Abstract
The anthropogenic climate crisis results in the gradual loss of tree species in locations where they were previously able to grow. This leads to increasing workloads and requirements for foresters and arborists as they are forced to restructure their forests and city parks. The advancements in computer vision (CV)—especially in supervised deep learning (DL)—can help cope with these new tasks. However, they rely on large, carefully annotated datasets to produce good and generalizable models. This paper presents BAMFORESTS: a dataset with 27,160 individually delineated tree crowns in 105 ha of very-high-resolution UAV imagery gathered with two different sensors from two drones. BAMFORESTS covers four areas of coniferous, mixed, and deciduous forests and city parks. The labels contain instance segmentations of individual trees, and the proposed splits are balanced by tree species and vitality. Furthermore, the dataset contains the corrected digital surface model (DSM), representing tree heights. BAMFORESTS is annotated in the COCO format and is especially suited for training deep neural networks (DNNs) to solve instance segmentation tasks. BAMFORESTS was created in the BaKIM project and is freely available under the CC BY 4.0 license.
1. Introduction
The anthropogenic climate crisis (ACC) is probably humanity’s most urgent and most significant problem to solve and adapt to in the coming years. Just as the years 2017–2022, 2023 was, according to the German Umweltbundesamt, the warmest year in Germany and worldwide [1]. This rapid increase in temperature puts much pressure on ecosystems worldwide, including forests and city trees [2,3,4,5]. As forests are natural carbon sinks, this is a self-reinforcing problem [6]. The effects are less resilient trees and increased pests and secondary pests such as bark beetles and mistletoes [7,8]. Especially in forests that are heavily used for forestry and, therefore, often structured as tree monocultures, this can lead to the loss of all trees in large areas [9].
To mitigate the effects of ACC on forests and city trees, arborists and foresters need to restructure their forests, city parks, and solitary city trees. Experience in the BaKIM (BaKIM is a cooperation project of the Cognitive Systems Chair of the University of Bamberg and the City of Bamberg. It is funded with EUR 450,000 by the Bavarian Ministry of Digital Affairs) project shows that, to accomplish this, German foresters and arborists rely on incomplete and often old data, which do not allow for a timely and regular evaluation. Furthermore, research shows that this is not a problem of Germany alone, but a European and probably worldwide one [10]. BaKIM shows that, to support foresters and arborists in their work, current data on the position of individual trees, tree species, tree vitality, and infestation with pests are needed.
Reliably detecting single trees is the underlying task, which helps all other previously mentioned tasks, making the information traceable on the single-tree level. Furthermore, foresters and arborists in the BaKIM project state that this information is also important and helpful for debating with local decision makers and politicians when it comes to the funding of city trees and forest areas. The rise of cheap UAV technology and deep learning in the past few years has built the base for solving this task of single tree detection. Overview papers such as Kattenborn et al. [11] showed that very-high-resolution UAV data with a ground sampling distance (GSD) of below 2 cm and deep neural networks (DNNs) from the computer vision domain yield good results. For example, Gan et al. [12] compared the widely used Python deep learning packages Detectree2 [13] and Deepforest [14] on different very-high-resolution GSDs. Xi et al. [15] tested different dimensionality reductions to overcome the drawbacks of multispectral images for ITCD approaches, which can yield better results than just RGB-imagery.
One remaining problem with supervised DNNs is the lack of high-quality training data. Annotating very-high-resolution UAV data on a single-tree level is time-consuming and expensive. Therefore, only a few small, openly accessible datasets exist. The Million Trees project [16] initiated by Weinstein et al. [17] tries to solve this by collecting and combining as many available datasets as possible. Nevertheless, this is an ongoing effort which, according to Ben Weinstein, will take place until fall 2025. To the best of our knowledge, none of the existing datasets come with a proposed dataset split to help compare the results of different methods and DNN architectures created by different researchers.
With this work, we publish BAMFORESTS: relative to other datasets with ITCD labels, it is a large benchmark dataset of very-high-resolution UAV imagery and individual delineated tree crowns (ITCD). To make it a benchmark dataset, we specify a fixed split of training, validation, and testing data. Furthermore, for ease of use, we publish the data in the COCO format in two different tile sizes so that researchers can easily test their methods and compare them with other results. BAMFORESTS will become part of the Million Trees benchmark dataset.
