Abstract
Rangelands represent about 25% of the Earth’s land surface but are under severe pressure. Rangeland degradation is a gradually increasing global environmental problem, resulting in temporary or permanent loss of ecosystem functions. Ecological rangeland studies aim to determine the productivity of rangelands as well as the severity of their degradation. Rigorous in situ assessments comprising visual identification of plant species are required as such assessments are perceived to be the most accurate way of monitoring rangeland degradation. However, in situ assessments are expensive and time-consuming exercises, especially when carried out over large areas. In situ assessments are also limited to areas that are accessible. This study aimed to evaluate the effectiveness of multispectral (MS) and hyperspectral (HS) remotely sensed, unmanned aerial vehicle (UAV)-based data and machine learning (random forest) methods to differentiate between 15 dominant Nama Karoo plant species to aid ecological impact surveys. The results showed that MS imagery is unsuitable, as classification accuracies were generally low (37.5%). In contrast, much higher classification accuracies (>70%) were achieved when the HS imagery was used. The narrow bands between 398 and 430 nanometres (nm) were found to be vital for discriminating between shrub and grass species. Using in situ Analytical Spectral Device (ASD) spectroscopic data, additional important wavebands between 350 and 400 nm were identified, which are not covered by either the MS or HS remotely sensed data. Using feature selection methods, 12 key wavelengths were identified for discriminating among the plant species with accuracies exceeding 90%. Reducing the dimensionality of the ASD data set to the 12 key bands increased classification accuracies from 84.8% (all bands) to 91.7% (12 bands). The methodology developed in this study can potentially be used to carry out UAV-based ecological assessments over large and inaccessible areas typical of Karoo rangelands.
1. Introduction
Rangelands are natural lands on which the indigenous vegetation is predominantly grasses, forbs, or woody shrubs that are used for livestock or game animal grazing [1,2]. Rangelands represent a prominent land resource covering about 25% of the Earth’s land surface [2,3]. Land degradation is a major challenge for sustainable rangeland use and management. Rangeland degradation is an increasing global environmental problem, resulting in temporary or permanent loss of ecosystem functions [4,5]. It is estimated that the net primary production of a quarter of the world’s total land area is decreasing, and this may impact as many as 1.5 billion people [6]. Rural areas in developing countries are most affected by this decreasing productivity [7,8,9]. In South Africa, nearly 60% of rangelands are either degraded [10,11] or prone to degradation [12,13].
Since the Industrial Revolution the world has seen rapid increases in global carbon dioxide (CO2) emissions due to ever larger population numbers. The CO2 concentrations in the atmosphere have reached levels that are 50% higher than when the large-scale burning of fossil fuels began. Atmospheric CO2 levels are driving ongoing climate change. As a result, more persistent and severe droughts are predicted for dryland systems around the world. It is anticipated that arid regions will receive less precipitation and experience higher temperatures in the future [14,15].
Climate change, poverty, and food insecurity all have significant impacts on how land users manage and use their land [9]. The main aim of ecological rangeland studies is to determine the condition and productivity of rangelands as well as the severity of their degradation. Rigorous in situ assessments involving the visual identification of plant species and soil cover are perceived to be the most accurate way of monitoring rangeland degradation. Traditional methods for sampling terrestrial vegetation are expensive and time-consuming exercises when carried out over large areas [16] and, moreover, they are limited to areas that are easily accessible [17]. Plant ecologists maintain that small-scale or discontinuous observations are inadequate for accurately determining the spatial distribution of vegetation species over large spatial scales [18]. Remote sensing (RS) technologies, on the other hand, enable more precise monitoring of changes in the vegetation layer and species composition across large and inaccessible areas [19,20,21].
It is advisable for rangeland sciences to exploit RS to assist farmers and rangeland managers to adapt to changing and variable climates. Routine monitoring of vegetation conditions using RS data allows rangeland management practises to be adapted and implemented promptly, thereby increasing rangeland productivity and reducing degradation risks [18]. A need exists to integrate the spatial approach of geographers and the ‘functional’ approach of plant ecologists [22]. Techniques need to be developed that bridge the gap between ground-based assessments and the longitudinal observations offered by remotely sensed data [23]. These techniques will facilitate the modelling of the distribution of plant species now and in the future to assess the extent of rangeland degradation.
A number of studies have made use of satellite imagery and historical aerial photography to quantify and monitor encroaching plant species [24,25,26,27,28]. These studies have also demonstrated a high degree of accuracy. However, the value of satellite RS for rangeland sciences is limited by the high spatial (0.3 m or better) and spectral (multiple narrow bands) resolution needed for species identification. For instance, Landsat-8 images have a relatively low spatial resolution of 30 m. Although Sentinel-2 images have a higher (10 m) spatial resolution, it is not high enough to distinguish between individual shrubs and grasses. Both imagery sources suffer from the mixed-pixel problem, where a pixel value represents a combination of plant species present within the pixel area rather than just one individual plant [29,30,31].
Machine learning has been successfully applied to multispectral and hyperspectral imagery for classifying plant species. Adam et al. [19], Abdel-Rahman et al. [32], Kumar et al. [33], and Mudereri et al. [34] used the random forest (RF) algorithm on high-dimensional HS data for plant species classification, achieving good accuracy.
Feature selection identifies important features for tasks like plant species classification. The GRRF algorithm distinguishes plant species by selecting the best spectral bands [19]. Feature selection reduces data set dimensionality, addressing the “curse of dimensionality” and improving classification accuracy by removing redundant features. The Hughes effect, where classification accuracy declines with more features, can be mitigated by feature selection [35]. Mureriwa et al. [19] showed that GRRF enhances spectral feature selection and classification accuracy. Other studies, like Li et al. [36], found that feature selection increased classification accuracy by up to 30%, highlighting its value in plant species identification. The RF method identifies features corresponding to specific physical or chemical properties of plant species. For instance, certain spectral bands indicate chlorophyll content, affecting leaf reflectance, while others relate to water content, influencing absorption in specific spectral regions. These key features enable the RF method to differentiate species based on unique spectral signatures, providing insights into biological processes and environmental factors, thus enhancing the model’s interpretability and application.
Sensors mounted on unmanned aerial vehicles (UAVs) provide much higher spatial resolution imagery compared to satellites and can consequently be used to overcome the mixed-pixel problem. Most UAVs currently on the market are equipped with consumer-grade visible RGB (red, green, and blue) and MS (typically RGB combined with a near-infrared and/or red-edge band) cameras. Such low-cost sensors mounted on UAVs have successfully been used to predict grain yield [37], detect plant diseases [38], and differentiate between aquatic plants and water [39,40]. However, applications that assess plants at the species level remain scant, mainly due to the inadequate spectral resolution of the available sensors [41,42,43,44]. Hill et al. [45] investigated whether a commercially available, consumer-grade UAV system is capable of producing accurate maps depicting the extent of invasion by the yellow flag iris (Iris pseudacorus L.) and whether this could be accomplished more efficiently than with a traditional field survey. Their study methods included pixel-based RF classification of an orthoimage created from the UAV imagery using a random forest (RF) classifier. The results revealed that the manual interpretation of the UAV-acquired imagery produced the most accurate infestation maps for the yellow flag iris, with a false-positive and false-negative classification rate of less than one percent. De Castro et al. [46] used a UAV equipped with an RGB camera combined with decision tree classification and object-based image analysis (OBIA) to map a single grass species (i.e., Cynodon dactylon) within vineyards. The classification was complicated by the high spectral similarity of grape vines, cover crops, grass species, and bare soil. Height information from a digital surface model (DSM) overcame the spectral similarity limitation and allowed for more precise maps of Cynodon dactylon distributions to be produced. Various methods were developed and evaluated by Dash et al. [47] for the remote detection of exotic invasive trees, namely Pinus sylvestris and P. ponderosa, in New Zealand’s South Island. Their study examined the effectiveness of machine learning for classifying multispectral and laser scanning data collected from both a human-crewed aircraft and a UAV. The running logistic regression and RF models provided highly accurate (Kappa < 0.996) detection of the invasive conifers. Marques et al. [48] investigated multitemporal analysis to automatically detect and map individual trees. UAV imagery was used to calculate RGB-based and visible and near-infrared (VNIR)-based VIs, which were then combined with a canopy height model. Using RGB-based Vis, an overall segmentation accuracy greater than 95% was achieved. Nevalainen et al. [21] investigated the performance of UAV-based photogrammetry and HS imaging for individual tree detection and tree species classification in boreal forests. They used a UAV equipped with a frame-format hyperspectral camera and an RGB camera. RF and multilayer perceptron (MLP) models produced the best results in the study, with a 95% OA and an F-score of 0.93.
Most of the existing studies that evaluated UAV systems aimed to identify and map invasive and exotic species, whereas others considered plantations with set distances between individual plants which are quite easily distinguishable from natural vegetation. However, the manual image interpretation used in some other studies is not suited to large spatial domains, especially those with complex structures [45,49]. Manual image interpretation can be particularly slow to carry out and is a less time-efficient method.
In situ spectroscopy studies carried out in South Africa [19] and Australia [50] showed that hyperspectral (HS) data could be used to distinguish some invasive and co-existing species [19,51]. This suggests that HS data can potentially be used to differentiate and map indicator rangeland species and assist in rangeland ecological assessments. The Nama Karoo covers a large part of the south-western part of South Africa and a small portion of Namibia. It is the third largest biome in South Africa, covering a total of 277,000 km2 [50]. Mapping plant species over such a large area is unfeasible using in situ assessments only. The vegetation layer in the Nama Karoo is dominated by shrub species less than 1 metre in height. The low rainfall results in plants growing a fair distance away from each other to reduce competition. Low plant density and a shrub layer create the ideal environment for testing UAV data for the classification of plant species.
The principle aim of this study was to evaluate to what extent machine learning, applied to HS data measured in situ using a full-spectrum spectroradiometer, can differentiate between common Karoo shrubs and grass species. The secondary aim was to compare the models produced from the in situ HS data to those produced using UAV HS and MS data. An assessment of guided regularised random forests (GRRFs) for improving spectral discrimination of common Karoo shrubs and grass species was also carried out.
Successful outcomes would allow ecological impact surveys to be carried out over larger and more inaccessible areas across semi-arid rangelands in South Africa (and potentially similar environments). They would also provide safer, less expensive, and more time-efficient methods for the classification of vegetation species as these surveys are required to assess the sustainability of existing rangeland management practises.
2. Materials and Methods
2.1. Study Area
The study site is located in the Carnarvon Agricultural Research Station, approximately 25 km west of the town of Carnarvon in the Northern Cape province of South Africa (31.0086°S, 21.8939°E) at an altitude of about 1310 m above mean sea level. This area is representative of the north-western Karoo rangelands which, over the last century, have been exploited mainly for sheep farming [52]. The long-term (1927–2019) mean annual rainfall is 201 mm, with most rainfall occurring during late summer and early autumn. The average daily temperature is 23.3 °C in the summer and 8.5 °C in the winter.
