Acquisition Time of Airborne Hyperspectral and On-Ground Reference Data for Classification of Highly Invasive Annual Vine Echinocystis lobata (Michx.) Torr. & A. Gray. GIScience Remote Sens. 2023, 60, 2204682. [Google Scholar] [CrossRef]
- Bradley, B.A. Remote Detection of Invasive Plants: A Review of Spectral, Textural and Phenological Approaches. Biol. Invasions 2014, 16, 1411–1425. [Google Scholar] [CrossRef]
- Duncan, P.; Podest, E.; Esler, K.J.; Geerts, S.; Lyons, C. Mapping Invasive Herbaceous Plant Species with Sentinel-2 Satellite Imagery: Echium Plantagineum in a Mediterranean Shrubland as a Case Study. Geomatics 2023, 3, 328–344. [Google Scholar] [CrossRef]
- Theron, K.J.; Pryke, J.S.; Latte, N.; Samways, M.J. Mapping an Alien Invasive Shrub within Conservation Corridors Using Super-Resolution Satellite Imagery. J. Environ. Manag. 2022, 321, 116023. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Huang, Y.; Liu, Q.; Fan, W.; Sun, Z.; Dong, H.; Wan, F.; Qiao, X. UAV and a Deep Convolutional Neural Network for Monitoring Invasive Alien Plants in the Wild. Comput. Electron. Agric. 2020, 174, 105519. [Google Scholar] [CrossRef]
- Bakacsy, L.; Tobak, Z.; van Leeuwen, B.; Szilassi, P.; Biró, C.; Szatmári, J. Drone-Based Identification and Monitoring of Two Invasive Alien Plant Species in Open Sand Grasslands by Six RGB Vegetation Indices. Drones 2023, 7, 207. [Google Scholar] [CrossRef]
- Cierniewski, J.; Ceglarek, J.; Karnieli, A.; Królewicz, S.; Kaźmierowski, C.; Zagajewski, B. Predicting the diurnal blue-sky albedo of soils using their laboratory reflectance spectra and roughness indices. J. Quant. Spectrosc. Radiat. Transf. 2017, 200, 25–31. [Google Scholar] [CrossRef]
- Zagajewski, B.; Kycko, M.; Tømmervik, H.; Bochenek, Z.; Wojtuń, B.; Bjerke, J.W.; Kłos, A. Feasibility of Hyperspectral Vegetation Indices for the Detection of Chlorophyll Concentration in Three High Arctic Plants: Salix Polaris, Bistorta Vivipara, and Dryas Octopetala. Acta Soc. Bot. Pol. 2018, 87, 3604. [Google Scholar] [CrossRef]
- Kycko, M.; Zagajewski, B.; Zwijacz-Kozica, M.; Cierniewski, J.; Romanowska, E.; Orłowska, K.; Ochtyra, A.; Jarocińska, A. Assessment of Hyperspectral Remote Sensing for Analyzing the Impact of Human Trampling on Alpine Swards. Mt. Res. Dev. 2017, 37, 66–74. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Gamon, J.A.; Townsend, P.A. (Eds.) Remote Sensing of Plant Biodiversity; Springer International Publishing: Cham, Switzerland, 2020; ISBN 978-3-030-33156-6. [Google Scholar]
- Kopeć, D.; Sabat-Tomala, A.; Michalska-Hejduk, D.; Jarocińska, A.; Niedzielko, J. Application of Airborne Hyperspectral Data for Mapping of Invasive Alien Spiraea tomentosa L.: A Serious Threat to Peat Bog Plant Communities. Wetl. Ecol. Manag. 2020, 28, 357–373. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Jarocińska, A.; Bzdęga, K.; Tokarska-Guzik, B. Classification of Expansive Grassland Species in Different Growth Stages Based on Hyperspectral and LiDAR Data. Remote Sens. 2018, 10, 2019. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Yang, R.; Wang, F.; Li, Y.; Zhang, S.; Wan, F.; Qiao, X.; Qian, W. Hyperspectral Imaging for Identification of an Invasive Plant Mikania Micrantha Kunth. Front. Plant Sci. 2021, 12, 626516. [Google Scholar] [CrossRef]
- Papp, L.; van Leeuwen, B.; Szilassi, P.; Tobak, Z.; Szatmári, J.; Árvai, M.; Mészáros, J.; Pásztor, L. Monitoring Invasive Plant Species Using Hyperspectral Remote Sensing Data. Land 2021, 10, 29. [Google Scholar] [CrossRef]
- Gite, H.R.; Solankar, M.M.; Surase, R.R.; Kale, K.V. Comparative Study and Analysis of Dimensionality Reduction Techniques for Hyperspectral Data. In Communications in Computer and Information Science; Springer: Singapore, 2019; Volume 1035, pp. 534–546. ISBN 9789811391804. [Google Scholar]
