Abstract
Urban tree classification enables informed decision-making processes in urban planning and management. This paper introduces a novel data reformation method, pseudo tree crown (PTC), which enhances the feature difference in the input layer and results in the improvement of the accuracy and efficiency of urban tree classification by utilizing artificial intelligence (AI) techniques. The study involved a comparative analysis of the performance of various machine learning (ML) classifiers. The results revealed a significant enhancement in classification accuracy, with an improvement exceeding 10% observed when high spatial resolution imagery captured by an unmanned aerial vehicle (UAV) was utilized. Furthermore, the study found an impressive average classification accuracy of 93% achieved by a classifier built on the PyTorch framework, with ResNet50 leveraged as its convolutional neural network layer. These findings underscore the potential of AI-driven approaches in advancing urban tree classification methodologies for enhanced urban planning and management practices.
1. Introduction
Urban trees play a vital role in fostering sustainable development within cities. They enhance the quality of the living environment, supply oxygen for organisms, and clean urban air [1]. Typically, urban trees grow in isolation amidst various urban facilities. Therefore, examining specific tree species becomes a fundamental aspect of urban management [2]. Traditional methods of classifying tree species heavily depend on on-site observation, relying on the visual recognition of inspectors, which is subjective and prone to errors. This approach is time-consuming and incurs significant costs [3]. The rapid advancement of unmanned aerial vehicle (UAV) remote-sensing technology has introduced high-resolution and multi-source data in a timely, effective, and cost-efficient manner. Developing a suitable algorithm to harness this valuable data is essential for delivering accurate results promptly in individual tree classification [4].
Recently, numerous advancements in machine learning (ML) approaches have emerged in classifying tree species, encompassing various data sources such as hyperspectral images, LiDAR data, and high-resolution images [5]. Out of the many well-established ML algorithms, one is the deep learning (DL) approach, with convolutional neural networks (CNNs), graph neural networks (GNNs), and their variations being among the most popular [6,7]. The results are usually comparable to more traditional ML approaches, such as random forest (RF) and support vector machine (SVM) [8,9,10].
Research into machine learning methodologies have extensively documented findings on tree species’ classification and segmentation. Nonetheless, most studies have primarily focused on evaluating the efficacy of specific ML approaches when applied to remote-sensing data. In the tree classification, at an individual or block level, the emphasis has predominantly been on imagery captured from a nadir view perspective. The outcomes of such analyses exhibit notable variability contingent upon the source data and its spatial resolution. We are actively enhancing the feature differences within the input data, adopting a paradigm shift towards reforming the pseudo tree crowns (PTCs) from nadir view images.
1.1. Major Achievements from Convolutional Neural Network (CNN), Graph Neural Network (GNN), and Their Variations
Since 2017, there has been a notable surge in the application of artificial intelligence (AI) and machine learning (ML) techniques for tree species classification, marking a significant trend in current research endeavours. For example, In 2017, Krizhevsky et al. [11] introduced the deeper CNN architecture, AlexNet, to the ImageNet dataset. This model, trained extensively on GPU, achieved breakthrough results on the dataset. In 1998, LeCun et al. [12] utilized CNN to classify two-dimensional shape changes, highlighting the significant advantages of CNN in automatically extracting features, capturing various visual structures, and facilitating classification. Qin et al. [13] developed a system employing deep neural networks to classify high-resolution images, significantly reducing manual costs. In 2019, Marrs et al. [14] applied machining learning approaches on LiDAR and hyperspectral and thermal imagers for a mixed coniferous-deciduous forest with 67% accuracy. In 2000, Egli [15] utilised a shallow CNN for tree species classification based on high-resolution drone images, demonstrating consistent classification results exceeding 92%, regardless of external conditions. Li et al. [16] applied three CNN models to classify single tree images in high-resolution images, revealing a substantial advantage over RF and SVM methods. Liang et al. [17] employed a spectral-spatial parallel convolutional neural network (SSPCNN) for feature classification in hyperspectral images, demonstrating strong competitiveness. Nezami et al. [18] investigated tree species classification using a 3D convolutional neural network (3D-CNN), with the best 3D-CNN classifier achieving approximately 5% higher accuracy than multilayer perception (MLP) at all levels. Pleesoiamu et al. [19] also used the deep learning approach for tree delineation. In 2021, Shi et al. [20] employed an adjusted asymmetric convolution transfer learning model for hyperspectral image classification, significantly improving local tree species classification accuracy. Chen et al. [21] integrated deep learning algorithms with UAV LIDAR data to segment single tree crowns, offering a comprehensive framework for accurate tree-crown segmentation in forest conditions.
However, the past two years of 2022 and 2023, has witnessed remarkable advancements in this field, with substantial progress reported. The key accomplishments and breakthroughs in this domain are summarized and detailed in Table 1 for comprehensive reference and analysis.

Table 1.
Major achievements from convolutional neural network (CNNs), graph neural network (GNNs) and their variations.
1.2. Major Achievements from RF and SVM
The research into RF and SVM has a long history and is still very active. In 2012, Freeman et al. [10] investigated the performance of RF in addressing species imbalances across strata in Nevada. They proposed strategies to mitigate imbalances in sampling intensity and target species within training data. In 2013, Adelabu et al. [39] employed RF and SVM based on EnMap Box for the tree species classification of five-band RapidEye satellite data. Their study revealed that SVM outperformed RF when dealing with limited training samples. In 2014, Rochdi et al. [40] utilized RF and SVM to assess the classification performance of LiDAR and RapidEye data, individually and in combination. Their results indicated a significant enhancement in classification accuracy with the joint use of LiDAR and RapidEye data, with the RF classifier outperforming SVM in tree species classification. In 2020, Zhao et al. [41] applied SVM and spectrum angle mapper (SAM) on natural, needle-leaves and broadleaved mixed forests and reported overall 70% consistency. We have summarized the recent major achievements reported in 2022 and 2023 in Table 2.

Table 2.
Major achievements from RF and SVM.
1.3. The Longitudinal Profile and Pseudo Tree Crown (PTC)
Upon meticulous examination and assessment of the documented research findings, it becomes evident that CNN emerges as being highly effective in individual tree classification. Nevertheless, the varied outcomes associated with different CNN variations and the inherent challenges of utilising two-dimensional image data for tree species classification prompted us to explore alternative approaches. We are delighted to announce a successful breakthrough in addressing these challenges. We have undertaken an innovative approach to data feature reformation, referred to as pseudo tree crown (PTC), and rigorously tested its efficacy across four distinct machine learning classifiers.
The exploration of PTC traces back to earlier efforts, notably highlighted in Fourier et al. [49] and Zhang and Hu [50], identified a promising avenue by leveraging the longitudinal profile of tree crowns, marking an initial attempt to correlate physical crown attributes with their optical nadir views. Results underscored a robust correlation. However, this approach lacked flexibility across various contexts due to computational constraints and reliance on parametrized equations and failed to gain widespread adoption. In 2017, Balkenhol and Zhang [51] further delved into the potential expansion of three-dimensional correlations between the physical tree crown and the tree crown greyscale three-dimensional view. Limited by classification methods of the time, their study primarily showcased individual physical crown attributes alongside their 3D greyscale representations, laying the groundwork for the PTC concept. Another prototype was reported in Miao et al. [52] in 2023.
To test the effectiveness of PTC, we selected four ML image classifiers: (1) YOLOv5 objective detection model with CSPDarknet53 (YOLOv5) [53]; (2) simple PyTorch framework classifier with ResNet50 [54] (referred to as PyTorch); (3) Tenserflow 2.0 framework classifier with ResNet50 (referred to as TF2); and (4) random forest (RF). The four classifiers reflect the current mainstream tree classification, providing acceptable accuracy. Adopting PTC can enhance the classification results across all image classifiers tested by at least 10%. For instance, the PyTorch+ResNet50 image classifier, with nadir RGB, obtained an acceptable 85% and achieved a remarkable improvement to 97% with PTC.
We aimed to investigate the reconstruction of the complete three-dimensional tree structure utilising PTC, incorporating LiDAR data for both model development and validation purposes in a future study.