The study area is covered by the Western Upper Karoo vegetation type within the Nama Karoo Biome. This biome sustains mainly low shrublands with a fluctuating grass component accredited by rainfall quantity and seasonality [53,54]. Grass dominates in the aeolian sand patches. The vegetation consists primarily of dwarf shrubs such as Pentzia spp., Eriocephalus spp., and Ruschia intricata, and grass species such as Stipagrostis spp. The soils are very shallow, with the A-horizon between 10 and 500 mm deep and mainly consisting of Glenrosa and Mispa soils. The A-horizon has an average clay content of between 10 and 12% [55].
2.2. Data Collection and Preparation
In situ data collection was conducted across three sites at the end of the rainy season from 1 to 15 May 2021 between 10:00 and 14:00 on sunny, cloudless days. Data collection took place at a time when the plants were fully grown, had reached their peak productivity level, and were experiencing little to no moisture stress. The 15 most dominant plant species were selected for sampling (Table 1). These species represent 90% of the plant species composition at the study sites and represent a wide range of palatable to less palatable species used to indicate veld condition.

Table 1.
Plant species selected for sampling.
The palatability of each plant species selected for this study was determined using literature sources, specifically the book by David Shearing on the Karoo: South African Wild Flower Guide. This source provides comprehensive information on the palatability of plant species based on their chemical composition, physical traits, and also the observed grazing preferences by livestock. Thus, the classification of species palatability is grounded on established botanical and ecological research, to ensure an accurate representation of various veld conditions.
Other potentially problematic species such as Senecio, Galenia, Heliotropium, Chrysocoma, Tylecodon, Dichapetalum, Vachellia, and Drosanthemum were not included in the analysis due to their relatively lower abundance and coverage in the specific study sites during the sampling period. While these species are present in the Nama Karoo, their representation was insignificant in the study sites. Additionally, the lower abundance of the grass species Enneapogon did not justify its inclusion in the list of the 15 dominant species.
2.3. Plant Survey
The dominant plant species at the study area were identified through visual identification. A Trimble® R8 survey-grade real-time kinematic (RTK) global positioning system (GPS) with a base station (Trimble Inc., Westminster, CO, USA) was used to record the exact centre position of the plants. A base station is set up at a known location and continuously broadcasts correction data to the GPS receiver. The GPS receiver, mounted on a survey pole, receives the correction data and uses it to calculate its position in real time with high accuracy. When the receiver is moved to the location of each plant, it records the coordinates of the plant’s location. The base station and the receiver communicate through radio frequencies, allowing the correction data to be transmitted in real time, resulting in a higher accuracy level than traditional GPS systems [56]. Using post-processing, up to 8 mm horizontal and 11 mm vertical positional accuracy can be achieved [56], but in practise, horizontal error estimates of 32 mm were attained. This error is low compared to the spacing of the plants (>200 mm).
2.4. In Situ Hyperspectral Data Acquisition (Spectroradiometer)
In situ HS data were collected using the FieldSpec4® Analytical Spectral Device (ASD) spectroradiometer (Analytical Spectral Devices Inc., Boulder, CO, USA). This spectroradiometer collects reflectance in the 350 to 2500 nm spectral range and uses the respective bandwidths of 1.1 nm and 1.4 nm in the spectral ranges of 350 to 1000 nm and 1001 to 2500 nm, respectively. Scans occurred between 10:00 and 14:00 local time under clear skies and stable wind conditions, as recommended by Sibanda et al. [57]. The spectral response of each plant was collected at canopy level by holding the detectors at a nadir-looking angle approximately 25 cm above the plant or in relation to the height of the plant so that its field of view (FOV) did not exceed the area of the plant. A bare fibre-optic cable was used, and the FOV was approximately 10 cm. The spectroradiometer was held at arm’s length from the observer to avoid scattered light from surrounding objects, such as the operator’s clothing or instrument [33,34]. The spectroradiometer was set to automatically collect and average 20 spectral measurements for each sample spectrum. After every 10 measured spectra, the instrument was optimised and calibrated, or whenever necessary; for example, when the instrument became saturated due to changing atmospheric conditions or irradiance of the sun [34]. The instrument was calibrated and optimised using a Spectralon white reference surface plate of 100% reflectance. Table 1 records the total averaged spectra for each plant species used in this study.
2.5. Camera Used in the Multispectral Remote Sensing Surveys
The Parrot Sequoia camera used in this study has four separate MS sensors with global shutters. The camera captures surface reflectance in the green (480–520 nm), red (640–680 nm), red-edge (730–740 nm), and NIR (770–810 nm) wavelength bands [58]. The horizontal FOV was set to 61.9°, and the vertical FOV to 48.5°. The images are 1280 × 960 pixels in size and are saved in RAW format. This camera’s maximum frame rate is one frame per second, and recordings were made on the internal memory. The camera contains a separate sunshine sensor with a hemispherical FOV that measures solar irradiance in the same spectral bands as the four image sensors. A RAW file is the uncompressed and unedited image data collected by the sensors of a digital camera or scanner, capturing a high level of visual detail while maintaining lossless quality. The camera was mounted to a fixed-wing Sensefly eBee UAV. An altitude of 50 m above the ground with the MS was maintained, resulting in <8 cm of resolution imagery.
The imagery was processed in Pix4Dmapper (v.4.6.4, Pix4D, Lausanne, Switzerland) using a standard Agriculture (Ag) MS workflow. The images for each site were processed to form a single orthomosaic. The ground control points (GCPs) were manually identified in the images and georeferenced using RTK GPS coordinates collected for reference plates at the site. A GPS receiver is included with the MS camera’s sunshine sensor and an inertial measurement unit (IMU) to measure the position and orientation of the sensor when capturing images [58]. Radiometric calibration was conducted in Pix4D using the reflectance target images from the Parrot Sequoia camera and the metadata from the sunshine sensor.
2.6. Camera Used in the Hyperspectral Remote Sensing Surveys
A Specim FX10 HS camera was used to collect remotely sensed hyperspectral images. The camera collects reflectance in the visible and near-infrared (VNIR) (400 to 1000 nm) wavelengths at 2.9 nm bandwidths. The camera offers wavelength selection from 224 bands, built-in image correction, and unified spectral calibration. Regarding spatial resolution, the sensor produces 1025 × 1025-pixel images at a maximum of 327 frames per second (FPS). The FX10 was mounted to a multirotor DJI Wind 4 UAV. A flight altitude of 70 m above the ground was maintained, resulting in <8 cm resolution imagery. The HS flights were carried out 10 min after the MS flights to minimise the impact of changing atmospheric conditions and solar irradiance.
The choice of UAVs was based on each sensor’s specific requirements. The fixed-wing UAV provided stable and efficient coverage for the MS camera and was set up for this specific sensor. The HS camera weighs more than the fixed-wing UAV, so it cannot carry such a heavy camera. The multirotor UAV offered greater manoeuvrability, lifting capability, and stability for the HS camera, reducing the risk of image blur.
2.7. Spectroscopy and UAV Data Analysis
The remotely sensed spectra were extracted from the HS and MS imagery at using the centroids of each plant as surveyed by the GPS. The UAV spectra were extracted using a buffer method that returns the average value from all pixels within a specified 3 × 3-pixel buffer area around the centroid [59].
Variable importance is a useful by-product of random forest (RF), used for feature ranking. Thus, RF variable importance reveals which wavebands (or waveband set) are most relevant for classification [60]. In this study, Gini importance was used as a measure of waveband importance. The Gini importance score quantifies the predictive ability of each variable; hence, it gained importance measures over all the trees in the RF [60]. The most essential features from a data set for classifying plant species were identified through feature selection. The best spectral bands were chosen using the feature selection algorithm known as guided regularised random forest (GRRF).
The accuracy of the species classifications based on the in situ spectroradiometer (henceforth referred to as ASD) and remotely sensed (HS and MS) data were determined by comparing their confusion matrices, overall accuracies (OAs), Kappa scores, and McNemar tests produced from an independent test data set (30% of samples). OA is a ratio (%) between the number of correctly classified samples and test samples [19]. The McNemar test assessed the statistical significance of differences between the RF and GRRF classification accuracies of each assessment method [61]. McNemar’s test first assesses a 2 × 2 contingency table and then considers only correct and incorrect points [20]. The RF OOB accuracies, which provide an unbiased estimate of the internal RF error, were used to assess misclassifications.
3. Results
3.1. Spectral Responses of Plant Species
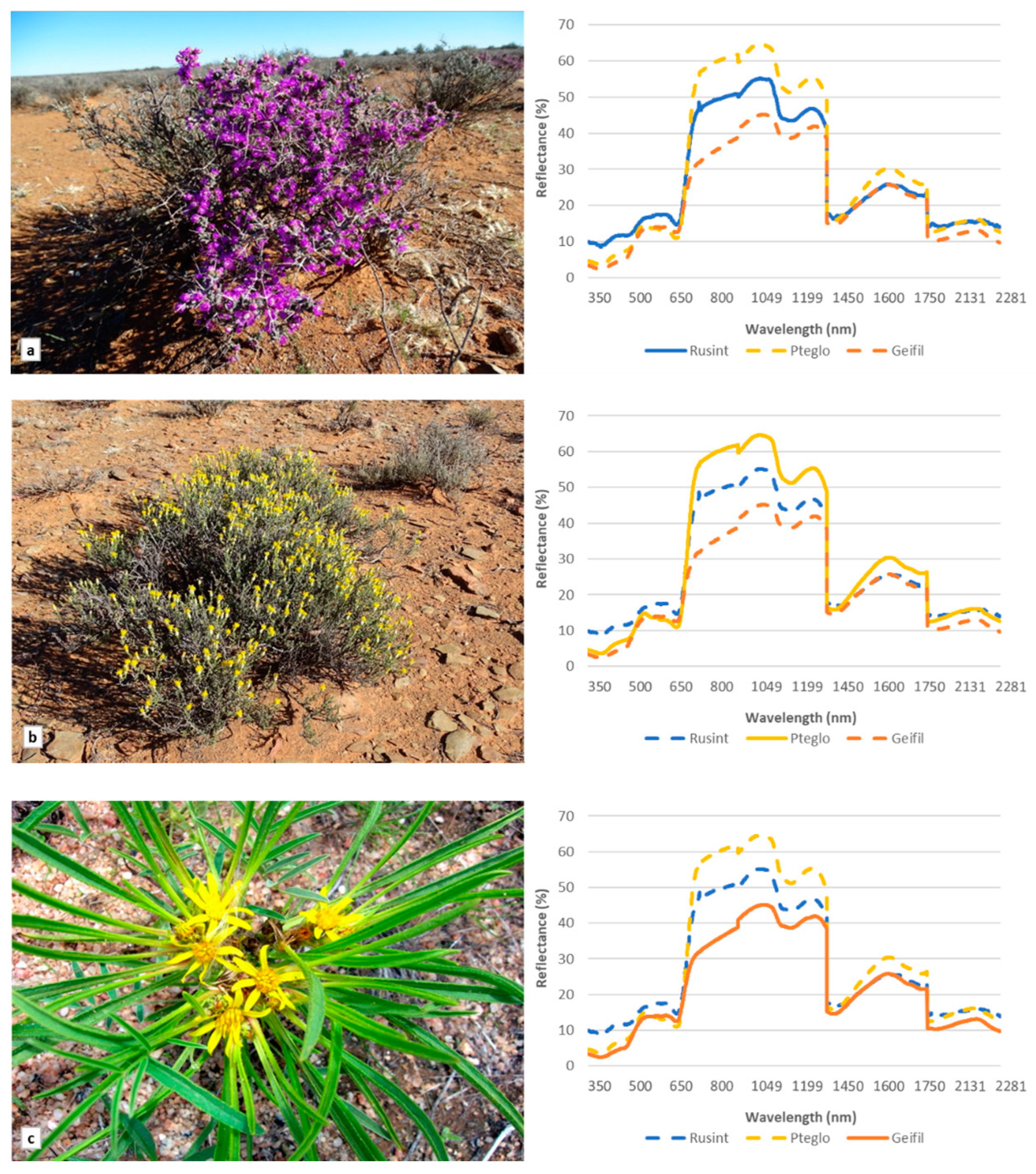
Spectral signatures for all 15 identified species were extracted from the ASD data (included in Supplementary Materials). Three species, Ruschia intricata, Pteronia glomerata, and Geigeria filifolia, are shown in Figure 1 as examples to demonstrate the variation in the spectral responses among some of the species. Multiple scattering occurred in the NIR (750–1075 nm) region and significant differences in magnitude were noted among the different species. High variation in reflectance magnitude among species was also noted in the blue (446–500 nm) and red (620–680 nm) regions.