2. Theoretical Background
Deep learning in the form of deep convolutional neural networks (DCNNs) brought great advancement to different tasks in computer vision. Image classification, object detection, semantic segmentation, and instance segmentation are all tasks that extract information from image data [18,19]. Kattenborn et al. [11] provided a detailed overview on the following aspects of computer vision in the forest domain. While image classification assigns a label to a whole image, the latter tasks generate spatial information in differing detail. Object detection produces bounding boxes for multiple objects in an image. These bounding boxes are represented by pixel coordinates and describe the outer bounds of detected objects. As multiple objects can be separately detected, these objects can be counted. For example, with a working object detection, it is possible to count trees in an image but not possible to obtain the exact area covered by the tree crowns. Semantic segmentation, on the other hand, is not capable of differentiating between instances as it predicts a class for each pixel. If overlapping instances of the same class exist, semantic segmentation does not separate them, therefore making it impossible to count the instances. The improvement over object detection is that additional spatial information on the class occurrence is given, as the class of each pixel is predicted. Assuming correct classification, semantic segmentation for example enables the calculation of the area-related share of different tree species. Instance segmentation basically combines the two aforementioned tasks. It classifies each pixel in an image, but at the same time keeps information of single instances. If instances of the same class overlap, they can still be counted. Therefore, instance segmentation enables us to count trees, obtain their area-related share, and obtain their exact tree crown dimensions.
Image classification, due to its simple input–output structure of an image as the input and a single label as the output, was the first problem that greatly benefited from a large benchmark dataset, more powerful computer hardware, and DCNNs in 2012 [20,21]. Huge performance leaps in the more complex tasks of object detection, semantic segmentation, and instance segmentation followed in the years after with the release of the COCO benchmark dataset [22].
The following subsections will summarize the application of such DCNNs in the domain of forests and trees in aerial images and deal with the state of benchmark datasets in this domain.
2.1. Supervised Deep Learning on Forest Datasets
DCNNs are the state-of-the-art method for different computer vision tasks and are also applied in the forest domain. Recent publications show that, for tree detection and individual tree crown delineation (ITCD), technically an instance segmentation, DCNNs are the best-performing models. The widespread, successful use of these models in this domain is summarized by different overview papers and systematical literature reviews, such as Kattenborn et al. [11] and Zhao et al. [23]. Furthermore, Fan et al. [24] showed that deep learning approaches outperform statistical segmentation approaches such as marker-controlled watershed transformation (MCWST).
For example, Weinstein et al. [17] implemented DeepForest, a RetinaNet pre-trained on several datasets, capable of performing the object detection of trees, even in closed canopies. Schiefer et al. [25] used the U-Net architecture to map tree species via semantic segmentation. Ball et al. [13] developed Detectree2, an instance segmentation DCNN based on Facebook’s Detectron2 architecture. While Schiefer et al. [25] published their dataset and code, Deepforest and Detectree2 are publicly available Python packages, including pre-trained weights and pre- and post-processing steps. Therefore, they enable researchers without profound knowledge in programming and machine learning to utilize DCNNs for their aerial image datasets.
While all of these approaches and applied methods show that DCNNs are a very good solution to the task of tree detection and segmentation, the performance in the forest domain is still far behind when compared with the performance of the same methods on datasets like ImageNet and COCO, reaching over 90% accuracy in image classification and AP.50 scores of over 70% for object detection [26,27].
2.2. Forest Datasets
Benchmark datasets in computer vision, such as the COCO dataset or ImageNet, consist of hundreds of thousands or even millions of images and millions of annotations [22,28]. Compared with these, datasets in the forest domain are quite small, covering only tens of thousands of instances in very-high-resolution datasets with ITCD annotations, and there are at most a few hundred thousand instances in datasets with lower resolution and lower-quality labels like point annotations. Datasets in the forest domain can be divided into the following categories:
- Label completeness:
- –
- All tree instances are labeled;
- –
- Only some tree instances are labeled;
- Ground sampling distance (GSD):
- –
- Low resolution (satellite imagery): >1.2 m per pixel;
- –
- High resolution (airplane): 40 cm–6 cm per pixel;
- –
- Very-high resolution (UAVs): <5 cm per pixel;
- Label type:
- –
- Points: the center of each tree crown is labeled;
- –
- Bounding boxes: the outer extents of each tree crown are labeled;
- –
- Polygons:
- ∗
- Semantic segmentation: tree species are labeled on a pixel level;
- ∗
- Instance segmentation: each tree crown is delineated on a pixel level.