- Royimani, L.; Mutanga, O.; Odindi, J.; Dube, T.; Matongera, T.N. Advancements in Satellite Remote Sensing for Mapping and Monitoring of Alien Invasive Plant Species (AIPs). Phys. Chem. Earth Parts A/B/C 2019, 112, 237–245. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a Complex of Statistical Variables into Principal Components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Comon, P. Independent Component Analysis, A New Concept? Signal Process. 1994, 36, 287–314. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A Transformation for Ordering Multispectral Data in Terms of Image Quality with Implications for Noise Removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Murinto, M.; Dyah, N.R. Feature Reduction Using the Minimum Noise Fraction and Principal Component Analysis Transforms for Improving the Classification of Hyperspectral Images. Asia-Pac. J. Sci. Technol. 2017, 22, 1–15. [Google Scholar] [CrossRef]
- Zhang, Z.; Kazakova, A.; Moskal, L.; Styers, D. Object-Based Tree Species Classification in Urban Ecosystems Using LiDAR and Hyperspectral Data. Forests 2016, 7, 122. [Google Scholar] [CrossRef]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of Herbaceous Vegetation Using Airborne Hyperspectral Imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef]
- Müllerová, J.; Brůna, J.; Bartaloš, T.; Dvořák, P.; Vítková, M.; Pyšek, P. Timing Is Important: Unmanned Aircraft vs. Satellite Imagery in Plant Invasion Monitoring. Front. Plant Sci. 2017, 8, 887. [Google Scholar] [CrossRef]
- Schulze-Brüninghoff, D.; Wachendorf, M.; Astor, T. Potentials and Limitations of WorldView-3 Data for the Detection of Invasive Lupinus polyphyllus Lindl. in Semi-Natural Grasslands. Remote Sens. 2021, 13, 4333. [Google Scholar] [CrossRef]
- Mundt, J.; Glenn, N.; Weber, K.; Prather, T.; Lass, L.; Pettingill, J. Discrimination of Hoary Cress and Determination of Its Detection Limits via Hyperspectral Image Processing and Accuracy Assessment Techniques. Remote Sens. Environ. 2005, 96, 509–517. [Google Scholar] [CrossRef]
- Bustamante, J.; Aragonés, D.; Afán, I.; Luque, C.; Pérez-Vázquez, A.; Castellanos, E.; Díaz-Delgado, R. Hyperspectral Sensors as a Management Tool to Prevent the Invasion of the Exotic Cordgrass Spartina Densiflora in the Doñana Wetlands. Remote Sens. 2016, 8, 1001. [Google Scholar] [CrossRef]
- Andrew, M.E.; Ustin, S.L. The Role of Environmental Context in Mapping Invasive Plants with Hyperspectral Image Data. Remote Sens. Environ. 2008, 112, 4301–4317. [Google Scholar] [CrossRef]
- Routh, D.; Seegmiller, L.; Bettigole, C.; Kuhn, C.; Oliver, C.D.; Glick, H.B. Improving the Reliability of Mixture Tuned Matched Filtering Remote Sensing Classification Results Using Supervised Learning Algorithms and Cross-Validation. Remote Sens. 2018, 10, 1675. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping Invasive Plants Using Hyperspectral Imagery and Breiman Cutler Classifications (RandomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Arasumani, M.; Singh, A.; Bunyan, M.; Robin, V.V. Testing the Efficacy of Hyperspectral (AVIRIS-NG), Multispectral (Sentinel-2) and Radar (Sentinel-1) Remote Sensing Images to Detect Native and Invasive Non-Native Trees. Biol. Invasions 2021, 23, 2863–2879. [Google Scholar] [CrossRef]
- Jensen, T.; Seerup Hass, F.; Seam Akbar, M.; Holm Petersen, P.; Jokar Arsanjani, J. Employing Machine Learning for Detection of Invasive Species Using Sentinel-2 and AVIRIS Data: The Case of Kudzu in the United States. Sustainability 2020, 12, 3544. [Google Scholar] [CrossRef]
- Vapnik, V.; Lerner, A. Pattern Recognition Using Generalized Portrait Method. Autom. Remote Control 1963, 24, 774–780. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Masocha, M.; Skidmore, A.K. Integrating Conventional Classifiers with a GIS Expert System to Increase the Accuracy of Invasive Species Mapping. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 487–494. [Google Scholar] [CrossRef]
- Heydari, S.S.; Mountrakis, G. Meta-Analysis of Deep Neural Networks in Remote Sensing: A Comparative Study of Mono-Temporal Classification to Support Vector Machines. ISPRS J. Photogramm. Remote Sens. 2019, 152, 192–210. [Google Scholar] [CrossRef]