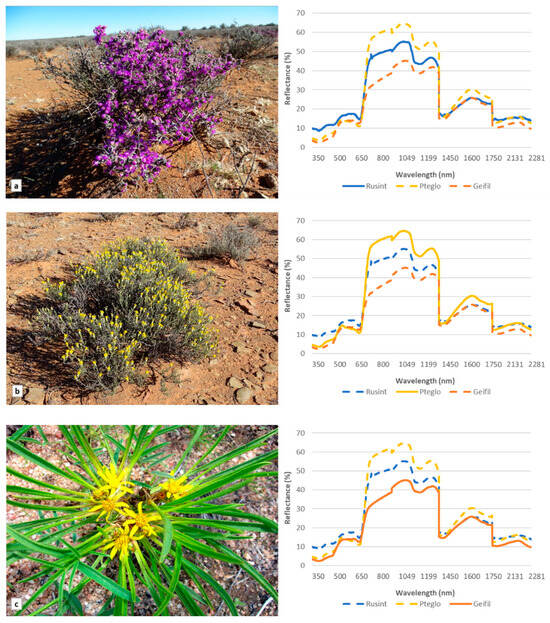
Figure 1.
Images and spectral reflectance based on the analytical spectral device of three species examined in the study: (a) Ruschia intricata, (b) Pteronia glomerata and (c) Geigeria filifolia.
3.2. Classification Accuracies and Variable Importance
3.2.1. Overall Accuracies
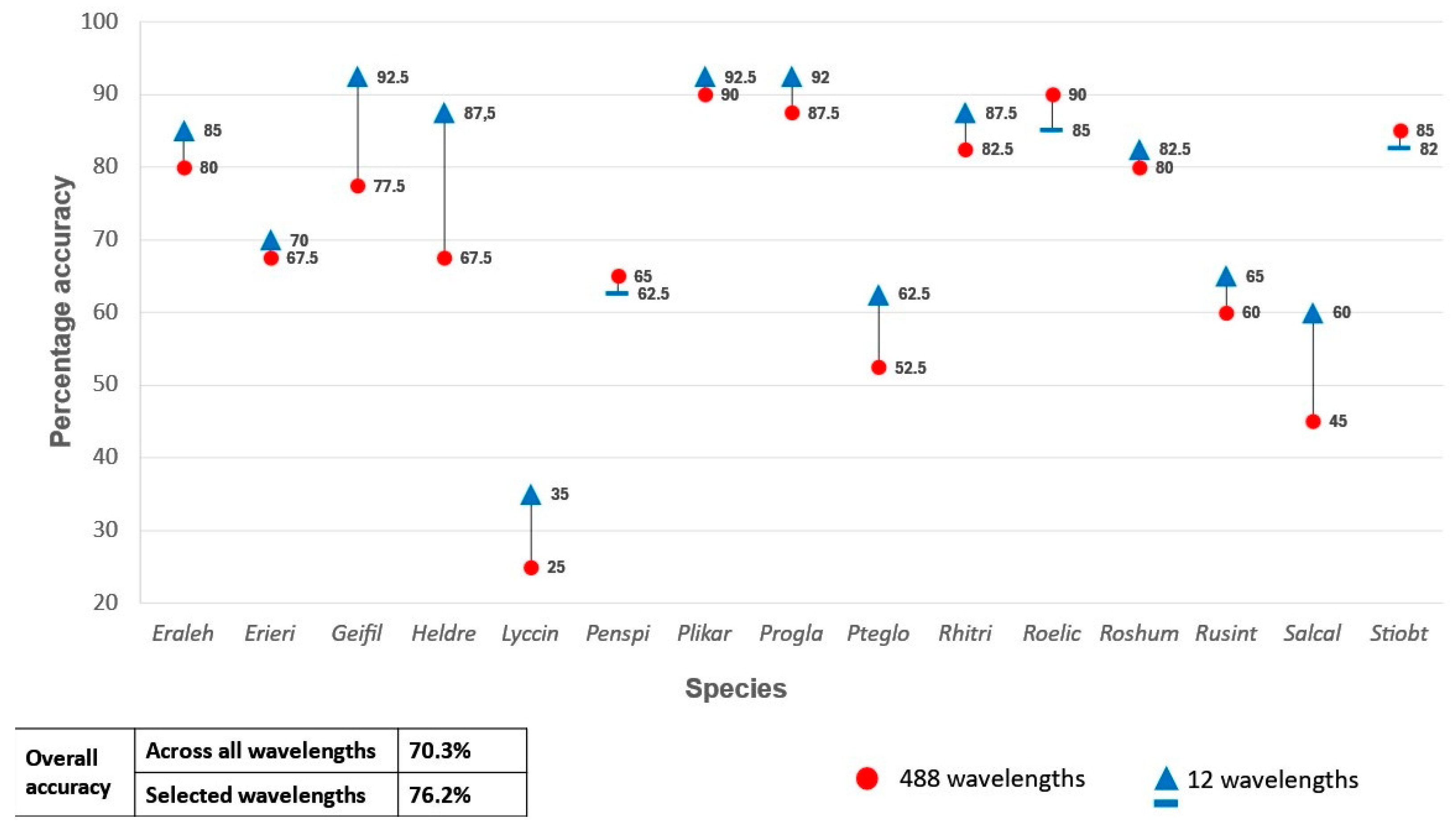
Table 2 shows that an overall species classification accuracy of 84.8% (0.83 Kappa) was achieved when all 1522 wavelengths from the in situ ASD measurements were used as predictor variables. Using all 224 wavelengths of the remotely sensed HS image as predictor variables yielded an OA of 70.3% (Table 3) and Kappa of 0.70. The lowest OA (37.5%) was achieved when the four bands from the remotely sensed MS data were used as inputs for the RF classifier (Table 4).

Table 2.
Confusion matrix of the overall classification and Kappa for discrimination between the 15 species using all 1522 wavelengths from the analytical spectral device data set. The error was calculated using the out-of-bag method and a test data set.

Table 3.
Confusion matrix of the overall classification and Kappa for discrimination between the 15 species using all 224 wavelengths from the unmanned aerial vehicle hyperspectral data set. The error was calculated using the out-of-bag method and a test data set.

Table 4.
Confusion matrix of the overall classification and Kappa for discrimination between the 15 species using all four wavelengths from the unmanned aerial vehicle multispectral data set. The error was calculated using the out-of-bag method and a test data set.
3.2.2. Species-Specific Accuracies
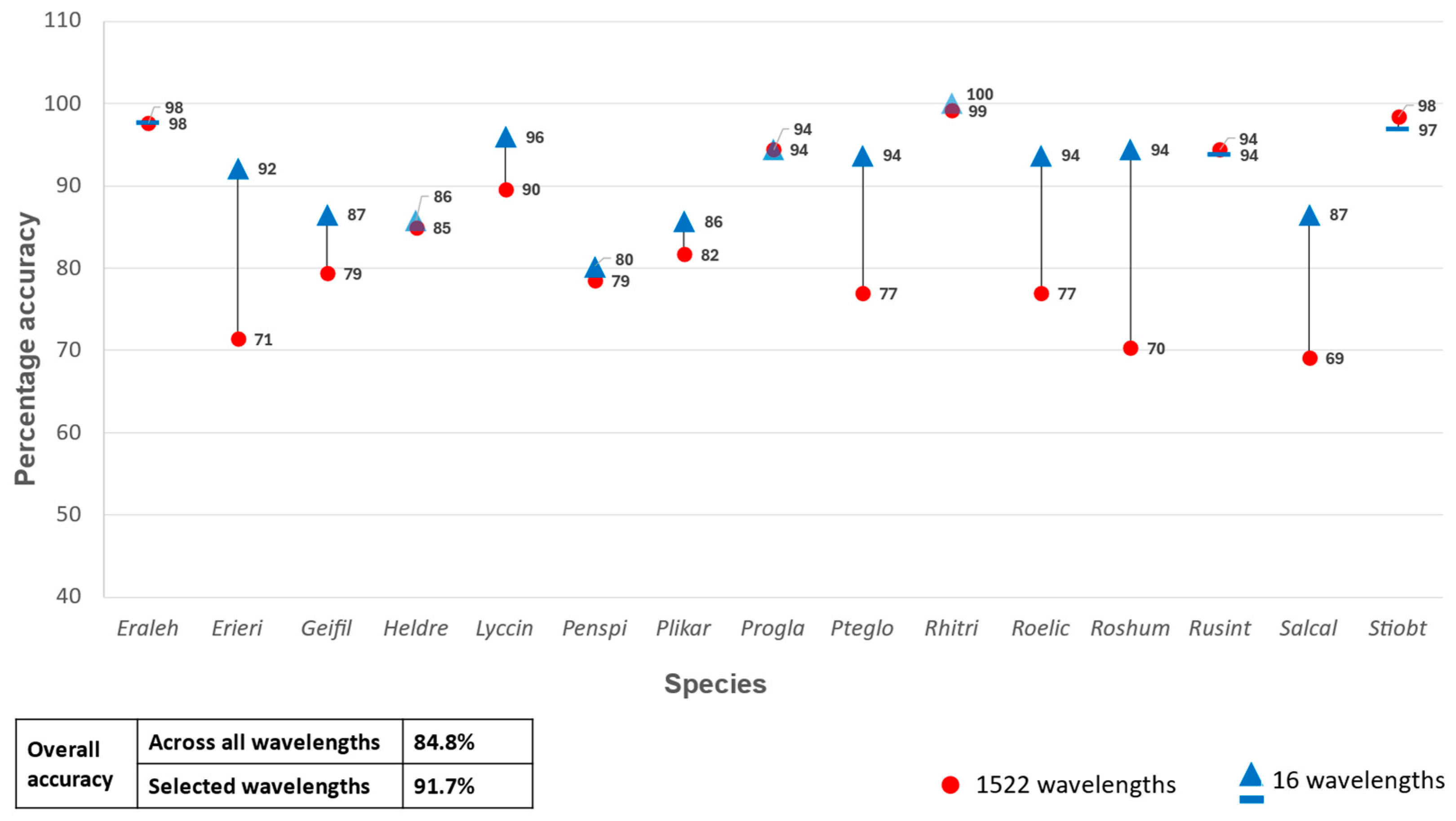
The highest overall classification accuracy was achieved for the species Rhigozum trichotomum (Rhitr) (99.1%), followed by Rosenia humilis (95.2%) and Salsola calluna (82.9%), when using the ASD data as input for the classifier. The species that obtained the lowest accuracy scores are Lycium cinereum (72.4%) and Pentzia spinescence (75.0%) (Table 2).