Furthermore, less clear categories such as forest density, geo-location, forest type, species distribution, and vegetation phase as well as weather conditions in the weeks before data acquisition should influence the transferability of models trained on a dataset, as research in land cover detection from satellite data has suggested [29].
The Million Trees project [16] aims to collect as many existing datasets covering all above-mentioned categories and creating a combined benchmark dataset of at least one million labeled trees. As BAMFORESTS falls into the category of a very-high-resolution datasets with complete polygon labels of single trees, we summarize similar datasets in Table 1.

Table 1.
Datasets in the forest domain similar to BAMFORESTS.
Cloutier et al. [32] recently published their dataset with 22,933 ITCs and image data from seven consecutive monthly flights. Their dataset is taken from two AOIs. One main AOI is used for training and validation, and a smaller second AOI, separated by a small body of water, serves as the test set. After BAMFORESTS, it is the second largest dataset freely available to the knowledge of the authors. Schiefer et al. [30] published a dataset for semantic segmentation, where not single trees, but patches of the same tree species, were labeled. Therefore, the number of ITCs is not known. Considering that FORTRESS consists of 47 ha, it is approximately half as big as BAMFORESTS. Due to the labels for semantic segmentation, it cannot be used to train and validate ITCD methods. Kruse et al. [33] made their dataset SiDroForest also publicly available. While it consists of a total of 19,342 shapes, only 872 of these are labeled by hand. The rest is generated automatically and is, therefore, of lower quality and probably incomplete. Jansen et al. [34] published a small unnamed dataset consisting of 2547 ITCs. Due to the small size of the dataset, it is, on its own, not suitable for the training of DCNNs.
3. Dataset
BAMFORESTS data come from four different AOIs, which are shown in Figure 1. All AOIs are located within a radius of 20 km in and around Bamberg, and the aerial surveys were carried out as part of the BaKIM project [36]. Figure 2 shows a detailed example view of each AOI. Figure 1 and Figure 2 are scaled equally so that the relative size of the AOIs and trees can be seen.
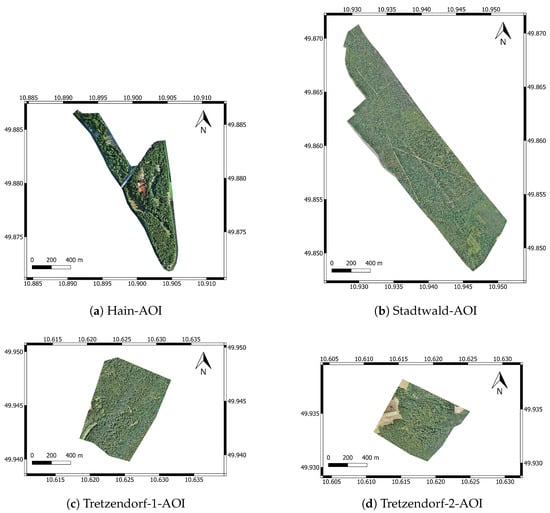
Figure 1.
Overview of all four AOIs in the BAMFORESTS benchmark dataset. Figure (a) shows the complete Hain-AOI, (b) shows the complete Stadtwald-AOI, (c) shows the complete Tretzendorf-1-AOI, and (d) shows the complete Tretzendorf-2-AOI. All images are scaled equally, allowing for comparison of the AOI sizes (EPSG:4326).


Figure 2.
Detailed view of all four AOIs in the BAMFORESTS benchmark dataset. Figure (a) shows a zoomed-in view of the Hain-AOI, (b) shows a zoomed-in view of the Stadtwald-AOI, (c) shows a zoomed-in view of the Tretzendorf-1-AOI, and (d) shows a zoomed-in view of the Tretzendorf-2-AOI. All images are scaled equally (EPSG:4326).
3.1. AOIs
The Hain-AOI is the cities forest-like park with a size of about 61 ha, and it is located south-east of the city center. As a park, it is heavily frequented, and arborists take a lot of measures to keep trees and reduce hazards of falling branches near paths. Furthermore, the Hain has the highest diversity of tree species because of its main functions as a recreational area and special area of conservation (SAC). It mainly consists of deciduous trees and, the resulting orthomosaic has a GSD of 1.82 cm.