2. Data and Preprocessing
2.1. Study Area, Instruments, and Data Acquisition
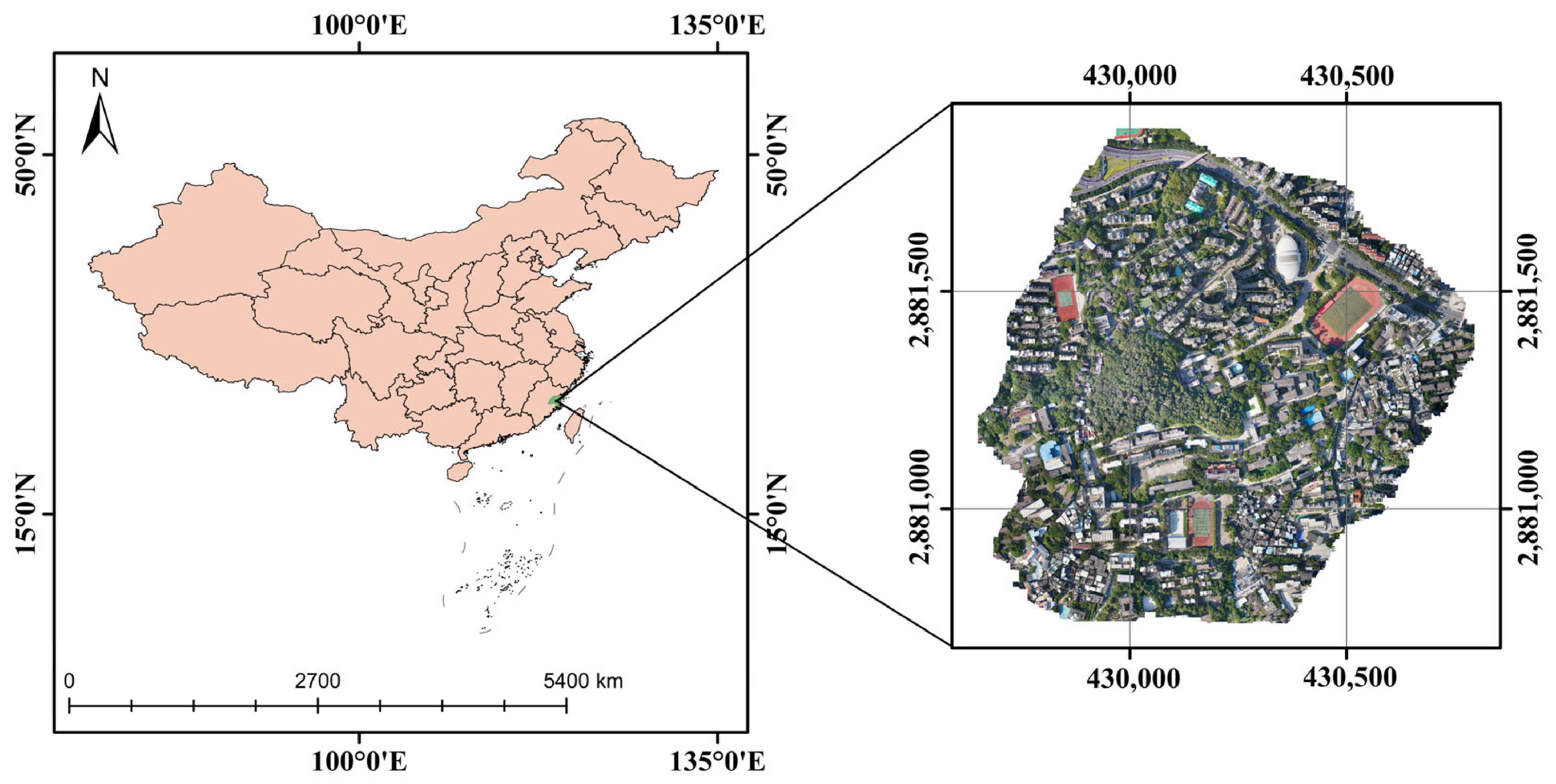
The research area of this study was located at the Cangshan Campus of Fujian Normal University (FNU) (26°02′05″N to 26°02′35″N and from 119°17′50″E to 119°18′29″E). It has a subtropical monsoon climate, as shown in Figure 1. The optical image was captured on 18 March 2022 under overcast weather conditions. The D2000 UAV from Feima Robotics (Shenzhen, China), equipped with Hesai XT32 LiDAR (Hesai Technology, Shanghai, China), was used for the survey at a flight altitude of 100 m. The camera model was SONY a6000 (Sony, Tokyo, Japan), with a sensor size of 23.5 mm by 15.6 mm Advanced Photo System type-C (APS-C), effective pixels of 24.3 million, and a 25 mm fixed-focus lens. The images were captured in the China Geodetic Coordinate System 2000 (CGCS2000) coordinate system and were in the Gauss–Krüger projection with the 3° zone and 100th band. The final image was saved in TIFF format with a resolution of 0.03 m.
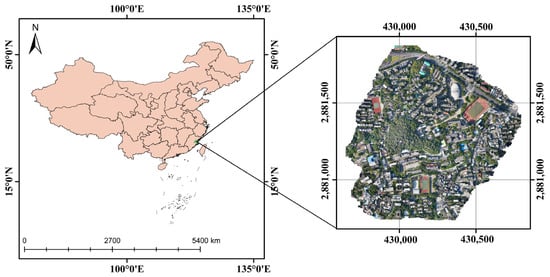
Figure 1.
Study area: Fujian Normal University Cangshan Campus, with the location map on the left and a drone orthophoto on the right.
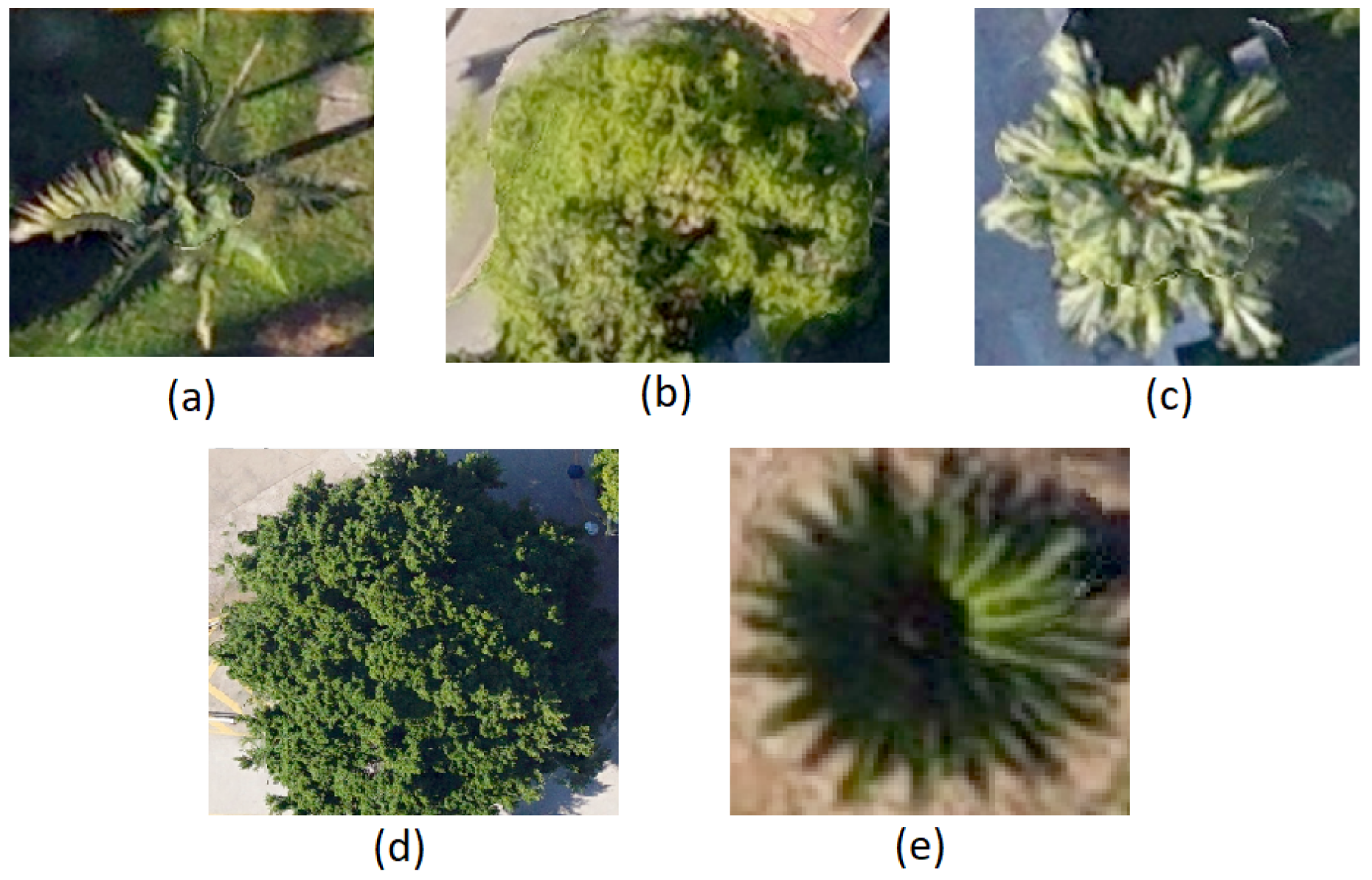
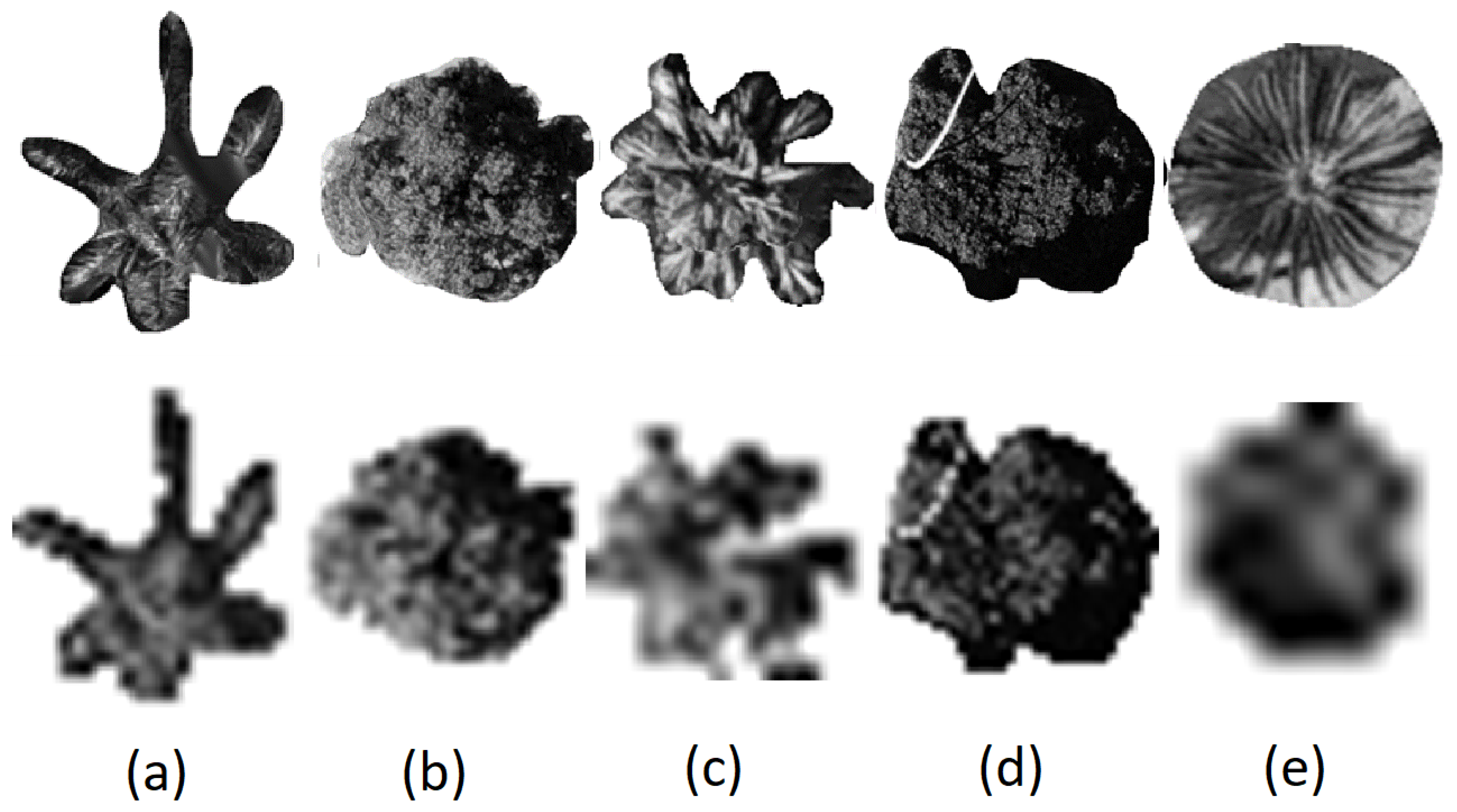