In terms of the remotely sensed data, the HS data set achieved the highest overall classification accuracy for Helichrysum rosum (84.4%) and the poisonous species Geigeria filifolia (83.8%). The two species that achieved the lowest accuracies using these data are Pteronia glomerata (41.8%) and Salsola calluna (44.0%). When the remotely sensed MS bands were used as predictor variables, the highest overall classification accuracy was achieved for the encroaching shrub species Prosopis glandulosa (58.1%) and Pentzia spinescence (59.1%). The discrimination between Helichrysum rosum (21.4%) and Salsola calluna (20.0%) was very low using MS data.
3.2.3. Most Important Bands
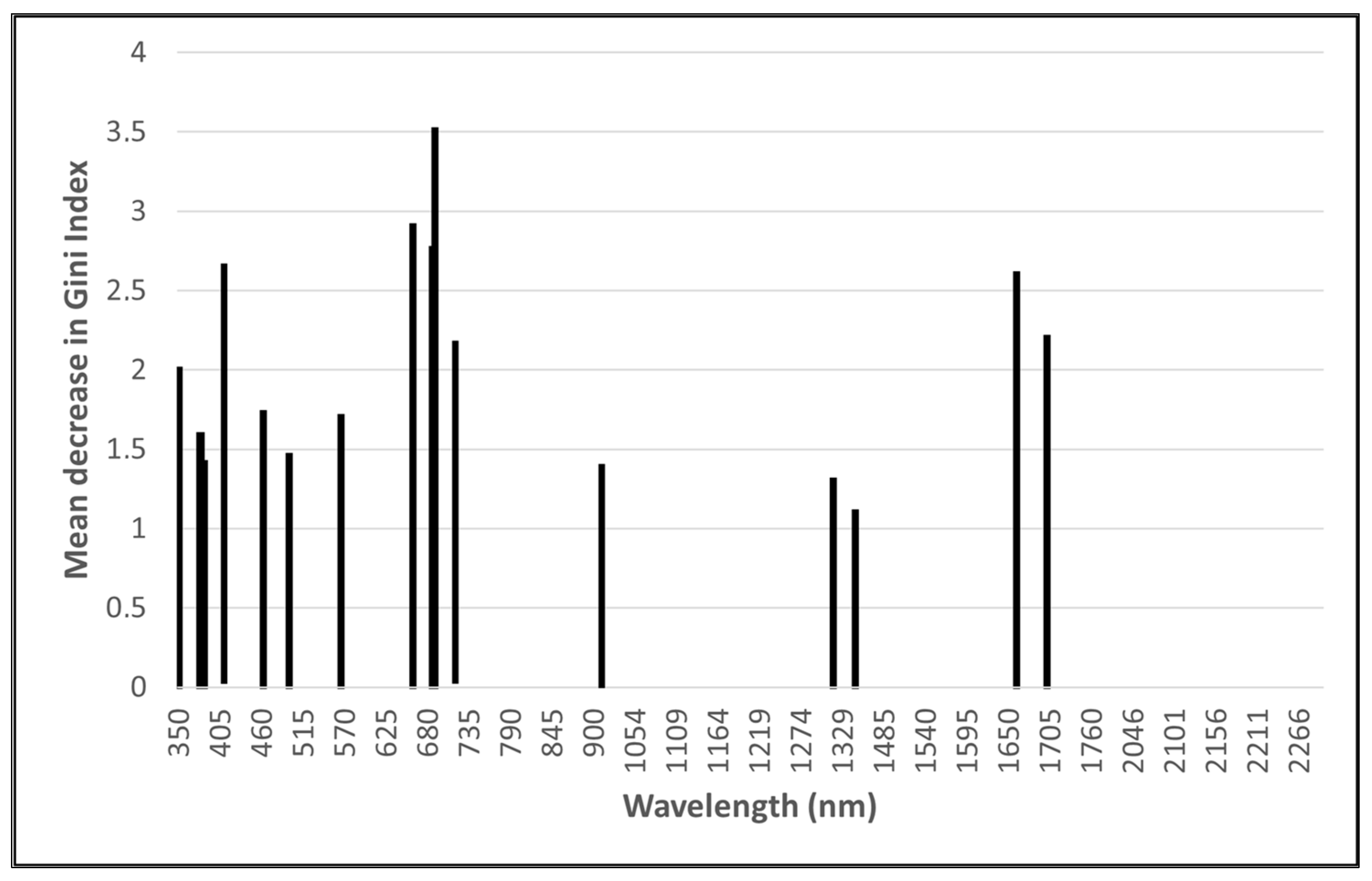
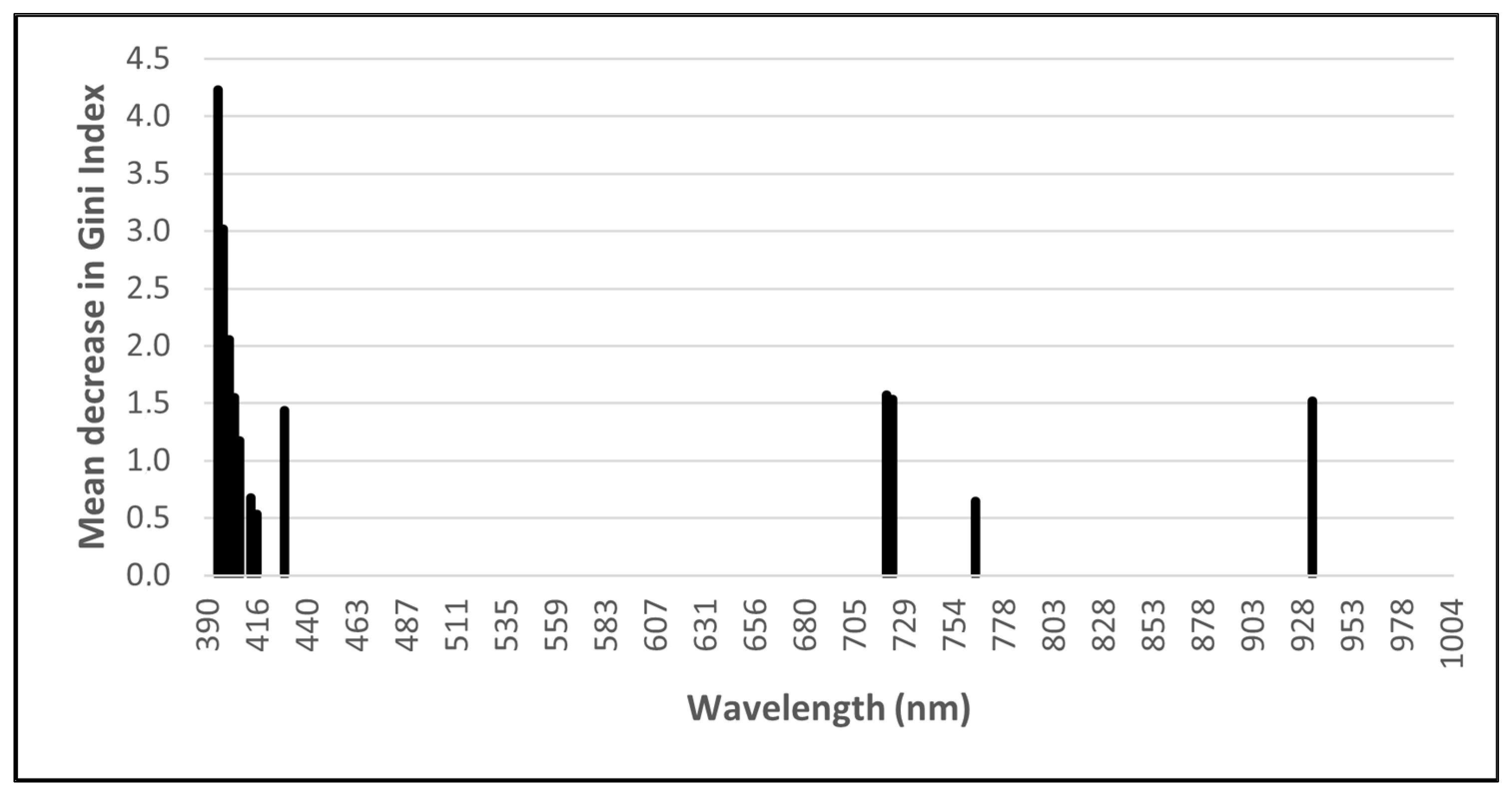
The RF importance scores were used to enable the GRRF selection of a subset of wavelengths to reduce the dimensionality of the input data and potentially improve classification accuracies. Using GRRF, 16 optimal wavelengths (Figure 2) for the ASD data and 12 optimal wavelengths (Figure 3) for the remotely sensed HS data were identified. Due to its low dimensionality (four bands), no feature selection was performed on the MS data.