The Stadtwald-AOI covers an area of 152 ha and is located about 5 km south-east of Bamberg’s city center. On the one hand, it is a managed forest, and on the other hand, it is a drinking water reservoir for parts of Bamberg and an SAC. In comparison with the city park, no measurements to keep single trees are taken, but if necessary, trees are removed and the timber is sold. It is a mainly coniferous forest, but in recent decades, second and third tree layers of deciduous trees were planted beneath the older coniferous trees. The resulting orthomosaic has a GSD of 1.70 cm.
The Tretzendorf-1-AOI and Tretzendorf-2-AOI share similar characteristics. The first one covers an area of 65 ha and the latter one an area of 47 ha. Both are located about 20 km west to north-west of Bamberg and are SACs. Because coniferous as well as deciduous trees grow in the Tretzendorf-AOIs, they can be described as managed mixed forests, and the resulting orthomosaics have a GSD of 1.61 cm and 1.79 cm.
3.2. UAVs and Sensors
Two different UAVs were used for data acquisition. A small DJI Phantom 4 (DJI-Shenzhen, China) with a 1” CMOS 20MP sensor and 84° angle of view (AOV), acquired images over the Hain region near the city center. For the forest areas, a larger Quantum Systems Trinity F90+ (Quantum Systems-Gilching, Germany) fixed-wing UAV with a Sony RX1 RII (Sony-Tokyo, Japan 42.4 MP sensor and 63° AOV was used. Both sensors are RGB sensors, and the resulting orthomosaics contain a red, green, and blue channel.
3.3. Data Acquisition
The data acquisition of all images in BAMFORESTS took place in the summer of 2022. Both Tretzendorf AOIs were captured on the fifth of July, the Stadtwald AOI was captured on the sixth of July, and the Hain AOI was captured on the third of August. While the flights of the Tretzendorf and Stadtwald AOIs were carried out at midday, due to legal reasons the flights of the Hain AOI took place after sunrise in the morning until early midday.
In all flights, we aimed for a forward overlap and sidelap of 80%. As the Trinity F90+ flies at fixed speeds, we compensated the lower forward overlap of about 65% with a higher sidelap of 85–90%. With the Trinity F90+, we always flew 120 m above ground level (AGL) and used a smoothed terrain follow mode in the hilly Tretzendorf-AOIs. With the DJI Phantom 4, we flew 85 m AGL to reach a similar ground sampling distance (GSD). The Hain-AOI was covered with a total of 3087 images, the Stadtwald-AOI with 3250 images, the Tretzendorf-1-AOI with 1577 images, and the Tretzendorf-2-AOI with 1229 images.
3.4. Orthomosaic Generation
For the generation of the orthomosaics, we chose the commercial software Agisoft Metashape (Version 1.8.4) after comparing the process and results to those of WebODM (Version 1.9.15). While WebODM is free and open-source, the setup is more difficult, and the flexibility in setting ground control points (GCPs) and influencing the resulting orthomosaic is lower. Nevertheless, the resulting orthomosaics can be described as of equal quality, as seen in Figure 3.

Figure 3.
Comparison of the resulting orthomosaics and artifacts. Figure (a) shows artifacts from the orthomosaic generation with Agisoft Metashape, whereas (b) shows artifacts from the orthomosaic generation with WebODM.
Agisoft Metashape uses a structure from motion algorithms to generate a georeferenced point cloud from the single overlapping images acquired with the UAVs. This tie point cloud is then cleaned with three statistical methods and a manual step where the remaining outliers below ground level and above the tree crowns are deleted by hand. For the statistical filtering, we found the best results were obtained by deleting about 20% of the tie points by reconstruction uncertainty, about 10% by reprojection error, and 10% by projection accuracy. After each filter step, we optimized the camera alignment. Based on the cleaned tie point cloud, we first generated the dense point cloud, then the digital surface model, and lastly the orthomosaic.
3.5. Labeling Process
The individual tree crowns were delineated and labeled by an experienced forester in 2023. To help the labeling process, they had additional access to orthomosaics taken in March and April for some of the AOIs. During and after labeling, they checked annotations on the sites. After this labeling, apprentices of the local forestry office randomly checked the labels onsite and corrected them if necessary. In addition to the tree species, the tree vitality was assessed from the imagery and categorized into vital, degrading, and dead. The tree species information and tree vitality information are used in this work to balance the dataset splits.