- Ge, H.; Wang, L.; Liu, M.; Zhu, Y.; Zhao, X.; Pan, H.; Liu, Y. Two-Branch Convolutional Neural Network with Polarized Full Attention for Hyperspectral Image Classification. Remote Sens. 2023, 15, 848. [Google Scholar] [CrossRef]
- Adamiak, M. Głębokie Uczenie w Procesie Teledetekcyjnej Interpretacji Przestrzeni Geograficznej—Przegląd Wybranych Zagadnień. Czas. Geogr. 2021, 92, 49–72. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Networks Accurately Predict Cover Fractions of Plant Species and Communities in Unmanned Aerial Vehicle Imagery. Remote Sens. Ecol. Conserv. 2020, 6, 472–486. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support Vector Machine Versus Random Forest for Remote Sensing Image Classification: A Meta-Analysis and Systematic Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Hasan, H.; Shafri, H.Z.M.; Habshi, M. A Comparison Between Support Vector Machine (SVM) and Convolutional Neural Network (CNN) Models For Hyperspectral Image Classification. IOP Conf. Ser. Earth Environ. Sci. 2019, 357, 012035. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Crabbe, R.A.; Lamb, D.; Edwards, C. Discrimination of Species Composition Types of a Grazed Pasture Landscape Using Sentinel-1 and Sentinel-2 Data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101978. [Google Scholar] [CrossRef]
- Gholami, R.; Fakhari, N. Support Vector Machine: Principles, Parameters, and Applications. In Handbook of Neural Computation; Elsevier: Amsterdam, The Netherlands, 2017; pp. 515–535. ISBN 9780128113196. [Google Scholar]
- Wang, L.; Silván-Cárdenas, J.L.; Yang, J.; Frazier, A.E. Invasive Saltcedar (Tamarisk spp.) Distribution Mapping Using Multiresolution Remote Sensing Imagery. Prof. Geogr. 2013, 65, 1–15. [Google Scholar] [CrossRef]
- Waśniewski, A.; Hościło, A.; Zagajewski, B.; Moukétou-Tarazewicz, D. Assessment of Sentinel-2 Satellite Images and Random Forest Classifier for Rainforest Mapping in Gabon. Forests 2020, 11, 941. [Google Scholar] [CrossRef]
- Kupková, L.; Červená, L.; Suchá, R.; Jakešová, L.; Zagajewski, B.; Březina, S.; Albrechtová, J. Classification of Tundra Vegetation in the Krkonoše Mts. National Park Using APEX, AISA Dual and Sentinel-2A Data. Eur. J. Remote Sens. 2017, 50, 29–46. [Google Scholar] [CrossRef]
- Nasiri, V.; Beloiu, M.; Asghar Darvishsefat, A.; Griess, V.C.; Maftei, C.; Waser, L.T. Mapping Tree Species Composition in a Caspian Temperate Mixed Forest Based on Spectral-Temporal Metrics and Machine Learning. Int. J. Appl. Earth Obs. Geoinf. 2023, 116, 103154. [Google Scholar] [CrossRef]
- Qiao, X.; Liu, X.; Wang, F.; Sun, Z.; Yang, L.; Pu, X.; Huang, Y.; Liu, S.; Qian, W. A Method of Invasive Alien Plant Identification Based on Hyperspectral Images. Agronomy 2022, 12, 2825. [Google Scholar] [CrossRef]
- Sabat-Tomala, A.; Raczko, E.; Zagajewski, B. Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data. Remote Sens. 2020, 12, 516. [Google Scholar] [CrossRef]
- Sabat-Tomala, A.; Raczko, E.; Zagajewski, B. Mapping Invasive Plant Species with Hyperspectral Data Based on Iterative Accuracy Assessment Techniques. Remote Sens. 2022, 14, 64. [Google Scholar] [CrossRef]
- Beuthin, M.M. Plant Guide for Bigleaf Lupine (Lupinus polyphyllus Lindl.). Available online: http://plants.usda.gov/ (accessed on 4 November 2023).
- Vinogradova, Y.K.; Tkacheva, E.V.; Mayorov, S.R. About Flowering Biology of Alien Species: 1. Lupinus polyphyllus Lindl. Russ. J. Biol. Invasions 2012, 3, 163–171. [Google Scholar] [CrossRef]
- Hansen, W.; Wollny, J.; Otte, A.; Eckstein, R.L.; Ludewig, K. Invasive Legume Affects Species and Functional Composition of Mountain Meadow Plant Communities. Biol. Invasions 2021, 23, 281–296. [Google Scholar] [CrossRef]
- HySpex. Available online: https://www.hyspex.com/ (accessed on 16 January 2024).
- PARGE Airborne Image Rectification. Available online: https://www.rese-apps.com/software/parge/index.html (accessed on 16 January 2024).