In our study area, we selected five dominant tree species, namely Archontophoenix alexandrae (Aa), Mangifera indica (Mi), Livistona chinensis (Lc), Ficus microcarpa (Fm), and Sago palm (Sp), as the main research objects, as shown in Figure 2.
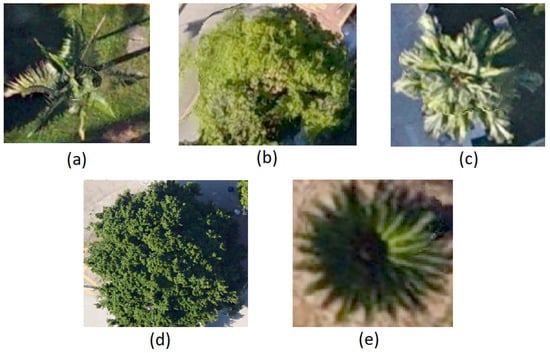
Figure 2.
The nadir view of (a) Archontophoenix alexandrae (Aa), (b) Mango indica (Mi), (c) Livistona chinensis (Lc), (d) Ficus microcarpa (Fm), and (e) Sago palm (Sp).
2.2. Data Pre-Processing and Pseudo Tree Crown (PTC) Generation
We first imported images into ArcGIS 10.2 and constructed an image pyramid to gather precise information for our experimental tree samples. We then manually identified and selected the shapefiles representing the target tree species, cropping corresponding image tiles accordingly. Validation of the tree species for every tree we used as a sample was conducted through on-site visits. Finally, we obtained a total of 696 samples, with Aa comprising 226, Mi 134, Lc 143, Fm 131, and Sp 62.
Despite the relatively confined geographical area and uniform growing conditions, it is important to acknowledge that these tree samples have demonstrated in-class variation. Various factors can influence their growth, such as sunlight exposure, weather patterns, and shooting angles. Consequently, discernible variations exist between different sample categories and within the same category.