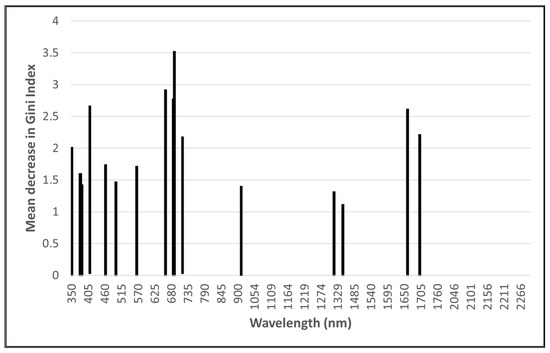
Figure 2.
Optimal analytical spectral device (ASD) wavelengths identified using guided regularised random forest.
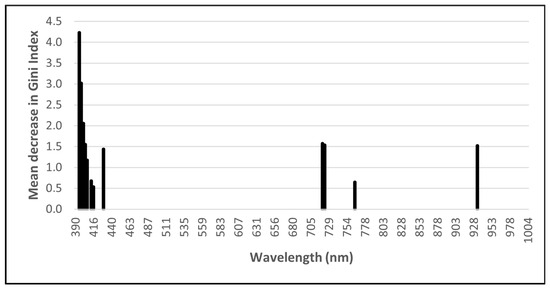
Figure 3.
Optimal remotely sensed hyperspectral (HS) wavelengths identified using guided regularised random forest.
The most important ASD wavebands are in the ultraviolet, visible (VIS), red-edge, and SWIR regions of the electromagnetic spectrum, with the majority occurring in the 350 to 450 nm region. Most of the wavebands selected from the remotely sensed HS imagery are in the VIS and red-edge regions, with the majority being in the 398 to 430 nm range.
3.2.4. Classification Results Using Selected Bands
A second set of classification experiments were carried out using only the selected ASD and HS bands as predictor variables in the RF classifier. When the 16 selected ASD bands were used as input, the classification model yielded an OA of 91.7% (Figure 4). This is a 6.9% increase in OA compared to when all the ASD bands were used as predictor variables. An OA of 76.2% was achieved when the selected 12 remotely sensed HS bands were used as predictor variables (Figure 5), which constitutes a 5.9% increase compared to when all the HS bands were used. Both increases (for the ASD and HS data) were statistically significant (p < 0.05) based the McNemar test.
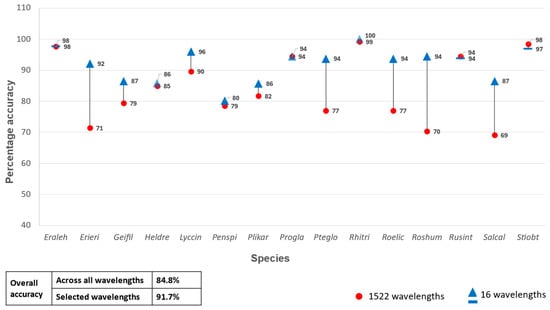
Figure 4.
Percentage accuracy for discriminating between the 15 species using all 1522 wavelengths from the analytical spectral device data set vs only the selected 16 wavelengths as predictor variables in the random forest classifier.
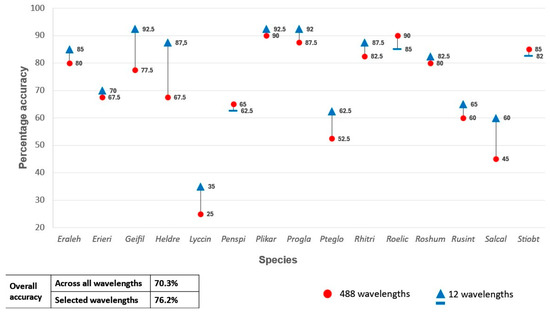
Figure 5.
Percentage accuracy for discriminating between the 15 species using all 224 wavelengths from the unmanned aerial vehicle hyperspectral data set vs only the selected 12 wavelengths as predictor variables in the random forest classifier.
Correlations among the selected predictor variables were calculated per data set to determine collinearity (see supplementary data). Pairs of highly correlated (R2 > 0.7) variables were identified, after which one was manually discarded. This resulted in a set of seven ASD bands, namely 351 nm, 366 nm, 386 nm, 394 nm 407 nm, 1431 nm, and 2031 nm. When these seven bands were used as the only RF predictor variables, an OA of 92.2% was achieved. Similarly, the predictor variables for the remotely sensed HS data were reduced to three bands, namely 397.66 nm, 720.87 nm, and 723.6 nm. However, using these bands as the only predictor variables produced an OA of 57.8%, which is substantially (18.4%) lower than when the 12-band subset was used.
4. Discussion
The purpose of collecting in situ ASD spectral measurements was (a) to develop a database of spectral signatures for the dominant plants found at the study site and (b) to establish a baseline against which the value of the two remotely sensed data sets for species identification and mapping can be compared.
4.1. Classification of ASD Measurements, HR Imagery and MS Imagery
As can be expected, the remotely sensed HS imagery was less effective (compared to the in situ ASD data) for discriminating among the 15 species. However, the level of accuracy achieved with the remotely sensed HS imagery (up to 91.7% OA) is satisfactory given the potential for wide-scale implementations. This level of accuracy is similar to that reported by Ishida et al. [62], in which remotely sensed (UAV) HS imaging was used in several vegetated areas [62], including mango orchards, to classify 14 different ground objects such as grass, soil, man-made objects, sugarcane, rice fields, mango, ash, and mahogany trees, and a few more plant species. Ishida et al. [62] found that shade had a large impact on OAs, but managed to increase accuracy by training the machine learning algorithm with shaded and illuminated samples separately [62]. Given the low height of the shrub layer and the sparseness of Karoo vegetation, shade was not an issue in our study. Our OAs are also similar to those of Yan et al. [63] in which a UAV multi-angle remote sensing method was used to classify contrasting vegetation species, including maise, soybeans, and weeds, and ash, mulberry and peach trees.
The remotely sensed MS imagery in this study was found to be ineffective for the classification of plant species in our study area. Our OAs were lower than those reported in comparable studies [64,65,66]. This is likely due to several factors, including the limited vegetation growth and low biomass at the study site caused by the low rainfall received during the study period. The reduced plant growth led to increased soil reflectance through the shrub foliage, making it harder to classify the species accurately. Additionally, the larger number of plant species in our study may have contributed to the lower OA compared to other studies, as the greater species diversity likely resulted in increased spectral overlap and reduced separability. Louargant et al. [67] also reported higher MS (also UAV) accuracies (OA = 55.5%) than in this study, but their focus was on crop–weed discrimination and not to classify a large number (>10) of species. Other studies on the mapping of forest species using MS UAV systems achieved OAs of between 84% and 92% [68,69]. However, these studies involved only five classes or fewer classes and the grouping of similar species, which likely accounts for the high accuracies. It is difficult to compare our results to those of other studies as the specific species types and number, the study sites, and the scales of implementation are very different.
The relatively large number (15) of plant species considered in this study resulted in much spectral confusion. This confusion will likely increase if the species are in similar phenological stages during the assessment period [66,69]. Following the example of Grybas and Congalton [66], this study considered the most common plant species found in Upper Karoo vegetation rather than a selection of a few very distinct species that exhibit the best separation [70].
4.2. Species-Specific Accuracies
By using remotely sensed (UAV) HS data, this investigation succeeded in distinguishing among 15 common species of Karoo shrubs with 76% accuracy. This holds attractive prospects with the continuous development and commercialisation of UAV-HS systems for vegetation mapping at the species level. The species Helichrysum rosum (84.4%) and Geigeria filifolia (83.8%) were very well differentiated using the remotely sensed HS data. This is promising for the automatic detection of encroaching and poisonous species. Sheep poisoned by ingesting Geigeria filifolia give rise to “vermeersiekte”, also known as vomiting disease. The poisoning occurs mainly in sheep and, to a lesser extent, in goats and cattle. Massive outbreaks of this disease in sheep were reported in the Karoo in the past. More than a million sheep died in 1929/30, and as many as 50,000 died in 1954. Another large outbreak of the disease occurred in 1981 [71]. It is vital for farmers to have access to tools that can assist in identifying and monitoring this deadly species across their rangelands.
The two species that achieved the lowest OAs are Pteronia glomerata and Salsola calluna, both of which have very small, semi-cylindrical or strongly keeled leaves attached close to the branch. The structure of the leaves seems to adversely affect classification accuracy when using the (HS and MS) UAV imagery. In contrast, when using the in situ ASD bands as predictor variables, relatively high classification accuracies for these two species were attained (Salsola calluna 83.3% and Pteronia glomerata 88.1%). More work is needed to investigate if lower UAV flying heights will improve classification accuracies of these species when remotely sensed (in particular HS) data are used as inputs for the classifier.
Other species that suffered from relatively low classification accuracies when the HS remotely sensed images were used are Lycium cinereum, Ruschia intricata and Roepera lichtensteiniana. All of these species are plants with succulent leaves, similar to Salsola calluna. Succulent plants have developed naturally elicited protective compounds to mitigate UV tissue damage and to protect them against UV stress [72,73,74]. The potential impacts of UV light on plants include increased leaf colouration and thickness [75]. This discolouration of leaves is often seen in Karoo plants such as Plintus karooicus, Ruschia intricata and Salsola species with a grey-purple colour and they do not have the typical bright green leaves of other plants [76]. Karoo plants have adapted to searing arid summers with leaves coated in a waxy reflective sunscreen [77,78]. It is noticeable from the classification results of the ASD data that the UV region in the 350 nm to 400 nm range is essential for distinguishing between these succulent species. This range is particularly significant because it encompasses the UV-A and part of the UV-B spectrum, where many protective compounds in succulent plants are most active. These compounds absorb UV radiation, leading to distinct spectral signatures that aid in species differentiation. The poor discrimination of these species by the remotely sensed HS data is attributed to the fact that the HS camera does not cover wavelengths shorter than 398 nm. Exploring an even broader UV range, including wavelengths below 350 nm, could potentially enhance species discrimination by capturing additional spectral features related to their UV protective mechanisms.
4.3. Most Important Bands
The GRRF method demonstrated its efficiency and effectiveness in improving the spectral discrimination of common Karoo shrub and grass species. The results showed that reducing the dimensionality of the in situ ASD or remotely sensed HS data to 12 and 16 bands, respectively, improved the spectral discrimination by up to 7%. This improvement in classification accuracy can be attributed to the reduction in dimensionality, which mitigates the risk of overfitting. Overfitting occurs when the number of predictor variables (bands) is large relative to the number of samples, a phenomenon known as the Hughes effect.