- ATCOR for Airborne Remote Sensing. Available online: https://www.rese-apps.com/software/atcor-4-airborne/index.html (accessed on 16 January 2024).
- Richter, R.; Schläpfer, D. Geo-Atmospheric Processing of Airborne Imaging Spectrometry Data. Part 2: Atmospheric/Topographic Correction. Int. J. Remote Sens. 2002, 23, 2631–2649. [Google Scholar] [CrossRef]
- Schläpfer, D.; Richter, R. Geo-Atmospheric Processing of Airborne Imaging Spectrometry Data. Part 1: Parametric Orthorectification. Int. J. Remote Sens. 2002, 23, 2609–2630. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Wojtuń, B.; Rogass, C.; Mielke, C.; Lavender, S. Subalpine and Alpine Vegetation Classification Based on Hyperspectral APEX and Simulated EnMAP Images. Int. J. Remote Sens. 2017, 38, 1839–1864. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A Framework for Mapping Tree Species Combining Hyperspectral and LiDAR Data: Role of Selected Classifiers and Sensor across Three Spatial Scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef]
- Zhang, J.; Yao, Y.; Suo, N. Automatic classification of fine-scale mountain vegetation based on mountain altitudinal belt. PLoS ONE 2020, 15, e0238165. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of Two Random Variables Is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Wijesingha, J.; Astor, T.; Schulze-Brüninghoff, D.; Wachendorf, M. Mapping Invasive Lupinus polyphyllus Lindl. in Semi-Natural Grasslands Using Object-Based Image Analysis of UAV-Borne Images. PFG—J. Photogramm. Remote Sens. Geoinf. Sci. 2020, 88, 391–406. [Google Scholar] [CrossRef]
- Thorsteinsdottir, A.B. Mapping Lupinus Nootkatensis in Iceland Using SPOT 5 Images; Lund University: Sweden, Switzerland, 2011. [Google Scholar]