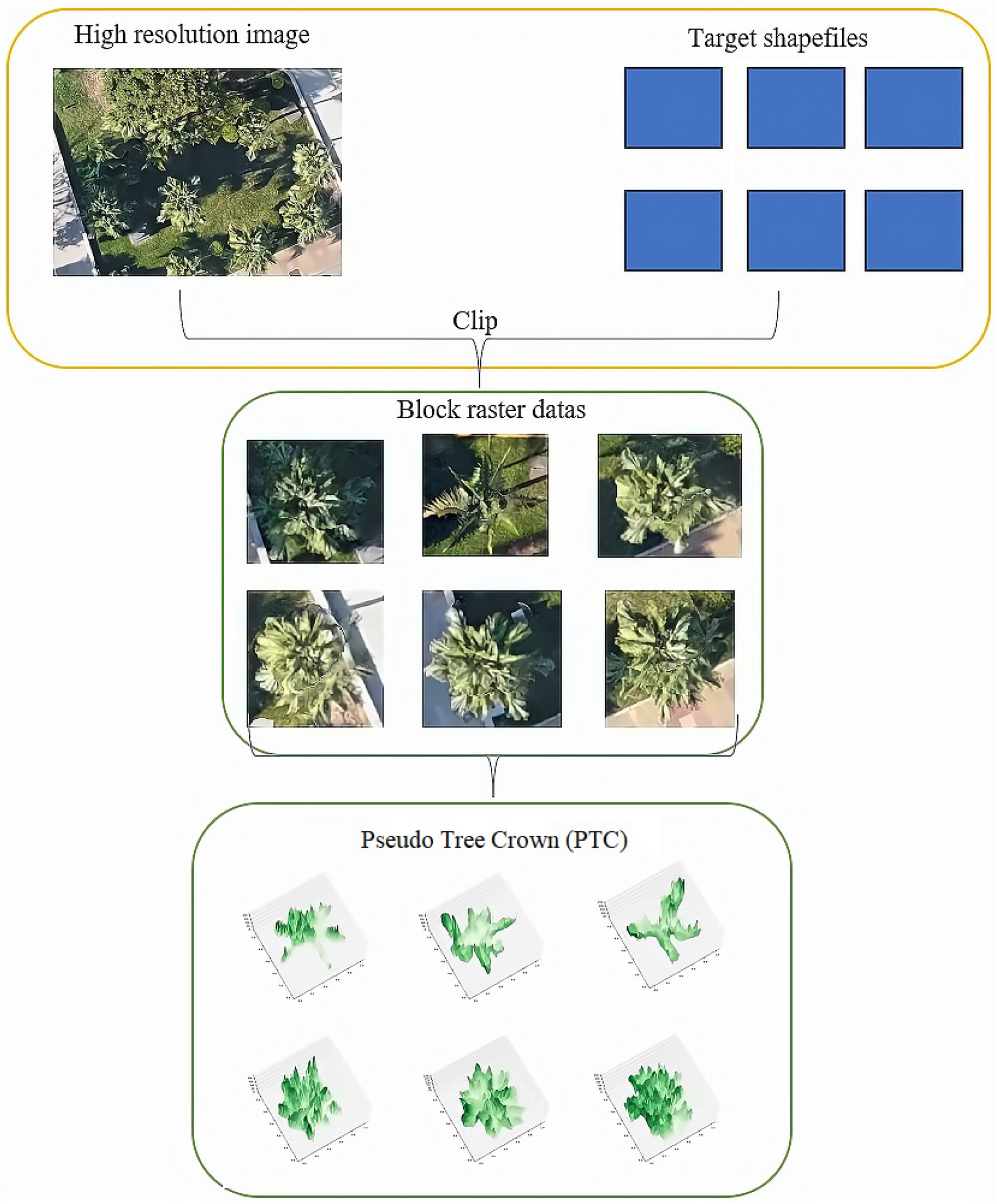
The general flowchart of pre-processing is illustrated in Figure 3.
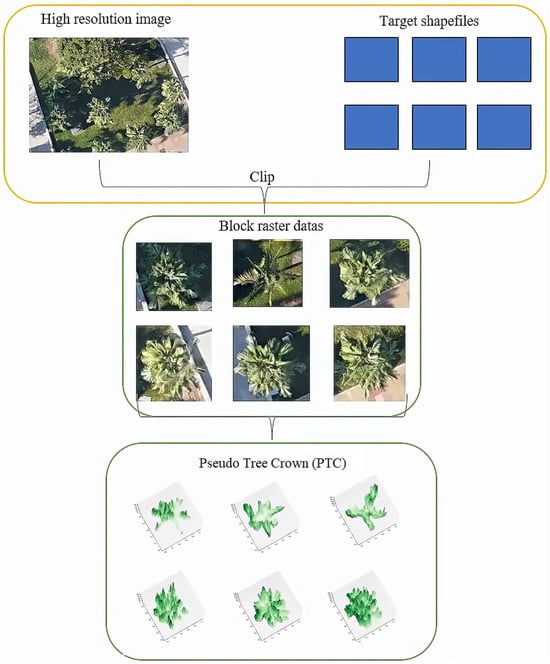
Figure 3.
The workflow of an individual tree sample in data pre-processing.
3. Methodology
Our study comprises two primary components that represent the main contributions of this research. Firstly, we introduced the pseudo tree crown (PTC) as a new input image instead of standard nadir view images, marking its first incorporation in the classification literature. Secondly, we compared four different deep learning (DL) image classifiers: simple PyTorch and TensorFlow 2.0 framework with default transform layer and ResNet50 neural network layer; You Look Only Once (YOLO), YOLOv5 object detection model built based on CSPDarknet53 [53]; and traditional ML random forest (RF), which is an ensemble of decision tree, but generally trained via the bagging method. Due to the paper’s limited length, we will focus only on the methods we used.
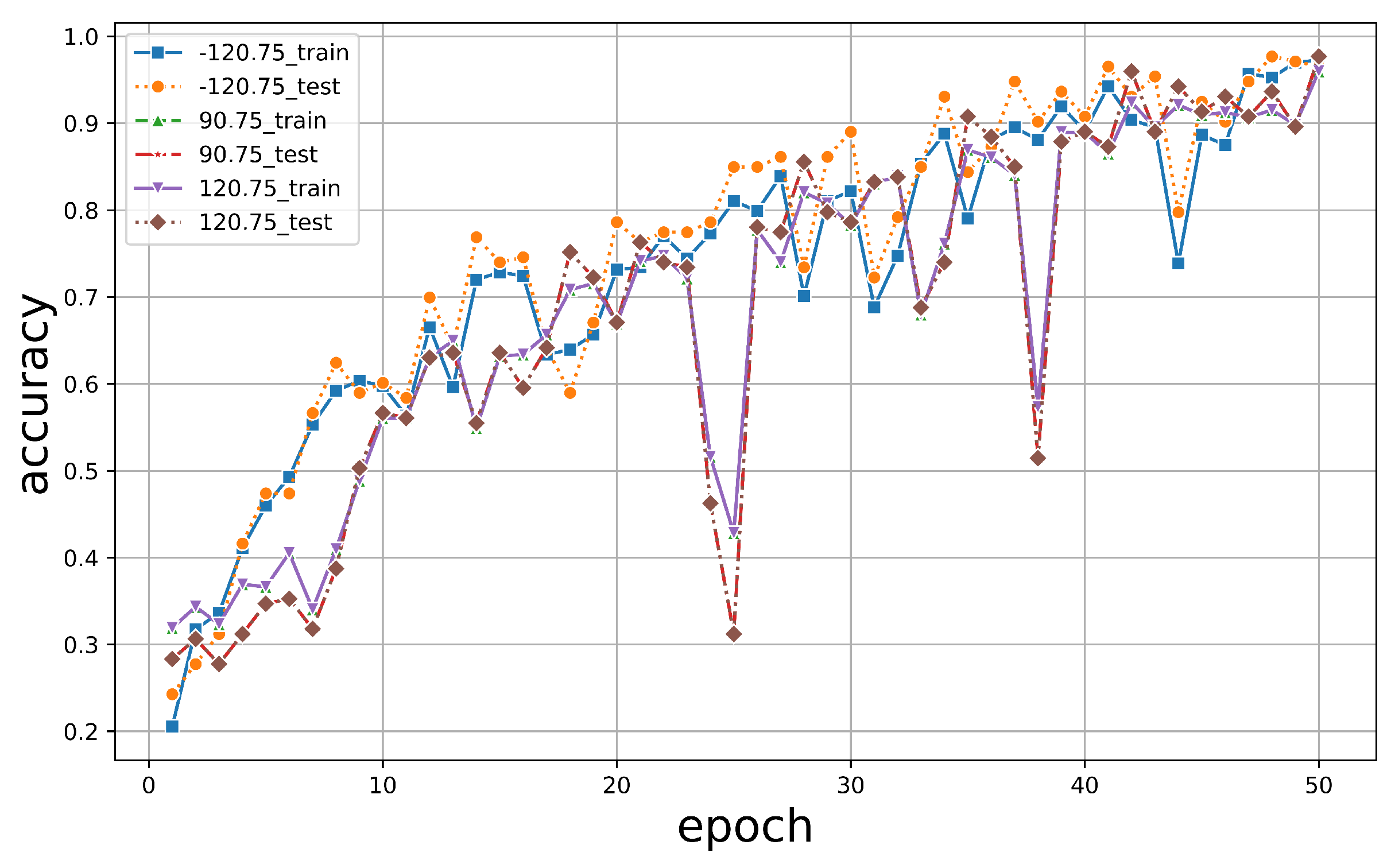
We set the input image size to 64 × 64 pixels for training using these three DL classifiers. During the training process, we utilized a batch size of 4, selected the SGD optimizer to optimize our algorithm, and employed shuffling to prevent overfitting and ensure the classifier captured more accurate features. The training was conducted for a total of 50 epochs. The initial learning rate was set to , and the momentum during training was set to . All the parameters obtained from the final trained model were recorded in Tensorboard, allowing us to monitor the model’s changing trends in real time. All data were split into a 1:4 ratio for testing and training.