By reducing the number of bands, the GRRF method ensured that the most crucial spectral information was retained while reducing the risk of overfitting. The subset of 12 ASD bands was further reduced to seven bands, maintaining a high classification accuracy of 92.2%. However, when the 12-band, remotely sensed HS subset was reduced, the classification accuracy deteriorated, suggesting that the 12 bands identified by GRRF are the optimal subset for the HS data. The GRRF method effectively addressed the issue of high-dimensional data by reducing the number of bands while retaining important spectral information, thereby improving the spectral discrimination of common Karoo shrub and grass species. The results also highlight the importance of considering the Hughes effect when working with high-dimensional data.
In comparison, Mureriwa et al. [19] found that the spectral separability of Prosopis glandulosa from co-existing species improved when using a subset of 20 bands from a field spectral measurement. While their study focused on different plant species, it is interesting to note that a larger number of bands was found to be sufficient for separability compared to our study. This highlights the importance of considering the unique characteristics of each study area when determining the optimal number of bands.
The GRRF method’s ability to reduce dimensionality and improve classification accuracy demonstrates its potential for broad applicability across various ecological and remote sensing studies. However, it is critical to recognise that the optimal number of bands may vary depending on each study area’s specific characteristics, such as species diversity, environmental conditions, seasonal changes in species, and the spectral properties of the target vegetation. This adaptability is a strength of the GRRF method, as it allows researchers to tailor the band selection process to the unique requirements of their study, thereby maximising classification accuracy and minimising overfitting.
Although the Karoo shrub and grass species were the focus of our study, other ecosystems and vegetation types can apply the principles underlying the GRRF method. Future research should explore the application of the GRRF method to different vegetation types to further validate its universal applicability and identify any necessary adaptations. By considering each study area’s unique features, researchers can ensure that the GRRF method remains a useful tool for spectral discrimination and classification in high-dimensional data situations.
4.4. Recommendations and Limitations
The methods used in this study can be applied to all Karoo rangelands, as the plant species considered have a wide distribution. To the authors knowledge, this is the first study to establish a database of spectral profiles for the dominant Karoo plant species. It is also the first to evaluate the performance of remotely sensed (HS-MS UAV) imagery for discriminating among these species. The classifications using the in situ ASD data provided a suitable benchmark against which the classifications of the remotely sensed data could be compared.
A limiting factor in the study is that vegetation growth was weak and biomass low at the study site because of the low rainfall received during the study period. The low rainfall curtailed plant growth, and so increased soil (background) reflectance through the shrub foliage. It is important to note that the more plant species involved, the lower the classification accuracy will typically decrease as the number of targeted plant species increases. The more diverse the vegetation, the more challenging it is to distinguish one species from another based on spectral data. This can result in overlapping spectral signatures and confusion in the classification process.
Additionally, the presence of many species can increase the background noise and reduce the spectral contrast, making it harder to distinguish one from another. The presence of many species can also increase the complexity of the underlying structure of the data, making it more challenging to find patterns that can be used to classify the different species accurately. By grouping plant species with similar nutritional values for animals, the OA can be improved. This approach is based on the idea that plant species with similar nutritional profiles will have more similar spectral signatures, reducing the complexity of the classification process. Instead of focussing on individual species, which can have overlapping spectral signatures, we can categorise plant species into broader value classes based on their nutritional content and grazing value. This can potentially reduce the classification process’s complexity and accuracy by focussing on broader categories rather than individual species.
To effectively implement this approach, one could categorise plant species into value classes based on their nutritional content and grazing value. This would involve grouping plant species that share similar nutritional profiles into the same value class. Spectral signature analysis can then be used to scrutinises the spectral signatures of these value classes to pinpoint distinct patterns suitable for classification. For instance, we can classify palatable grass species or Karoo shrub species based on their comparable grazing value and palatability values. This approach leverages the similarities within each value class to improve classification accuracy by reducing the spectral overlap and enhancing the distinctiveness of the spectral signatures within each class. As a safety precaution, most UAV systems are not operated below 50 metres above the ground. However, it is recommended that future studies evaluate whether lower than 50 metre flying heights will increase the classification accuracy of shrubs with tiny leaves, such as Stipagrostis obtusa. However, pilots ought to guard against varying flying heights. Such variations may occur due to wind gusts, changing elevation and flight-path corrections. Such changes impact resolution and result in orthorectification issues if the image overlap is insufficient [79,80]. At lower flying heights such variations might have a larger impact. Experience gained from this study suggests that a minimum frontal overlap of 80% and 70% side overlap at a flight height of 50 metres above the ground is recommendable.
Although UAV systems can overcome most of the limitations of satellites (e.g., lower resolution, timespans between images, and cloud cover), they are not without shortcomings. Due to the forward motion of a UAV platform, the captured images can be blurred due to strong winds or gusts [81,82,83]. Moreover, UAVs cannot operate on days with winds exceeding 12 m/s. The short battery life of UAVs is also a significant restriction, with only short 15 min flight times being achievable when carrying heavy payloads such as an HS camera. The use of fixed-wing UAVs which can cover larger areas in a single flight is advisable. There is, however, the prospect that new generations of HS cameras will be much smaller and lighter.
An inherent limitation of this study is that, due to the low rainfall during the study period, only one survey was carried out. The rainfall was insufficiently spread across the four seasons of the study period to consider plant phenology, as plants only reacted and showed growth after the summer rains. It is likely that the influence of plant phenology would increase OAs for certain species at certain times of the year. Eriocephalus spp. (completely white when in seed) and Ruschia spp. (bright pink flowers) are cases in point. These phenological changes significantly alter the spectral signatures of the plants, which can enhance or reduce the separability of species in remote sensing data.
Flowering and seeding stages can cause distinct spectral variations due to changes in colour, reflectance, and other optical properties. For example, the bright pink flowers of Ruschia spp. increase reflectance in the red and near-infrared regions, while the white seeds of Eriocephalus spp. reflect more light across the visible spectrum. These changes can be critical for improving discrimination accuracy among plant species during remote sensing assessments.
Assessments of plant phenological changes over the four climatic seasons could help to determine the ideal period for obtaining the highest discrimination accuracy among the plant species during normal rainfall conditions. This notwithstanding, Pfitzner et al. [51] used remotely sensed HS imagery to monitor non-native grasses and concluded that maximum separability was not achievable under a specific phenological or seasonal stage. They found that species responses to a significant rainfall event or to disturbances by grazing animals resulted in maximum separability.
It is vital that future cost-effective MS sensors for UAVs target wavebands in the VIS, red-edge, and SWIR regions. As determined in this study, wavebands between 350 and 450 nm are particularly useful for Karoo plant species differentiation. It is noteworthy that the wavebands between 350 nm and 397 nm are currently not covered by most HS or MS sensors that UAVs can carry.
In this study, we implemented a GRRF, filter feature selection method to reduce the dimensionality of the ASD and HS data. It would be worthwhile for future studies to test additional feature selection algorithms, such as the Boruta wrapper, which have been very effective in previous studies [84,85].
5. Conclusions
By employing spectroscopic methods, this study aimed to evaluate the effectiveness of remotely sensed, UAV-based data for differentiating among Karoo plant species. The ability to map plants using remotely sensed imagery will aid ecological impact surveys required to assess the sustainability of existing rangeland management practises. Using the ASD data, we identified important wavebands between 350 and 400 nm that are not covered by either the HS or MS sensor. Current UAV HS sensors generally only detect wavelengths of 400 nm or longer. This explains the lower (compared to the ASD data) OA achieved when the remotely sensed HS imagery was used as input for the classifier. Succulent plant species were especially poorly differentiated using the HS imagery. High (>90%) OAs are vital for operational implementations, particularly if the methodology developed in this study is replicated in the neighbouring succulent Karoo vegetation type, which consists primarily of succulent shrub species and is one of the world’s most diverse plant communities. The MS imagery was found to be unsuitable for species discrimination in the Karoo because it does not adequately represent the critical 398 and 430 nm region of the electromagnetic spectrum. New airborne MS sensors should preferably focus on these specific wavebands to increase the value of MS sensors for plant species differentiation in the Karoo environment.
The vegetation condition in the study site was not in optimal condition during the study period due to the meagre rainfall. Yet, despite the poor conditions and high soil background noise, the classifications performed remarkably well. Future assessments may provide clarity on whether if higher accuracies can be obtained under better climatic conditions. The high OAs achieved with only a few narrow bands underlines the potential of HS remote sensing as a tool for mapping indicator grass and Karoo shrub species.
Plant ecologists have long argued that small-scale or discontinuous observations are inadequate to determine the accurate spatial distribution of vegetation species over large spatial scales [18]. Vegetation classification and monitoring can now be carried out over larger and inaccessible areas by using UAVs. The remote monitoring of vegetation composition can assist in managing extensive Karoo rangelands more sustainably. This can help prevent livestock deaths by identifying outbreaks of poisonous plant species. It can also provide a safer, less expensive and less time-consuming method for land users and researchers to better understand the changes in vegetation composition due to land degradation, high stocking densities (overgrazing), locust outbreaks, and climate change.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/rs16203869/s1, Supplementary data.
Author Contributions
Conceptualization, C.J.H. and A.v.N.; methodology, C.J.H.; formal analysis, C.J.H.; investigation, C.J.H.; writing—original draft preparation, C.J.H.; writing—review and editing, A.v.N. and C.J.H.; visualisation, C.J.H.; supervision, A.v.N.; project administration, C.J.H.; funding acquisition, A.v.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work is based on the research supported in part by the National Research Foundation of South Africa (Grant Numbers: 142438).