3.6. Dataset Metrics
BAMFORESTS consists of a total of 27,160 individually delineated tree crowns (ITC). Of those, 1978 are located in the 15 labeled hectares of the Hain-AOI, 15,473 are located in the 46 labeled hectares of the Stadtwald-AOI, 5900 are located in the 29 labeled hectares of the Tretzendorf-1-AOI, and 3809 are located in the 15 labeled hectares of the Tretzendorf-2-AOI. For an overview of the species and vitality distribution of the whole dataset, see Figure 4. With this publication, as we focus on the ITCD task, we only publish the ITC shapes and not all existing labels. Nevertheless, the labels are important for comparability and the splitting of the dataset.
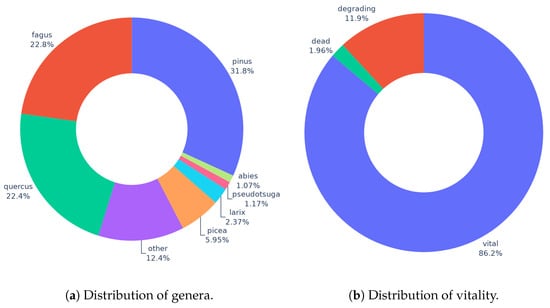
Figure 4.
Statistics of BAMFORESTS labels. (a) shows the distribution of all genera with a share of ≥1% in BAMFORESTS. Genera with a share below 1% were combined into the other class. (b) shows the distribution of the three vitality levels in BAMFORESTS.
3.7. Benchmark Dataset Split
Based on the AOI location, the tree species distribution, and vitality distribution, we define a fixed testing set for BAMFORESTS and propose a training set and validation set split. This ensures the comparability of different applied methods for ITCD and therefore helps the community in the forest domain to compare their models.
To ensure that spatial autocorrelation, as described by Kattenborn et al. [37], does not lead to an overestimation of the performance of methods, Test-Set-1 (15 ha, 14%) solely consists of the complete Hain-AOI. It is 5–20 km away from the other AOIs and is of severely different structure. Furthermore, it was captured with another UAV and sensor. All of these factors are good for estimating the real-world performance of methods. On the other hand, we want researchers to gain better insight into their methods performance on more alike data. This is why we define Test-Set-2 (14 ha, 13%), which is taken from the same AOIs as the training and validation set. To make sure that no redundant image information is in Test-Set-2, the split is based on complete hectare plots. This allows for overlapping tiles inside the hectare plots, thereby making the best use of the existing image data.
The validation set (Val-Set) consists of 16 ha (15%). Like Test-Set-2, it is taken from the AOIs Stadtwald and Tretzendorf. It follows the same split method based on the hectare plots. The training set (Train-Set) consists of the remaining 60 ha (57%) from the Stadtwald and Tretzendorf AOIs. Except for Test-Set-1, which is the Hain-AOI, all sets are as similar as possible, considering the distribution of classes. See Table 2 for the relative class share in each set.

Table 2.
Distribution of classes in the proposed split of sets in BAMFORESTS.
3.8. COCO Label Generation
The COCO dataset is a commonly used dataset for deep learning-based object detection. BAMFORESTS is released in the same data format as the COCO dataset [38] so that it can be easily used with state-of-the-art deep learning models. The images in the dataset are divided into the mentioned 57.14% Train-Set, 15.24% Val-Set, 14.29% Test-Set-1, and 13.33% Test-Set-2. Corresponding annotations are available in their respective .GeoJSON files.
Based on the division of the hectare plots, the orthomosaics and associated annotations are cropped and then divided and collectively converted into the COCO format. There is a 50% overlap between the images both horizontally and vertically. To meet the needs of the various scenes and available GPU-VRAM, the BAMFORESTS dataset provides two different image sizes (1024 × 1024 pixels and 2048 × 2048 pixels). At the edges of the images, individual crowns are cropped. In the center of the image, the individual crowns are completely recorded, as shown in Figure 5.
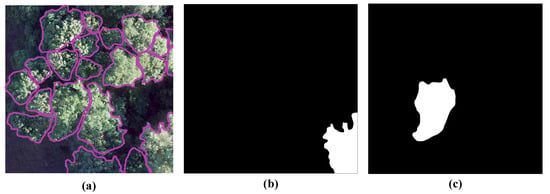
Figure 5.