3.1. Pseudo Tree Crown (PTC)
The first step involved generating the PTC from the nadir view tree crown images, as shown in Figure 4. All collected sample data underwent parsing to extract information regarding the number of rows, columns, and associated bands for each sample. Subsequently, an affine matrix was formulated, and the image’s projection details were extracted to retrieve pixel data for the green band. After these data were acquired, they were converted into an array format and organized into a grid. Any pixel values exceeding 255 were reset to nadir values, and the subplot’s projection mode was configured for three-dimensional representation.
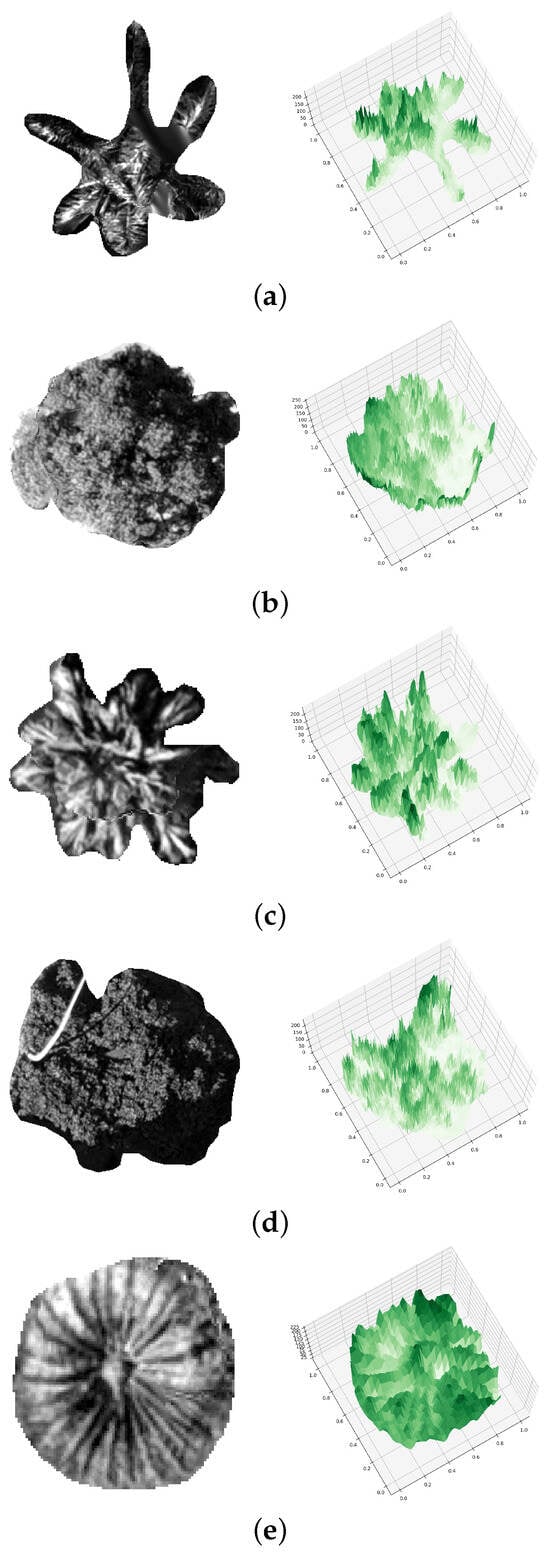
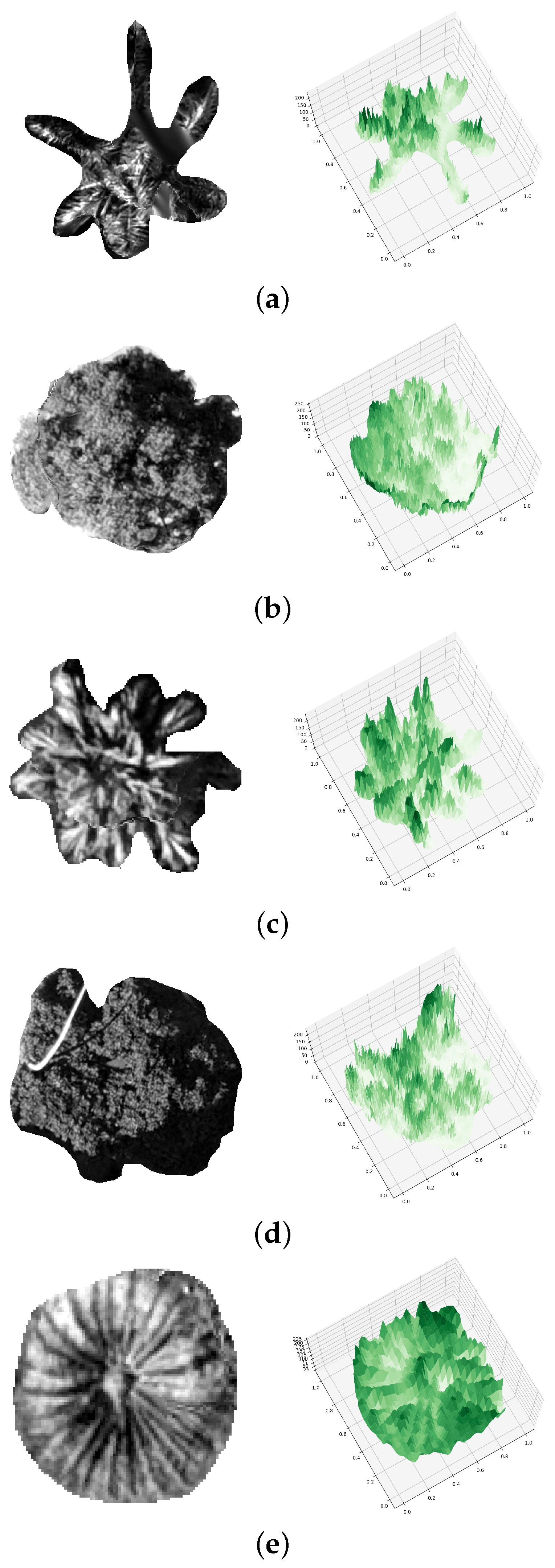
Figure 4.
PTC of (a) Aa (b) Mi (c) Lc (d) Fm, and (e) Sp.
For the creation of our PTC, azimuth angles of −120° and an elevation angle of 75° were used. We began with a conventional default setting from Python 3D plot. However, we conducted a study to verify the variance in azimuth and elevation angles, elaborated in Section 4.3. Additionally, a comparative analysis was undertaken using different spatial resolutions of PTC and original nadir view images instead of PTC, with the findings detailed in Section 4.4 and Section 4.5, respectively.