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
The authors thank the Department of Agriculture, Environmental Affairs, Land Reform and Rural Development (Northern Cape province) for maintaining and continuously monitoring this valuable long-term grazing trial. The care for and management of the trial by Tommie Buys, the research station manager, is also recognised. We also thank Pieter de Necker for assisting with language editing an early version of this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Allen, V.G.; Batello, C.; Berretta, E.J.; Hodgson, J.; Kothmann, M.; Li, X.; McIvor, J.; Milne, J.; Morris, C.; Peeters, A.; et al. An International Terminology for Grazing Lands and Grazing Animals. Grass Forage Sci. 2011, 66, 2–28. [Google Scholar] [CrossRef]
- Zerga, B. Rangeland Degradation and Restoration: A Global Perspective. Point J. Agric. Biotechnol. Res. 2015, 1, 37–54. [Google Scholar]
- Liebig, M.A.; Gross, J.R.; Kronberg, S.L.; Hanson, J.D.; Frank, A.B.; Phillips, R.L. Soil Response to Long-Term Grazing in the Northern Great Plains of North America. Agric. Ecosyst. Environ. 2006, 115, 270–276. [Google Scholar] [CrossRef]
- Stocking, M.A.; Mumaghan, N. Handbook for the Field Assessment of Land Degradation. London: Earthscan In (O’Higgin, RC, Eds), Savannah Woodland Degradation Assessments in Ghana: Integrating Ecological Indicators with Local Perceptions. Earth Environ. 2001, 3, 246–281. [Google Scholar]
- Schwilch, G.; Hessel, R.; Verzandvoort, S. (Eds.) Desire for Greener Land. In Options for Sustainable Land Management in Drylands; University of Bern, Centre for Development and Environment CDE: Bern, Switzerland; Alterra: Wageningen, The Netherlands; ISRIC—World Soil Information: Wageningen, The Netherlands; CTA—Technical Centre for Agricultural and Rural Cooperation: Wageningen, The Netherlands, 2012. [Google Scholar]
- Nachtergaele, F.; Petri, M.; Biancalani, R.; van Lynden, G.; van Velthuizen, H.; Bloise, M. Global Land Degradation Information System (GLADIS). Beta Version. An Information Database for Land Degradation Assessment at Global Level; Land Degradation Assessment in Drylands Technical Report; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2010; Volume 17. [Google Scholar]
- Von Braun, J.; Gatzweiler, F.W. Marginality: Addressing the Nexus of Poverty, Exclusion and Ecology; Springer Nature: Berlin, Germany, 2014. [Google Scholar]
- Barbier, E.B.; Hochard, J.P. Does Land Degradation Increase Poverty in Developing Countries? PLoS ONE 2016, 11, e0152973. [Google Scholar] [CrossRef]
- Barbier, E.B.; Hochard, J.P. Land Degradation and Poverty. Nat. Sustain. 2018, 1, 623–631. [Google Scholar] [CrossRef]
- Hoffmann, T.; Todd, S.; Ntshona, Z.; Turner, S. Land Degradation in South Africa; University of Cape Town: Cape Town, South Africa, 2014. [Google Scholar]
- Middleton, N.; Thomas, D. World Atlas of Desertification, 2nd ed.; Arnold, Hodder Headline, PLC: London, UK, 1997; ISBN 0340691662. [Google Scholar]
- Hoffman, T.; Ashwell, A. Nature Divided: Land Degradation in South Africa; University of Cape Town Press: Cape Town, South Africa, 2001; ISBN 1919713549. [Google Scholar]
- Mani, S.; Osborne, C.P.; Cleaver, F. Land Degradation in South Africa: Justice and Climate Change in Tension. People Nat. 2021, 3, 978–989. [Google Scholar] [CrossRef]
- Lioubimtseva, E.; Henebry, G.M. Climate and Environmental Change in Arid Central Asia: Impacts, Vulnerability, and Adaptations. J. Arid Environ. 2009, 73, 963–977. [Google Scholar] [CrossRef]
- MacKellar, N.; New, M.; Jack, C. Observed and Modelled Trends in Rainfall and Temperature for South Africa: 1960–2010. S. Afr. J. Sci. 2014, 110, 1–13. [Google Scholar] [CrossRef]
- Barry, P.S.; Mendenhall, J.; Jarecke, P.; Folkman, M.; Pearlman, J.; Markham, B. EO-1 Hyperion Hyperspectral Aggregation and Comparison with EO-1 Advanced Land Imager and Landsat 7 ETM+. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Toronto, ON, Canada, 24–28 June 2002; Volume 3, pp. 1648–1651. [Google Scholar]
- Johansen, K.; Phinn, S.; Dixon, I.; Douglas, M.; Lowry, J. Comparison of Image and Rapid Field Assessments of Riparian Zone Condition in Australian Tropical Savannas. For. Ecol. Manag. 2007, 240, 42–60. [Google Scholar] [CrossRef]
- Huylenbroeck, L.; Laslier, M.; Dufour, S.; Georges, B.; Lejeune, P.; Michez, A. Using Remote Sensing to Characterize Riparian Vegetation: A Review of Available Tools and Perspectives for Managers. J. Environ. Manag. 2020, 267, 110652. [Google Scholar] [CrossRef] [PubMed]
- Mureriwa, N.; Adam, E.; Sahu, A.; Tesfamichael, S. Examining the Spectral Separability of Prosopis Glandulosa from Co-Existent Species Using Field Spectral Measurement and Guided Regularized Random Forest. Remote Sens. 2016, 8, 144. [Google Scholar] [CrossRef]
- Crisóstomo de Castro Filho, H.; Abílio de Carvalho Júnior, O.; Ferreira de Carvalho, O.L.; Pozzobon de Bem, P.; dos Santos de Moura, R.; Olino de Albuquerque, A.; Silva, C.R.; Ferreira, P.H.G.; Guimarães, R.F.; Gomes, R.A.T. Rice Crop Detection Using LSTM, Bi-LSTM, and Machine Learning Models from Sentinel-1 Time Series. Remote Sens. 2020, 12, 2655. [Google Scholar] [CrossRef]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual Tree Detection and Classification with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef]
- Cord, A.F.; Meentemeyer, R.K.; Leitão, P.J.; Václavík, T. Modelling Species Distributions with Remote Sensing Data: Bridging Disciplinary Perspectives. J. Biogeogr. 2013, 40, 2226–2227. [Google Scholar] [CrossRef]
- Yang, D.; Meng, R.; Morrison, B.D.; McMahon, A.; Hantson, W.; Hayes, D.J.; Breen, A.L.; Salmon, V.G.; Serbin, S.P. A Multi-Sensor Unoccupied Aerial System Improves Characterization of Vegetation Composition and Canopy Properties in the Arctic Tundra. Remote Sens. 2020, 12, 2638. [Google Scholar] [CrossRef]
- van den Berg, E.C.; Kotze, I.; Beukes, H. Detection, Quantification and Monitoring of Prosopis in the Northern Cape Province of South Africa Using Remote Sensing and GIS. S. Afr. J. Geomat. 2013, 2, 68–81. [Google Scholar]
- Hudak, A.T.; Wessman, C.A. Textural Analysis of Historical Aerial Photography to Characterize Woody Plant Encroachment in South African Savanna. Remote Sens. Environ. 1998, 66, 317–330. [Google Scholar] [CrossRef]
- Symeonakis, E.; Higginbottom, T. Bush encroachment monitoring using multi-temporal landsat data and random forests. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, 10, 29–35. [Google Scholar] [CrossRef]
- Ludwig, A.; Meyer, H.; Nauss, T. Automatic Classification of Google Earth Images for a Larger Scale Monitoring of Bush Encroachment in South Africa. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 89–94. [Google Scholar] [CrossRef]
- Shekede, M.D.; Murwira, A.; Masocha, M. Wavelet-Based Detection of Bush Encroachment in a Savanna Using Multi-Temporal Aerial Photographs and Satellite Imagery. Int. J. Appl. Earth Obs. Geoinf. 2015, 35, 209–216. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, X.; Qin, Y.; Wang, J.; Xu, X.; Hu, Y.; Qiao, Z. Mapping Cropping Intensity in China Using Time Series Landsat and Sentinel-2 Images and Google Earth Engine. Remote Sens. Environ. 2020, 239, 111624. [Google Scholar] [CrossRef]
- Phiri, D.; Morgenroth, J. Developments in Landsat Land Cover Classification Methods: A Review. Remote Sens. 2017, 9, 967. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A Survey of Image Classification Methods and Techniques for Improving Classification Performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- AbdelRahman, M.A.E.; Afifi, A.A.; Scopa, A. A Time Series Investigation to Assess Climate Change and Anthropogenic Impacts on Quantitative Land Degradation in the North Delta, Egypt. ISPRS Int. J. Geoinf. 2021, 11, 30. [Google Scholar] [CrossRef]
- Kumar, A.; Manjunath, K.R.; Bala, R.; Sud, R.K.; Singh, R.D.; Panigrahy, S. Field Hyperspectral Data Analysis for Discriminating Spectral Behavior of Tea Plantations under Various Management Practices. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 352–359. [Google Scholar] [CrossRef]
- Mudereri, B.T.; Dube, T.; Niassy, S.; Kimathi, E.; Landmann, T.; Khan, Z.; Abdel-Rahman, E.M. Is It Possible to Discern Striga Weed (Striga Hermonthica) Infestation Levels in Maize Agro-Ecological Systems Using in-Situ Spectroscopy? Int. J. Appl. Earth Obs. Geoinf. 2020, 85, 102008. [Google Scholar] [CrossRef]
- Hughes, G. On the Mean Accuracy of Statistical Pattern Recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Zhang, B.; Lu, L. Object-Based Crop Classification with Landsat-MODIS Enhanced Time-Series Data. Remote Sens. 2015, 7, 16091–16107. [Google Scholar] [CrossRef]
- Walsh, O.S.; Marshall, J.M.; Nambi, E.; Jackson, C.A.; Ansah, E.O.; Lamichhane, R.; McClintick-Chess, J.; Bautista, F. Wheat Yield and Protein Estimation with Handheld and Unmanned Aerial Vehicle-Mounted Sensors. Agronomy 2023, 13, 207. [Google Scholar] [CrossRef]
- Neupane, K.; Baysal-Gurel, F. Automatic Identification and Monitoring of Plant Diseases Using Unmanned Aerial Vehicles: A Review. Remote Sens. 2021, 13, 3841. [Google Scholar] [CrossRef]
- Nebiker, S.; Lack, N.; Abächerli, M.; Läderach, S. Light-Weight Multispectral UAV Sensors and Their Capabilities for Predicting Grain Yield and Detecting Plant Diseases. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 963–970. [Google Scholar] [CrossRef]
- Song, B.; Park, K. Detection of Aquatic Plants Using Multispectral UAV Imagery and Vegetation Index. Remote Sens. 2020, 12, 387. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; Peña, J.M. An Automatic Object-Based Method for Optimal Thresholding in UAV Images: Application for Vegetation Detection in Herbaceous Crops. Comput. Electron. Agric. 2015, 114, 43–52. [Google Scholar] [CrossRef]