BAMFORESTS in the COCO format: (a) orthomosaics and its annotations of individual tree crowns; (b) partial crown at the edge; (c) full crown in the center.
3.9. COCO Label Stats
The data description of the BAMFORESTS dataset in the COCO format can be seen in Table 3. Due to image cropping and overlapping, a single tree crown may be captured several times in different images. Therefore, the number of annotated tree crowns is much higher than the actual number of individual trees in BAMFORESTS.

Table 3.
The stats of the BAMFORESTS dataset in the COCO format.
Test-Set-1 is based on the Hain plot, which is completely independent of the Train-Set and Val-Set. However, Test-Set-2 comes from the same forests as the Train-Set and Val-Set. As shown in Figure 6 and Table 2, the images and annotations from the dataset show noticeable differences, such as tree species, crown sizes, and planting densities between Test-Set-1 and Test-Set-2. The design of the test sets is based on the intention that the trained model will be tested in both similar and different situations. The different results of these two test sets in training models are valuable for researchers to better explore the robustness and transferability of training models.
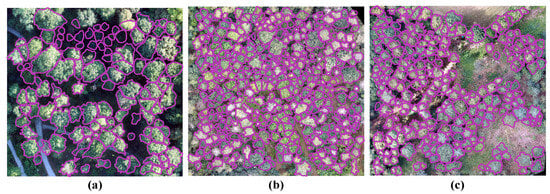
Figure 6.
Images and annotations of individual tree crowns from (a) the Hain plot of Test-Set-1, (b) the Stadtwald plot of Test-Set-2, and (c) the Tretzendorf plot of Test-Set-2.
4. Discussion
With BAMFORESTS, we do not present the first openly accessible dataset of individual delineated tree crowns but rather the first dataset in the forest domain that proposes a fixed dataset split; therefore, it serves as a benchmark dataset. Furthermore, with a total of 105 annotated hectares, a total of 27,160 trees, and a GSD of 1.6–1.8 cm, to the best of our knowledge, BAMFORESTS will be the single largest very-high-resolution dataset.
Nevertheless, BAMFORESTS is still a relatively small dataset for application in deep learning, but projects like Million Trees from Weinstein [16] will hopefully solve this in the future. Furthermore, the labels in BAMFORESTS are not of perfect quality. Especially in overlapping deciduous tree crowns, it is impossible to correctly label all trees. We solved this by controlling the labels with random samples, but it must be assumed that some trees are over- or under-segmented.
With two different test sets, we enable two different and important insights on method or model performance: Test-Set-2 gives good insights on the performance on similar data, covering applications where a model is trained or fine-tuned on labeled data of the same AOI and then applied to the remaining unlabeled area; meanwhile, Test-Set-1 gives good insights of the models real-world performance when it is applied to data from severely different AOIs and other sensors. With a ready-to-train dataset, split into tiles and made available with COCO annotation jsons, we make it as easy as possible for other researchers to train and test their methods on BAMFORESTS. Therefore, BAMFORESTS is an important and very much-needed contribution to deep learning in the forest domain and ITCD task.
5. Outlook and Future Work
To further improve BAMFORESTS, we are working on the following improvements: first, we will apply and update the labels from 2022 to orthomosaics from other sensors but the same AOIs in 2023; and second, as BaKIM and BAMFORESTS is open source, we plan to gather datasets and labels from other municipalities and cities in Germany. With these measures, we plan to reach 50,000 labeled ITCs in 2024. Additionally, we plan to publish the tree species and vitality information with BAMFORESTS-2.
Author Contributions
Conceptualization, J.T. (Jonas Troles) and J.T. (Jiaojiao Tian); Data curation, W.F.; Writing—original draft, J.T. (Jonas Troles) and W.F.; Writing—review & editing, U.S. and J.T. (Jiaojiao Tian). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Bavarian Ministry of Digital Affairs, grant number 450,000€.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Trends der Lufttemperatur. Available online: https://www.umweltbundesamt.de/daten/klima/trends-der-lufttemperatur (accessed on 13 March 2024).