3.2. Image Classifiers
3.2.1. YOLOv5 with CSPDarknet53
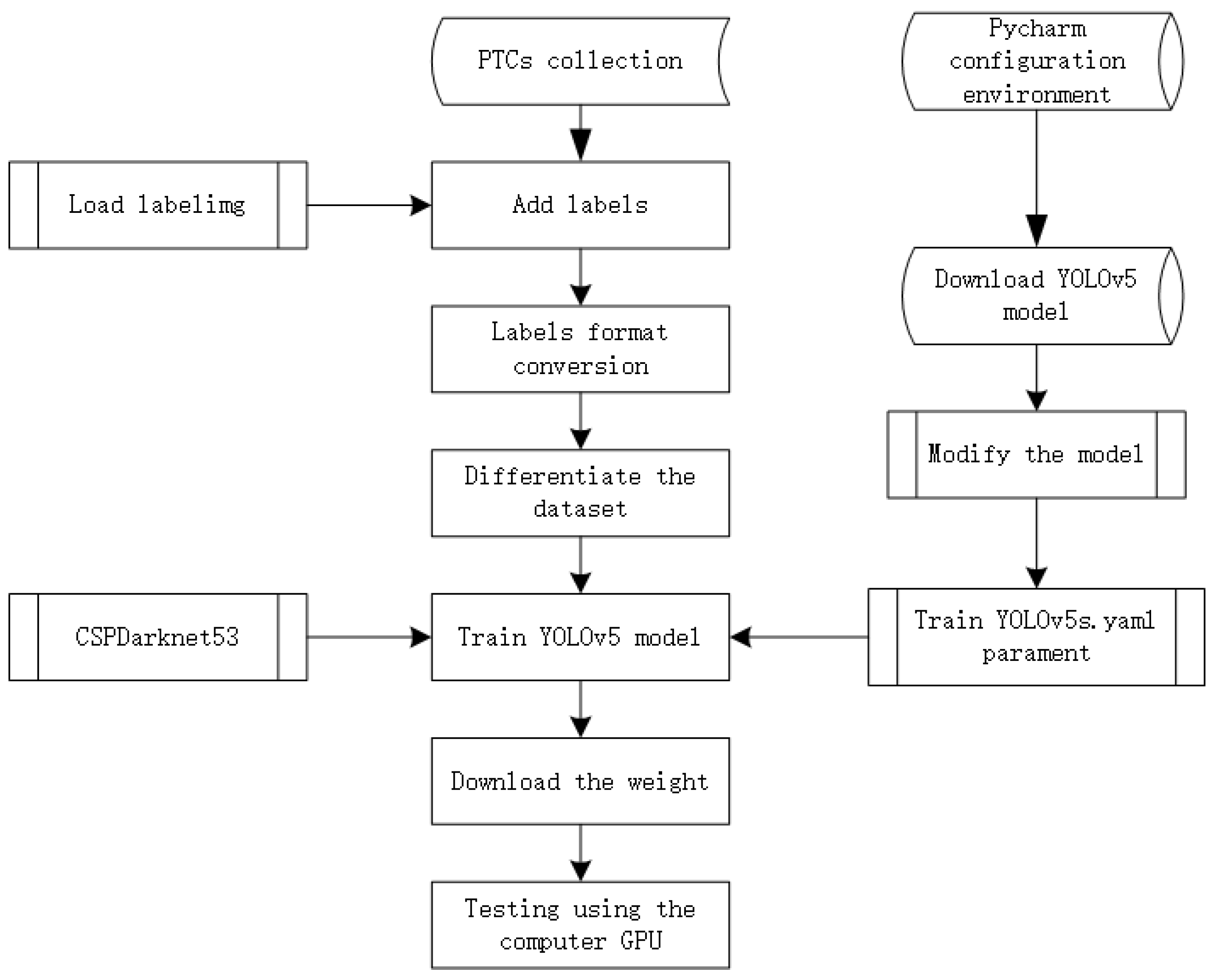
The YOLOv5 model [53], has demonstrated notable advancements in both detection accuracy and inference speed. Its reduced weight file, approximately 90% smaller than that of YOLOv4, renders it well-suited for real-time detection tasks on embedded devices. YOLOv5 distinguishes itself through its combination of high detection accuracy, lightweight design, swift detection times, and mature technology. This model encompasses four variations: YOLOv5s, YOLOv5m, YOLOv5l, and YOLOv5x, each accompanied by distinct weight files. Variances among these architectures are rooted in their feature extraction modules and convolutional kernels. Also, model size and parameter count differences exist among the four variants. For training purposes, we selected the YOLOv5s.pt weight file, with the training process outlined in Figure 5.
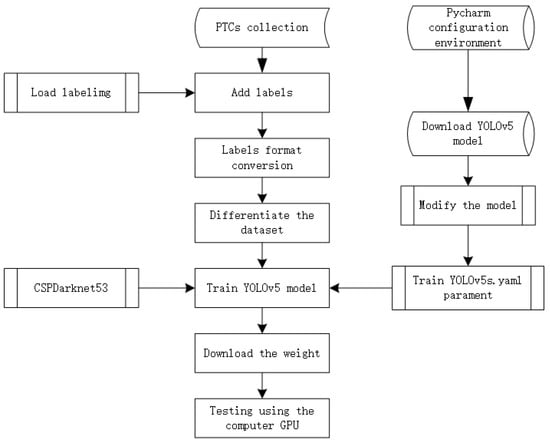
Figure 5.
YOLOv5 with CSPDarknet53 classifier flowchart.
Although YOLOv9, introduced in 2024 [55], represents the latest iteration in the YOLO series, it, alongside YOLOv6 and YOLOv7, builds upon the robust codebase established by the stable version of YOLOv5. While YOLOv5 may not be the most recent version, it remains the most stable. YOLOv8 shares significant similarities with YOLOv5, given their common developer group. In our research focusing on the efficacy of PTC across various ML classifiers, we used YOLOv5 to ensure precise and resilient outcomes. Future investigations may explore the potential for marginally quicker results with YOLOv8, as noted in [56].
Since PyTorch and TF2 can be used as the backbone of YOLOv5, we selected CSPDarknet53 as the feature layer. CSPDarknet53 enhances feature extraction efficiency by integrating cross-stage partial connections, facilitating more accurate object localization and classification.
3.2.2. PyTorch with ResNet50
In most cases, DL frameworks tend to focus on usability or speed, making it challenging to balance the two. PyTorch is an ML framework that indicates that these two goals are somewhat compatible. It provides an imperative and Python programming style, supporting code as a model, making debugging easy, and maintaining compatibility with other popular scientific computing libraries. Additionally, it remains efficient and supports GPU acceleration for computations. Previous efforts recognized the value of dynamic eager execution in deep learning, and some recent frameworks have implemented this run-time-defined approach. However, they either sacrifice performance or use faster but less expressive languages, limiting their applicability. Through careful implementation and design choices, PyTorch achieves dynamic, eager execution without sacrificing performance. It enables dynamic tensor computations using automatic differentiation and GPU acceleration, maintaining performance comparable to the fastest deep learning libraries. This combination has gained popularity in the research community. PyTorch provides an array-based programming model accelerated by GPUs and allows differentiation through automatic differentiation integrated into the Python ecosystem.
One of the highlights of PyTorch is its simple and efficient interoperability, opening up possibilities to leverage the rich Python library ecosystem as part of user programs. PyTorch allows bidirectional data exchange with external libraries. It provides a mechanism using the torch.from_numpy() function and .numpy(), facilitating the conversion of tensors between NumPy arrays and PyTorch tensors. These exchanges occur without any data copying, making these operations very convenient and efficient, regardless of the size of the converted arrays.
Another significant advantage of PyTorch is that users can freely replace any component in PyTorch that does not meet their project requirements or performance needs. These components are designed to be completely interchangeable, allowing users to adjust them based on their temporary needs.