- Benevides, P.J.; Costa, H.; Moreira, F.D.; Caetano, M.R. Mapping Annual Crops in Portugal with Sentinel-2 Data. In Remote Sensing for Agriculture, Ecosystems, and Hydrology XXIV; Neale, C.M., Maltese, A., Eds.; SPIE: Bellingham, WA, USA, 2022; Volume 12262, p. 20. [Google Scholar]
- Kopeć, D.; Zakrzewska, A.; Halladin-Dąbrowska, A.; Wylazłowska, J.; Kania, A.; Niedzielko, J. Using Airborne Hyperspectral Imaging Spectroscopy to Accurately Monitor Invasive and Expansive Herb Plants: Limitations and Requirements of the Method. Sensors 2019, 19, 2871. [Google Scholar] [CrossRef]
- Mirik, M.; Ansley, R.J.; Steddom, K.; Jones, D.C.; Rush, C.M.; Michels, G.J.; Elliott, N.C. Remote Distinction of a Noxious Weed (Musk Thistle: Carduus Nutans) Using Airborne Hyperspectral Imagery and the Support Vector Machine Classifier. Remote Sens. 2013, 5, 612. [Google Scholar] [CrossRef]
- Iqbal, I.M.; Balzter, H.; Firdaus-e-Bareen; Shabbir, A. Mapping Lantana Camara and Leucaena Leucocephala in Protected Areas of Pakistan: A Geo-Spatial Approach. Remote Sens. 2023, 15, 1020. [Google Scholar] [CrossRef]
- Barbosa, J.; Asner, G.; Martin, R.; Baldeck, C.; Hughes, F.; Johnson, T. Determining Subcanopy Psidium Cattleianum Invasion in Hawaiian Forests Using Imaging Spectroscopy. Remote Sens. 2016, 8, 33. [Google Scholar] [CrossRef]
- Adugna, T.; Xu, W.; Fan, J. Comparison of Random Forest and Support Vector Machine Classifiers for Regional Land Cover Mapping Using Coarse Resolution FY-3C Images. Remote Sens. 2022, 14, 574. [Google Scholar] [CrossRef]


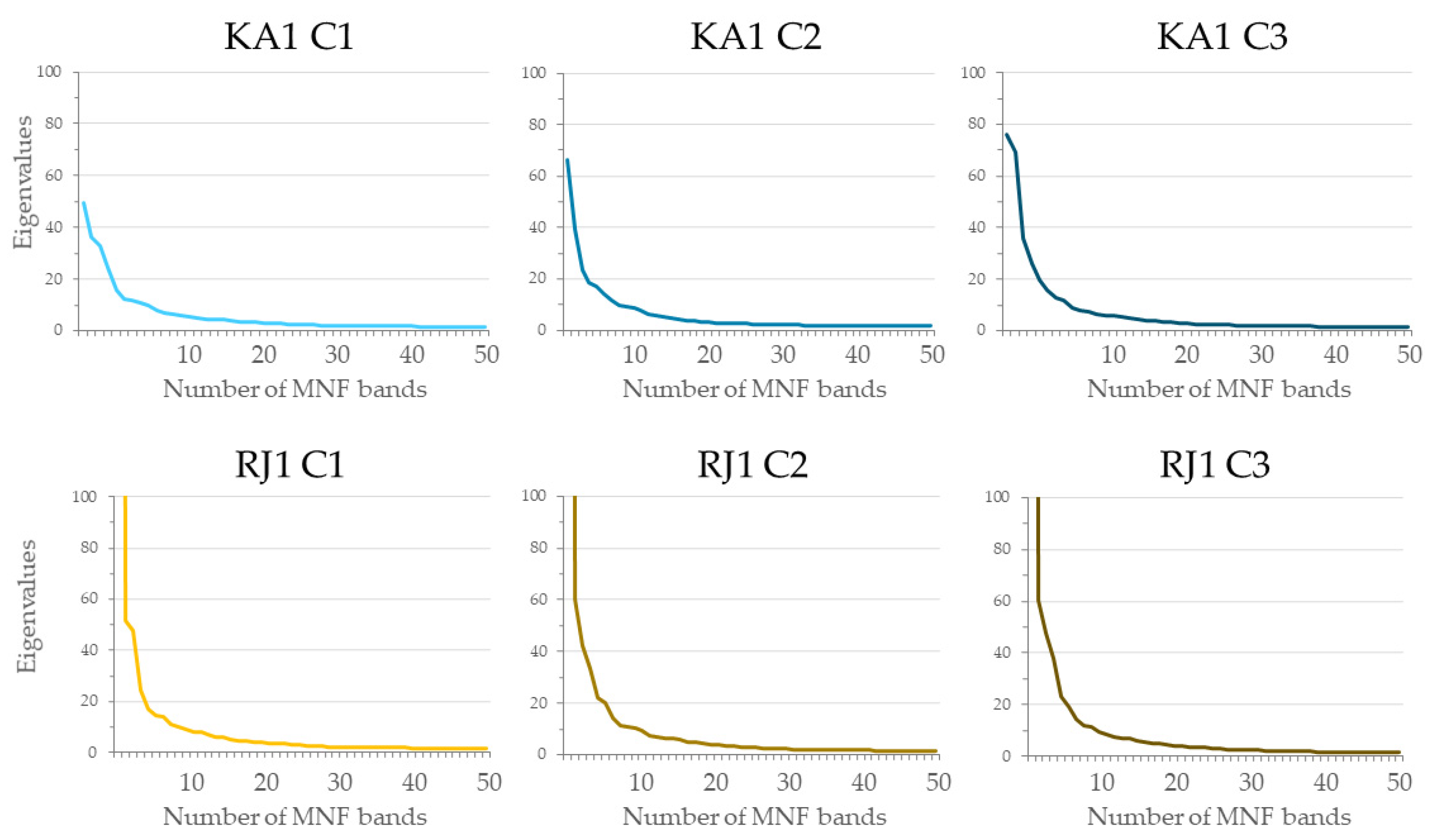
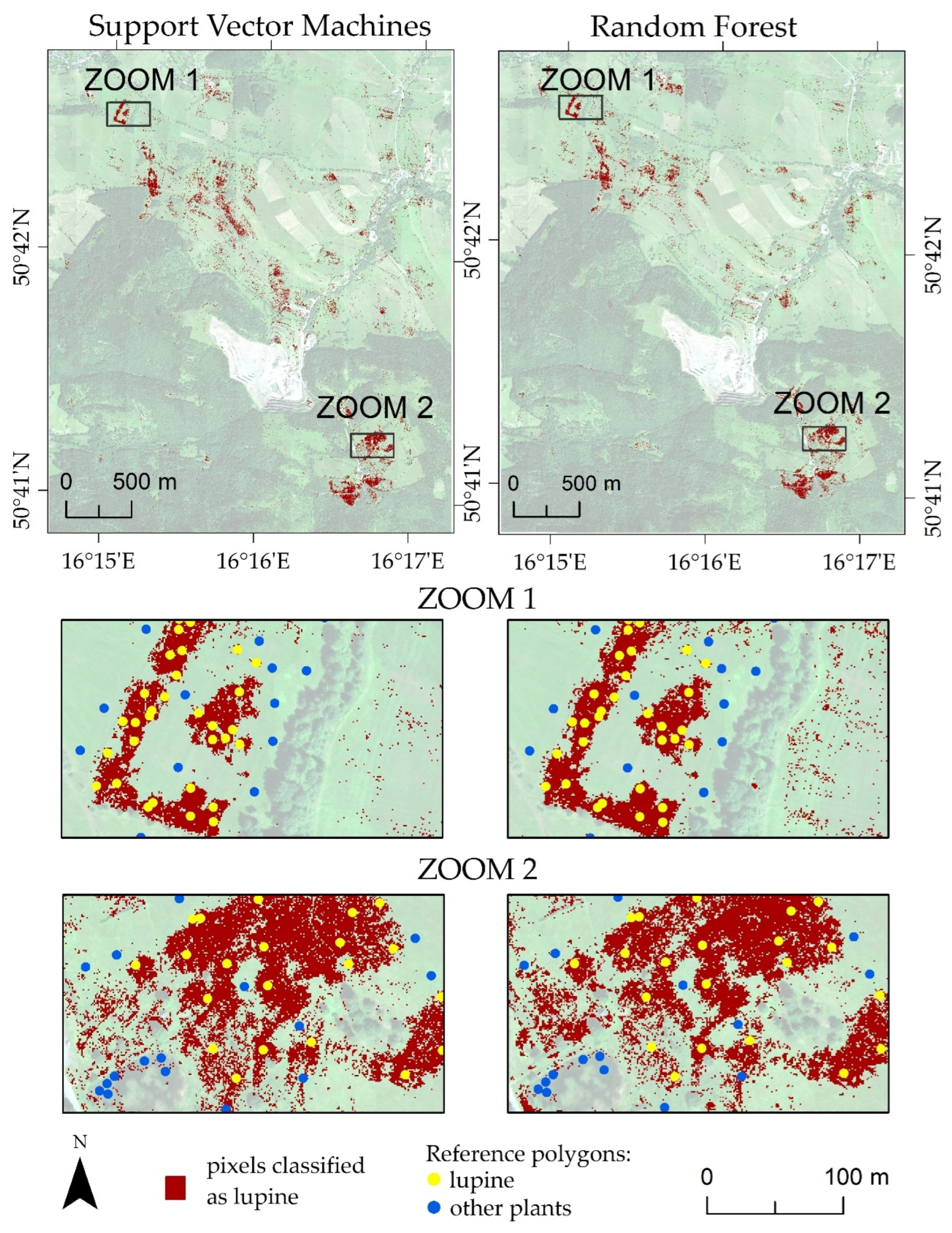
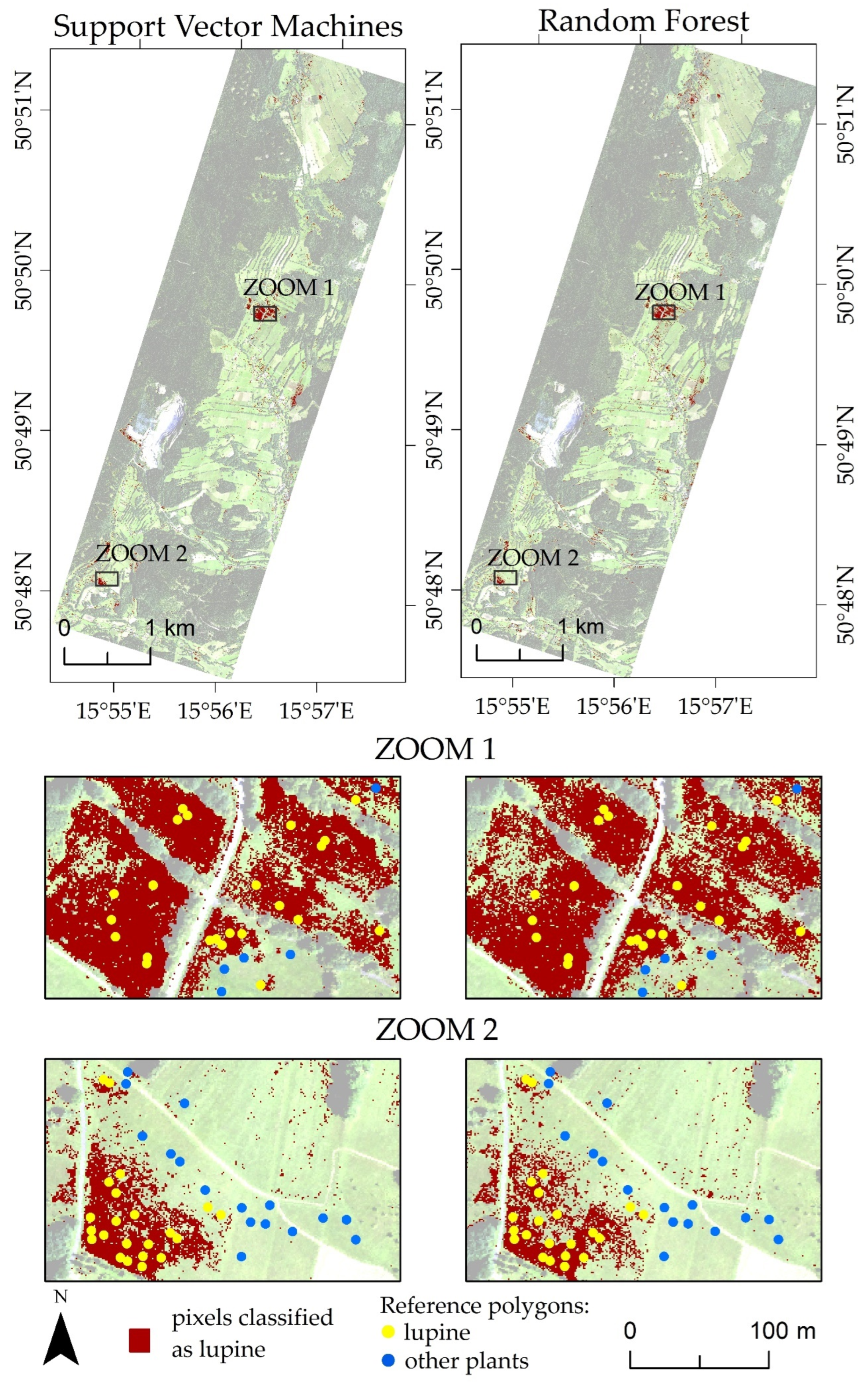
| Number of Campaign | Date of Flight Campaigns | Date of Field Measurements | |
|---|---|---|---|
| Kamienne Mountains | Rudawy Janowickie | ||
| C1 | 21 May 2016 | 21 May 2016 | May/June 2016 |