- Forzieri, G.; Dakos, V.; McDowell, N.G.; Ramdane, A.; Cescatti, A. Emerging signals of declining forest resilience under climate change. Nature 2022, 608, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Seidl, R.; Schelhaas, M.J.; Rammer, W.; Verkerk, P.J. Increasing forest disturbances in Europe and their impact on carbon storage. Nat. Clim. Change 2014, 4, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Treml, V.; Mašek, J.; Tumajer, J.; Rydval, M.; Čada, V.; Ledvinka, O.; Svoboda, M. Trends in climatically driven extreme growth reductions of Picea abies and Pinus sylvestris in Central Europe. Glob. Change Biol. 2022, 28, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Kehr, R. Possible effects of drought stress on native broadleaved tree species—Assessment in light of the 2018/19 drought. Jahrb. Baumpflege 2020, 2020, 103–107. [Google Scholar]
- Friedlingstein, P.; O’sullivan, M.; Jones, M.W.; Andrew, R.M.; Gregor, L.; Hauck, J.; Le Quéré, C.; Luijkx, I.T.; Olsen, A.; Peters, G.P.; et al. Global carbon budget 2022. Earth Syst. Sci. Data Discuss. 2022, 2022, 1–159. [Google Scholar] [CrossRef]
- Sturrock, R.; Frankel, S.; Brown, A.; Hennon, P.; Kliejunas, J.; Lewis, K.; Worrall, J.; Woods, A. Climate change and forest diseases. Plant Pathol. 2011, 60, 133–149. [Google Scholar] [CrossRef]
- Turner, R.; Smith, P. Mistletoes increasing in eucalypt forest near Eden, New South Wales. Aust. J. Bot. 2016, 64, 171–179. [Google Scholar] [CrossRef]
- Hartmann, H.; Bastos, A.; Das, A.J.; Esquivel-Muelbert, A.; Hammond, W.M.; Martínez-Vilalta, J.; McDowell, N.G.; Powers, J.S.; Pugh, T.A.; Ruthrof, K.X.; et al. Climate change risks to global forest health: Emergence of unexpected events of elevated tree mortality worldwide. Annu. Rev. Plant Biol. 2022, 73, 673–702. [Google Scholar] [CrossRef]
- EFISCEN Inventory Database. Available online: https://efi.int/knowledge/models/efiscen/inventory (accessed on 13 March 2024).
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Gan, Y.; Wang, Q.; Iio, A. Tree crown detection and delineation in a temperate deciduous forest from UAV RGB imagery using deep learning approaches: Effects of spatial resolution and species characteristics. Remote Sens. 2023, 15, 778. [Google Scholar] [CrossRef]
- Ball, J.G.; Hickman, S.H.; Jackson, T.D.; Koay, X.J.; Hirst, J.; Jay, W.; Archer, M.; Aubry-Kientz, M.; Vincent, G.; Coomes, D.A. Accurate delineation of individual tree crowns in tropical forests from aerial RGB imagery using Mask R-CNN. Remote Sens. Ecol. Conserv. 2023, 9, 641–655. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Aubry-Kientz, M.; Vincent, G.; Senyondo, H.; White, E.P. DeepForest: A Python package for RGB deep learning tree crown delineation. Methods Ecol. Evol. 2020, 11, 1743–1751. [Google Scholar] [CrossRef]
- Xi, X.; Xia, K.; Yang, Y.; Du, X.; Feng, H. Evaluation of dimensionality reduction methods for individual tree crown delineation using instance segmentation network and UAV multispectral imagery in urban forest. Comput. Electron. Agric. 2021, 191, 106506. [Google Scholar] [CrossRef]
- Weinstein, B. A Benchmark Dataset for Airborne Machine Learning. Available online: https://milliontrees.idtrees.org/ (accessed on 13 March 2024).
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual tree-crown detection in RGB imagery using semi-supervised deep learning neural networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Ioannidou, A.; Chatzilari, E.; Nikolopoulos, S.; Kompatsiaris, I. Deep learning advances in computer vision with 3d data: A survey. ACM Comput. Surv. (CSUR) 2017, 50, 1–38. [Google Scholar] [CrossRef]
- Liu, B.; Yu, L.; Che, C.; Lin, Q.; Hu, H.; Zhao, X. Integration and Performance Analysis of Artificial Intelligence and Computer Vision Based on Deep Learning Algorithms. arXiv 2023, arXiv:2312.12872. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25. Available online: https://proceedings.neurips.cc/paper_files/paper/2012/file/c399862d3b9d6b76c8436e924a68c45b-Paper.pdf (accessed on 13 March 2024). [CrossRef]
- Lin, T.; Maire, M.; Belongie, S.J.; Bourdev, L.D.; Girshick, R.B.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. arXiv 2014, arXiv:1405.0312. [Google Scholar]
- Zhao, H.; Morgenroth, J.; Pearse, G.; Schindler, J. A systematic review of individual tree crown detection and delineation with convolutional neural networks (CNN). Curr. For. Rep. 2023, 9, 149–170. [Google Scholar] [CrossRef]
- Fan, W.; Tian, J.; Troles, J.; Döllerer, M.; Kindu, M.; Knoke, T. Comparing Deep Learning and MCWST Approaches for Individual Tree Crown Segmentation. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2024, X-1-2024, 67–73. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Rafael Padilla, A.R.; the Hugging Face Team. Open Object Detection Leaderboard. 2023. Available online: https://huggingface.co/spaces/rafaelpadilla/object_detection_leaderboard (accessed on 13 March 2024).