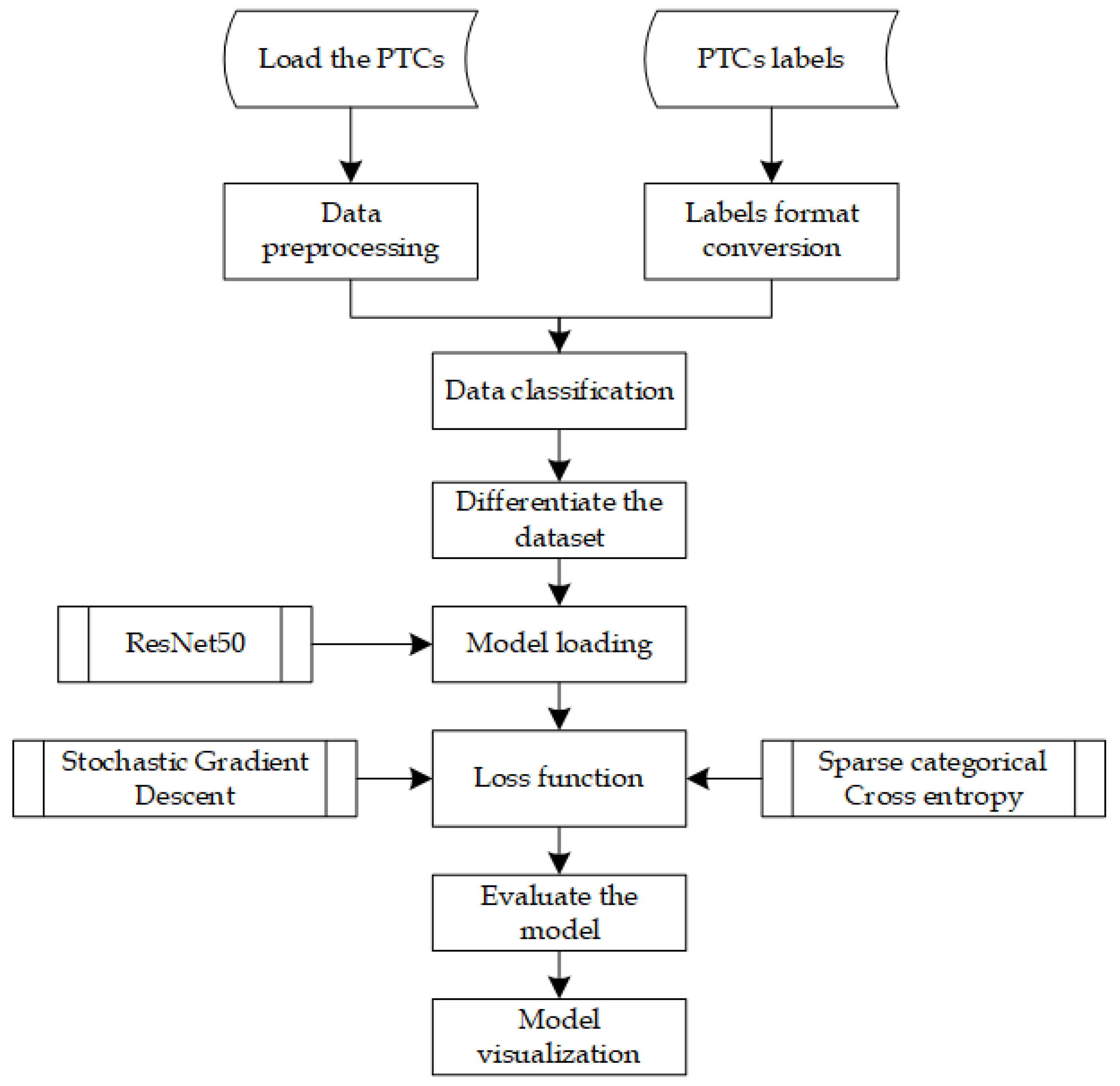
Efficiently running DL algorithms from the Python interpreter is currently one of the biggest challenges in this field. However, PyTorch addresses this issue differently by carefully optimizing various aspects of deep learning execution while allowing users to easily leverage additional optimization strategies. The simple PyTorch-based classifier is illustrated in Figure 6.
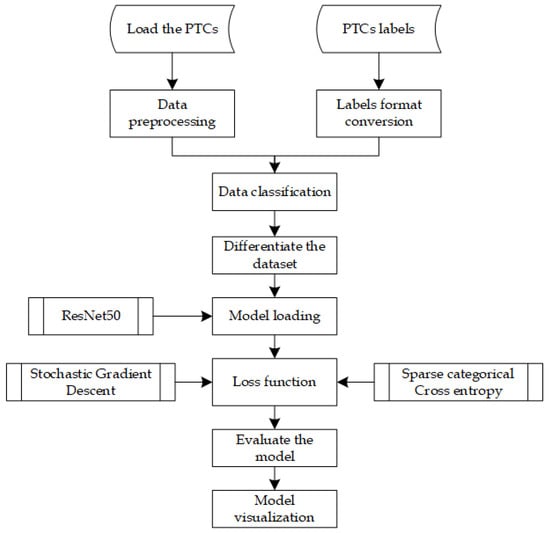
Figure 6.
PyTorch with ResNet50 classifier flowchart.
3.2.3. Tensorflow 2.0 (TF2) with ResNet50
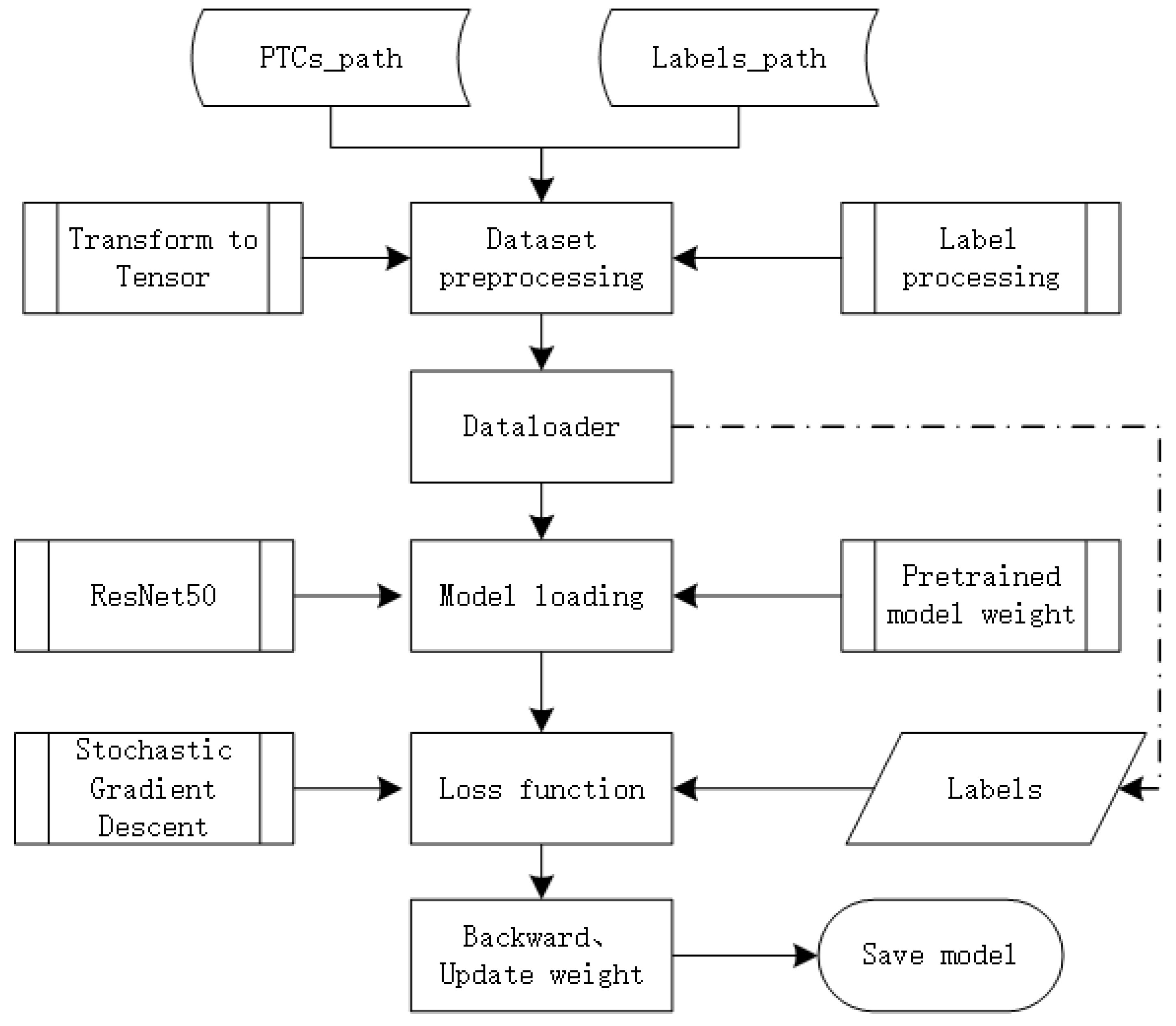
TensorFlow 2.11.0 is a numerical computing software library based on data flow graphs, providing interfaces and computational frameworks for implementing DL algorithms. It combines flexibility and scalability, supporting various commonly used programming languages. TensorFlow ensures its efficiency and stability by utilising CUDA, among other technologies. It allows mapping computation results to different hardware or operating systems, such as Windows, Linux, Android, iOS, and even large-scale GPU clusters. This significantly reduces development challenges. TensorFlow provides large-scale distributed training methods, enabling users to update model parameters using different devices, thus saving development costs. This flexibility allows users to implement model designs and train models on massive datasets rapidly.