| C2 | 7 August 2016 | 7 August 2016 | August 2016 |
| C3 | 12 September 2016 | 11 September 2016 | September 2016 |
| Research Area | Campaign | Number of Reference Polygons | ||
|---|---|---|---|---|
| Lupine (Training/Validation Polygons) | Co-Occurring Plants | Land Cover Classes | ||
| KA1 | C1 | 180 (90/90) | 250 | 200 (50 for each class: buildings, soil, trees, water) |
| C2 | 170 (80/90) | 250 | 200 (50 for each class: buildings, soil, trees, water) | |
| C3 | 145 (55/90) | 250 | 200 (50 for each class: buildings, soil, trees, water) | |
| RJ1 | C1 | 100 (50/50) | 200 | 150 (50 for each class: buildings, soil, trees) |
| C2 | 96 (46/50) | 200 | 150 (50 for each class: buildings, soil, trees) | |
| C3 | 98 (48/50) | 200 | 150 (50 for each class: buildings, soil, trees) | |
| Area | Raster Datasets | Median F1 Score Accuracy for Lupine (25 Iterations) | |||||
|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | |||||
| RF | SVM | RF | SVM | RF | SVM | ||
| Kamienne Mountains (KA1) | 430 spectral bands | 0.64 * | 0.76 * | 0.75 * | 0.80 * | 0.70 * | 0.77 * |
| 10 MNFs | 0.75 * | 0.74 * | 0.80 * | 0.76 * | 0.77 * | 0.76 * | |
| 20 MNFs | 0.80 * | 0.77 * | 0.83 * | 0.80 * | 0.79 * | 0.76 * | |
| 30 MNFs | 0.81 | 0.78 | 0.82 | 0.82 | 0.80 | 0.78 | |
| 40 MNFs | 0.81 | 0.79 | 0.82 | 0.83 | 0.80 | 0.78 | |
| 50 MNFs | 0.81 | 0.78 * | 0.82 | 0.82 | 0.80 | 0.77 * | |
| Rudawy Janowickie (RJ1) | 430 spectral bands | 0.70 * | 0.81 * | 0.70 * | 0.79 * | 0.62 * | 0.72 * |
| 10 MNFs | 0.69 * | 0.70 * | 0.79 * | 0.79 * | 0.72 * | 0.72 * | |
| 20 MNFs | 0.77 * | 0.77 | 0.82 * | 0.82 * | 0.78 * | 0.77 * | |
| 30 MNFs | 0.79 | 0.77 | 0.84 * | 0.85 | 0.80 | 0.79 * | |
| 40 MNFs | 0.79 | 0.77 | 0.83 | 0.84 | 0.79 | 0.78 * | |
| 50 MNFs | 0.79 | 0.77 | 0.83 | 0.82 * | 0.79 | 0.73 * | |