- Carvajal-Ramírez, F.; da Silva, J.R.M.; Agüera-Vega, F.; Martínez-Carricondo, P.; Serrano, J.; Moral, F.J. Evaluation of Fire Severity Indices Based on Pre- and Post-Fire Multispectral Imagery Sensed from UAV. Remote Sens. 2019, 11, 993. [Google Scholar] [CrossRef]
- Guan, S.; Fukami, K.; Matsunaka, H.; Okami, M.; Tanaka, R.; Nakano, H.; Sakai, T.; Nakano, K.; Ohdan, H.; Takahashi, K. Assessing Correlation of High-Resolution NDVI with Fertilizer Application Level and Yield of Rice and Wheat Crops Using Small UAVs. Remote Sens. 2019, 11, 112. [Google Scholar] [CrossRef]
- González-Jaramillo, V.; Fries, A.; Bendix, J. AGB Estimation in a Tropical Mountain Forest (TMF) by Means of RGB and Multispectral Images Using an Unmanned Aerial Vehicle (UAV). Remote Sens. 2019, 11, 1413. [Google Scholar] [CrossRef]
- Hill, D.J.; Tarasoff, C.; Whitworth, G.E.; Baron, J.; Bradshaw, J.L.; Church, J.S. Utility of Unmanned Aerial Vehicles for Mapping Invasive Plant Species: A Case Study on Yellow Flag Iris (Iris pseudacorus L.). Int. J. Remote Sens. 2017, 38, 2083–2105. [Google Scholar] [CrossRef]
- de Castro, A.I.; Peña, J.M.; Torres-Sánchez, J.; Jiménez-Brenes, F.M.; Valencia-Gredilla, F.; Recasens, J.; López-Granados, F. Mapping Cynodon Dactylon Infesting Cover Crops with an Automatic Decision Tree-OBIA Procedure and UAV Imagery for Precision Viticulture. Remote Sens. 2020, 12, 56. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Paul, T.S.H.; Morgenroth, J.; Pearse, G.D. Early Detection of Invasive Exotic Trees Using UAV and Manned Aircraft Multispectral and LiDAR Data. Remote Sens. 2019, 11, 1812. [Google Scholar] [CrossRef]
- Marques, P.; Pádua, L.; Adão, T.; Hruška, J.; Peres, E.; Sousa, A.; Sousa, J.J. UAV-Based Automatic Detection and Monitoring of Chestnut Trees. Remote Sens. 2019, 11, 855. [Google Scholar] [CrossRef]
- Müllerová, J.; Pergl, J.; Pyšek, P. Remote Sensing as a Tool for Monitoring Plant Invasions: Testing the Effects of Data Resolution and Image Classification Approach on the Detection of a Model Plant Species Heracleum Mantegazzianum (Giant Hogweed). Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 55–65. [Google Scholar] [CrossRef]
- Mucina, L.; Rutherford, M.C.; Palmer, A.R.; Milton, S.J.; Scott, L.; Lloyd, J.W.; Van der Merwe, B.; Hoare, D.B.; Bezuidenhout, H.; Vlok, J.H.J. Nama-Karoo Biome. The vegetation of South Africa, Lesotho and Swaziland. Strelitzia 2006, 19, 324–347. [Google Scholar]
- Pfitzner, K.; Bartolo, R.; Whiteside, T.; Loewensteiner, D.; Esparon, A. Hyperspectral Monitoring of Non-Native Tropical Grasses over Phenological Seasons. Remote Sens. 2021, 13, 738. [Google Scholar] [CrossRef]
- Meyer, T.C. Weikapasiteitstudies Op Veld in Die Ariede Karoo. Master’s Thesis, University of the Orange Free State, Bloemfontein, South Africa, 1992, unpublished. [Google Scholar]
- O’connor, T.G.; Roux, P.W. Vegetation Changes (1949–71) in a Semi-Arid, Grassy Dwarf Shrubland in the Karoo, South Africa: Influence of Rainfall Variability and Grazing by Sheep. J. Appl. Ecol. 1995, 32, 612–626. [Google Scholar] [CrossRef]
- Milton, S.J.; Dean, W.R.J. Anthropogenic Impacts and Implications for Ecological Restoration in the Karoo, South Africa. Anthropocene 2021, 36, 100307. [Google Scholar] [CrossRef]
- Van der Merwe, H.; Du Toit, J.C.O.; Van den Berg, L.; O’Connor, T.G. Impact of Sheep Grazing Intensity on Vegetation at the Arid Karoo Stocking Rate Trial after 27 Years, Carnarvon, South Africa. J. Arid Environ. 2018, 155, 36–45. [Google Scholar] [CrossRef]
- Trimble Trimble R8 GNSS System. In Trimble Datasheet; Trimble Navigation Limited: Westminster, CO, USA, 2012.
- Sibanda, M.; Mutanga, O.; Rouget, M.; Odindi, J. Exploring the Potential of in Situ Hyperspectral Data and Multivariate Techniques in Discriminating Different Fertilizer Treatments in Grasslands. J. Appl. Remote Sens. 2015, 9, 096033. [Google Scholar] [CrossRef]
- Olsson, P.-O.; Vivekar, A.; Adler, K.; Garcia Millan, V.E.; Koc, A.; Alamrani, M.; Eklundh, L. Radiometric Correction of Multispectral Uas Images: Evaluating the Accuracy of the Parrot Sequoia Camera and Sunshine Sensor. Remote Sens. 2021, 13, 577. [Google Scholar] [CrossRef]
- Thomson, E.R.; Spiegel, M.P.; Althuizen, I.H.J.; Bass, P.; Chen, S.; Chmurzynski, A.; Halbritter, A.H.; Henn, J.J.; Jónsdóttir, I.S.; Klanderud, K.; et al. Multiscale Mapping of Plant Functional Groups and Plant Traits in the High Arctic Using Field Spectroscopy, UAV Imagery and Sentinel-2A Data. Environ. Res. Lett. 2021, 16, 055006. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- McNemar, Q. Note on the Sampling Error of the Difference between Correlated Proportions or Percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kurihara, J.; Viray, F.A.; Namuco, S.B.; Paringit, E.C.; Perez, G.J.; Takahashi, Y.; Marciano, J.J., Jr. A Novel Approach for Vegetation Classification Using UAV-Based Hyperspectral Imaging. Comput. Electron. Agric. 2018, 144, 80–85. [Google Scholar] [CrossRef]
- Yan, Y.; Deng, L.; Liu, X.; Zhu, L. Application of UAV-Based Multi-Angle Hyperspectral Remote Sensing in Fine Vegetation Classification. Remote Sens. 2019, 11, 2753. [Google Scholar] [CrossRef]
- Franklin, S.E.; Ahmed, O.S. Deciduous Tree Species Classification Using Object-Based Analysis and Machine Learning with Unmanned Aerial Vehicle Multispectral Data. Int. J. Remote Sens. 2018, 39, 5236–5245. [Google Scholar] [CrossRef]
- Gini, R.; Sona, G.; Ronchetti, G.; Passoni, D.; Pinto, L. Improving Tree Species Classification Using UAS Multispectral Images and Texture Measures. ISPRS Int. J. Geoinf. 2018, 7, 315. [Google Scholar] [CrossRef]
- Grybas, H.; Congalton, R.G. A Comparison of Multi-Temporal RGB and Multispectral UAS Imagery for Tree Species Classification in Heterogeneous New Hampshire Forests. Remote Sens. 2021, 13, 2631. [Google Scholar] [CrossRef]
- Louargant, M.; Villette, S.; Jones, G.; Vigneau, N.; Paoli, J.-N.; Gée, C. Weed Detection by UAV: Simulation of the Impact of Spectral Mixing in Multispectral Images. Precis. Agric. 2017, 18, 932–951. [Google Scholar] [CrossRef]
- Lisein, J.; Michez, A.; Claessens, H.; Lejeune, P. Discrimination of Deciduous Tree Species from Time Series of Unmanned Aerial System Imagery. PLoS ONE 2015, 10, e0141006. [Google Scholar] [CrossRef]
- Michez, A.; Piégay, H.; Jonathan, L.; Claessens, H.; Lejeune, P. Mapping of Riparian Invasive Species with Supervised Classification of Unmanned Aerial System (UAS) Imagery. Int. J. Appl. Earth Obs. Geoinf. 2016, 44, 88–94. [Google Scholar] [CrossRef]
- Weil, G.; Lensky, I.M.; Resheff, Y.S.; Levin, N. Optimizing the Timing of Unmanned Aerial Vehicle Image Acquisition for Applied Mapping of Woody Vegetation Species Using Feature Selection. Remote Sens. 2017, 9, 1130. [Google Scholar] [CrossRef]
- Joubert, J.P.J. Section of Toxicology on Geigeria ornativa. J. S. Afr. Vet. Assoc. 1983, 54, 255. [Google Scholar] [PubMed]
- Stapleton, A.E. Ultraviolet Radiation and Plants: Burning Questions. Plant Cell 1992, 4, 1353. [Google Scholar] [CrossRef]
- Brosché, M.; Strid, Å. Molecular Events Following Perception of Ultraviolet-B Radiation by Plants. Physiol. Plant. 2003, 117, 1–10. [Google Scholar] [CrossRef]
- Fedina, I.; Velitchkova, M.; Georgieva, K.; Demirevska, K.; Simova, L. UV-B Response of Green and Etiolated Barley Seedlings. Biol. Plant. 2007, 51, 699–706. [Google Scholar] [CrossRef]
- Valenta, K.; Dimac-Stohl, K.; Baines, F.; Smith, T.; Piotrowski, G.; Hill, N.; Kuppler, J.; Nevo, O. Ultraviolet Radiation Changes Plant Color. BMC Plant Biol. 2020, 20, 253. [Google Scholar] [CrossRef]
- Court, D. Succulent Flora of Southern Africa (Revised Edition), 3rd ed.; Struik Nature: Cape Town, South Africa, 2010. [Google Scholar]
- Gibson, A.C. Succulent Photosynthetic Organs. In Structure-Function Relations of Warm Desert Plants; Springer: Berlin/Heidelberg, Germany, 1996; pp. 117–142. [Google Scholar]
- Jacobs, J.F.; Koper, G.J.M.; Ursem, W.N.J. UV Protective Coatings: A Botanical Approach. Prog. Org. Coat. 2007, 58, 166–171. [Google Scholar] [CrossRef]
- Colomina, I.; Molina, P. Unmanned Aerial Systems for Photogrammetry and Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef]
- Dandois, J.P.; Olano, M.; Ellis, E.C. Optimal Altitude, Overlap, and Weather Conditions for Computer Vision UAV Estimates of Forest Structure. Remote Sens. 2015, 7, 13895–13920. [Google Scholar] [CrossRef]
- Feroz, S.; Abu Dabous, S. Uav-Based Remote Sensing Applications for Bridge Condition Assessment. Remote Sens. 2021, 13, 1809. [Google Scholar] [CrossRef]
- Oktay, T.; Celik, H.; Turkmen, I. Maximizing Autonomous Performance of Fixed-Wing Unmanned Aerial Vehicle to Reduce Motion Blur in Taken Images. Proc. Inst. Mech. Eng. Part I J. Syst. Control Eng. 2018, 232, 857–868. [Google Scholar] [CrossRef]
- Sieberth, T.; Wackrow, R.; Chandler, J.H. UAV Image Blur–Its Influence and Ways to Correct It. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 40, 33–39. [Google Scholar] [CrossRef]
- Poona, N.K.; Ismail, R. Developing Optimized Spectral Indices Using Machine Learning to Model Fusarium Circinatum Stress in Pinus Radiata Seedlings. J. Appl. Remote Sens. 2019, 13, 34515. [Google Scholar] [CrossRef]
- Poona, N.K.; Ismail, R. Using Boruta-Selected Spectroscopic Wavebands for the Asymptomatic Detection of Fusarium Circinatum Stress. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3764–3772. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).