- Image Classification on ImageNet. 2024. Available online: https://paperswithcode.com/sota/image-classification-on-imagenet (accessed on 13 March 2024).
- Yang, K.; Yau, J.; Fei-Fei, L.; Deng, J.; Russakovsky, O. A Study of Face Obfuscation in ImageNet. arXiv 2021, arXiv:2103.06191. [Google Scholar]
- Qin, R.; Liu, T. A Review of Landcover Classification with Very-High Resolution Remotely Sensed Optical Images—Analysis Unit, Model Scalability and Transferability. Remote Sens. 2022, 14, 646. [Google Scholar] [CrossRef]
- Schiefer, F.; Frey, J.; Kattenborn, T. FORTRESS—Forest Tree Species Segmentation in Very-High Resolution UAV-Based Orthomosaics; Karlsruhe Institute of Technology: Karlsruhe, Germany, 2022. [Google Scholar] [CrossRef]
- Cloutier, M.; Germain, M.; Laliberté, E. Quebec Trees Dataset; Zenodo: Geneva, Switzerland, 2023. [Google Scholar] [CrossRef]
- Cloutier, M.; Germain, M.; Laliberté, E. Influence of Temperate Forest Autumn Leaf Phenology on Segmentation of Tree Species from UAV Imagery Using Deep Learning. bioRxiv 2023, 1–45. [Google Scholar] [CrossRef]
- Kruse, S.; Farkas, L.; Brieger, F.; Geng, R.; Heim, B.; Pestryakova, L.A.; Zakharov, E.S.; Herzschuh, U.; van Geffen, F. SiDroForest: Orthomosaics, SfM point clouds and products from aerial image data of expedition vegetation plots in 2018 in Central Yakutia and Chukotka, Siberia, 2022. Earth Syst. Sci. Data 2022, 14, 4967–4994. [Google Scholar] [CrossRef]
- Jansen, A.; Nicholson, J.; Esparon, A.; Whiteside, T.; Welch, M.; Tunstill, M.; Paramjyothi, H.; Gadhiraju, V.; van Bodegraven, S.; Bartolo, R. A Deep Learning Dataset for Savanna Tree Species in Northern Australia; Zenodo: Geneva, Switzerland, 2022. [Google Scholar] [CrossRef]
- Jansen, A.J.; Nicholson, J.D.; Esparon, A.; Whiteside, T.; Welch, M.; Tunstill, M.; Paramjyothi, H.; Gadhiraju, V.; van Bodegraven, S.; Bartolo, R.E. Deep Learning with Northern Australian Savanna Tree Species: A Novel Dataset. Data 2023, 8, 44. [Google Scholar] [CrossRef]
- Troles, J.; Nieding, R.; Simons, S.; Schmid, U. Task Planning Support for Arborists and Foresters: Comparing Deep Learning Approaches for Tree Inventory and Tree Vitality Assessment Based on UAV-Data. In Innovations for Community Services; Krieger, U.R., Eichler, G., Erfurth, C., Fahrnberger, G., Eds.; Springer: Cham, Switzerland, 2023; pp. 103–122. [Google Scholar]
- Kattenborn, T.; Schiefer, F.; Frey, J.; Feilhauer, H.; Mahecha, M.D.; Dormann, C.F. Spatially autocorrelated training and validation samples inflate performance assessment of convolutional neural networks. ISPRS Open J. Photogramm. Remote Sens. 2022, 5, 100018. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Proceedings, Part V 13. Springer: Berlin/Heidelberg, Germany, 2014; pp. 740–755. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).