| The frequency of occurrence of a median F1 score above 0.8 | 3 | 1 | 8 | 7 | 0 | 0 | |
| Kamienne Mountains | |||||
|---|---|---|---|---|---|
| Support Vector Machines | |||||
| Class | Lupine | Background | Total | UA (%) | Commission (%) |
| Lupine | 784 | 69 | 853 | 91.91 | 8.09 |
| Background | 332 | 2751 | 3083 | 89.23 | 10.77 |
| Total | 1116 | 2820 | 3936 | ||
| PA (%) | 70.25 | 97.55 | |||
| Omission (%) | 29.75 | 2.45 | |||
| Random Forest | |||||
| Class | Lupine | Background | Total | UA (%) | Commission (%) |
| Lupine | 793 | 85 | 878 | 90.32 | 9.68 |
| Background | 323 | 2735 | 3058 | 89.44 | 10.56 |
| Total | 1116 | 2820 | 3936 | ||
| PA (%) | 71.06 | 96.99 | |||
| Omission (%) | 28.94 | 3.01 | |||
| Rudawy Janowickie | |||||
|---|---|---|---|---|---|
| Support Vector Machines | |||||
| Class | Lupine | Background | Total | UA (%) | Commission (%) |
| Lupine | 796 | 66 | 862 | 92.34 | 7.66 |
| Background | 193 | 3361 | 3554 | 94.57 | 5.43 |
| Total | 989 | 3427 | 4416 | ||
| PA (%) | 80.49 | 98.07 | |||
| Omission (%) | 19.51 | 1.93 | |||
| Random Forest | |||||
| Class | Lupine | Background | Total | UA (%) | Commission (%) |
| Lupine | 773 | 107 | 880 | 87.84 | 12.16 |
| Background | 216 | 3320 | 3536 | 93.89 | 6.11 |
| Total | 989 | 3427 | 4416 | ||
| PA (%) | 78.16 | 96.88 | |||
| Omission (%) | 21.84 | 3.12 | |||
| Author | Sensor | Algorithm | Invasive Species | F1 Score | OA (%) |
|---|---|---|---|---|---|
| Present paper | HySpex | RF | Lupinus polyphyllus | 0.80–0.83 | 89–93 |
| SVM | Lupinus polyphyllus | 0.80–0.86 | 89–94 | ||
| [72] | UAV (RGB and thermal cameras) | OBIA + RF | Lupinus polyphyllus | - | 78–97 |
| [32] | WorldView-3 | GBM | Lupinus polyphyllus | 0.76 | - |
| [73] | SPOT 5 | MLC | Lupinus nootkatensis | 0.76–0.92 | 64–94 |
| [19] | HySpex | RF | Spiraea tomentosa | 0.83 | 99 |
| [58] | HySpex | SVM | Calamagrostis epigejos | 0.87–0.9 | - |
| Rubus spp. | 0.89–0.98 | - | |||
| Solidago spp. | 0.96–0.99 | - | |||
| [20] | HySpex | RF | Molinia caerulea | 0.78–0.89 | - |
| Calamagrostis epigejos | 0.61–0.72 | - | |||
| [9] | HySpex | RF | Echinocystis lobata | 0.64–0.87 | 97 |
| [37] | PROBE-1 | RF | Centaurea maculosa | 0.67 | 84 |
| Euphorbia esula | 0.72 | 86 | |||
| [75] | HySpex | RF | Solidago gigantea | 0.73 | - |
| Phragmites australis | 0.79 | - | |||
| Molinia caerulea | 0.80 | - | |||
| Filipendula ulmaria | 0.80 | - | |||
| [76] | AISA | SVM | Carduus nutans | 0.74–0.88 | 79–91 |
| [52] | AISA | SVM | Tamarix spp. | 93–95 | 86–88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabat-Tomala, A.; Raczko, E.; Zagajewski, B. Airborne Hyperspectral Images and Machine Learning Algorithms for the Identification of Lupine Invasive Species in Natura 2000 Meadows. Remote Sens. 2024, 16, 580. https://doi.org/10.3390/rs16030580
Sabat-Tomala A, Raczko E, Zagajewski B. Airborne Hyperspectral Images and Machine Learning Algorithms for the Identification of Lupine Invasive Species in Natura 2000 Meadows. Remote Sensing. 2024; 16(3):580. https://doi.org/10.3390/rs16030580
Chicago/Turabian StyleSabat-Tomala, Anita, Edwin Raczko, and Bogdan Zagajewski. 2024. "Airborne Hyperspectral Images and Machine Learning Algorithms for the Identification of Lupine Invasive Species in Natura 2000 Meadows" Remote Sensing 16, no. 3: 580. https://doi.org/10.3390/rs16030580
APA StyleSabat-Tomala, A., Raczko, E., & Zagajewski, B. (2024). Airborne Hyperspectral Images and Machine Learning Algorithms for the Identification of Lupine Invasive Species in Natura 2000 Meadows. Remote Sensing, 16(3), 580. https://doi.org/10.3390/rs16030580