In recent years, TensorFlow has been widely applied in machine learning, deep learning, and other computational fields, including speech recognition, natural language processing, computer vision, robot control, information extraction, and others. In October 2019, Google released TensorFlow 2.0, and one of its significant changes was the official integration and comprehensive support for Keras. Keras is a high-level neural network API known for its highly modular, minimalist, and extensible features. It provides clear and actionable error messages and supports CNN. TensorFlow 2.0 uses the sequential, compile, and fit methods of Keras to build, compile, and train models. The TensorFlow 2.0 training process flowchart is illustrated in Figure 7.
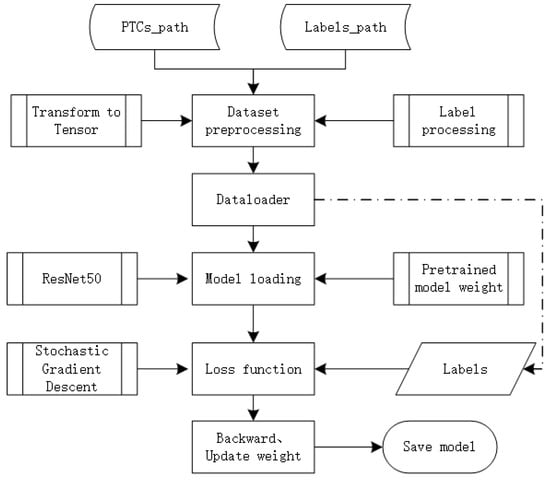
Figure 7.
TF2.0 with ResNet50 classifier flowchart.
3.2.4. Random Forest (RF)
Random Forest is a supervised learning algorithm employing an ensemble learning method consisting of numerous decision trees to generate a consensus output, representing the best answer to a given problem. RF can be used for classification or regression and is a popular ensemble learning algorithm. It implements the bagging (bootstrap aggregation) method in ensemble learning, serving as a homogeneous estimator composed of many decision trees. The individual decision trees in a random forest are not correlated. When random forest is used to perform classification, each sample is evaluated and classified by every decision tree in the forest. Each decision tree produces a classification result, and the final result of the random forest is determined by the majority result (mode) among all decision trees.
4. Results and Discussion
4.1. DL Classifier Comparison
The primary objective of this research was to assess how various models affect tree species classification using PTC. Three neural network classifiers, PyTorch, TF2, and YOLOv5, were selected for analysis. The results are summarized in Table 3 in terms of average training time and average classification accuracy. We found that PyTorch is more flexible and user-friendly than are the other two methods. It provides an intuitive Python API, making it more convenient for users. While TF2 has seen improvements in API design since its 1.0 version, it can still feel relatively complex in certain situations. YOLOv5, being specialized in object detection tasks, has its model and structure fixed towards specific objectives in object detection and may not be as flexible as PyTorch in image classification. PyTorch boasts strong community support and a wealth of third-party libraries, offering various pre-trained models and tools for rapidly developing image classification applications. Although TF2 also has robust community support, its ecosystem is relatively complex compared to PyTorch. YOLOv5, on the other hand, has fewer model weights and options than do the first two. In achieving the same goal, PyTorch often achieves the desired results with less effort, while TF2 may require more work for similar objectives. YOLOv5, being less widely used in image classification and having a relatively smaller scale, may face limitations in support and issue resolution compared to the broader availability of PyTorch.

Table 3.
The average training time and average classification accuracy of different ML models.
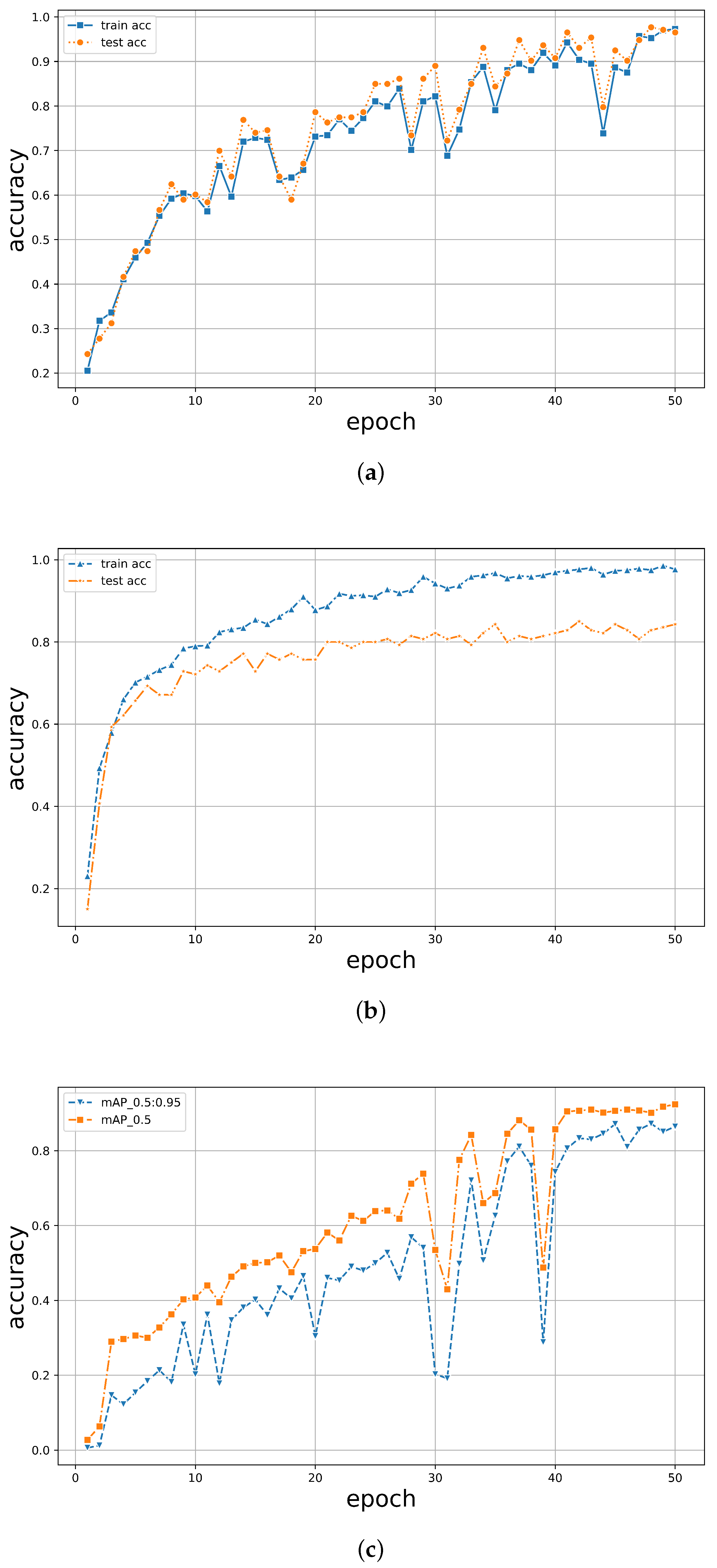
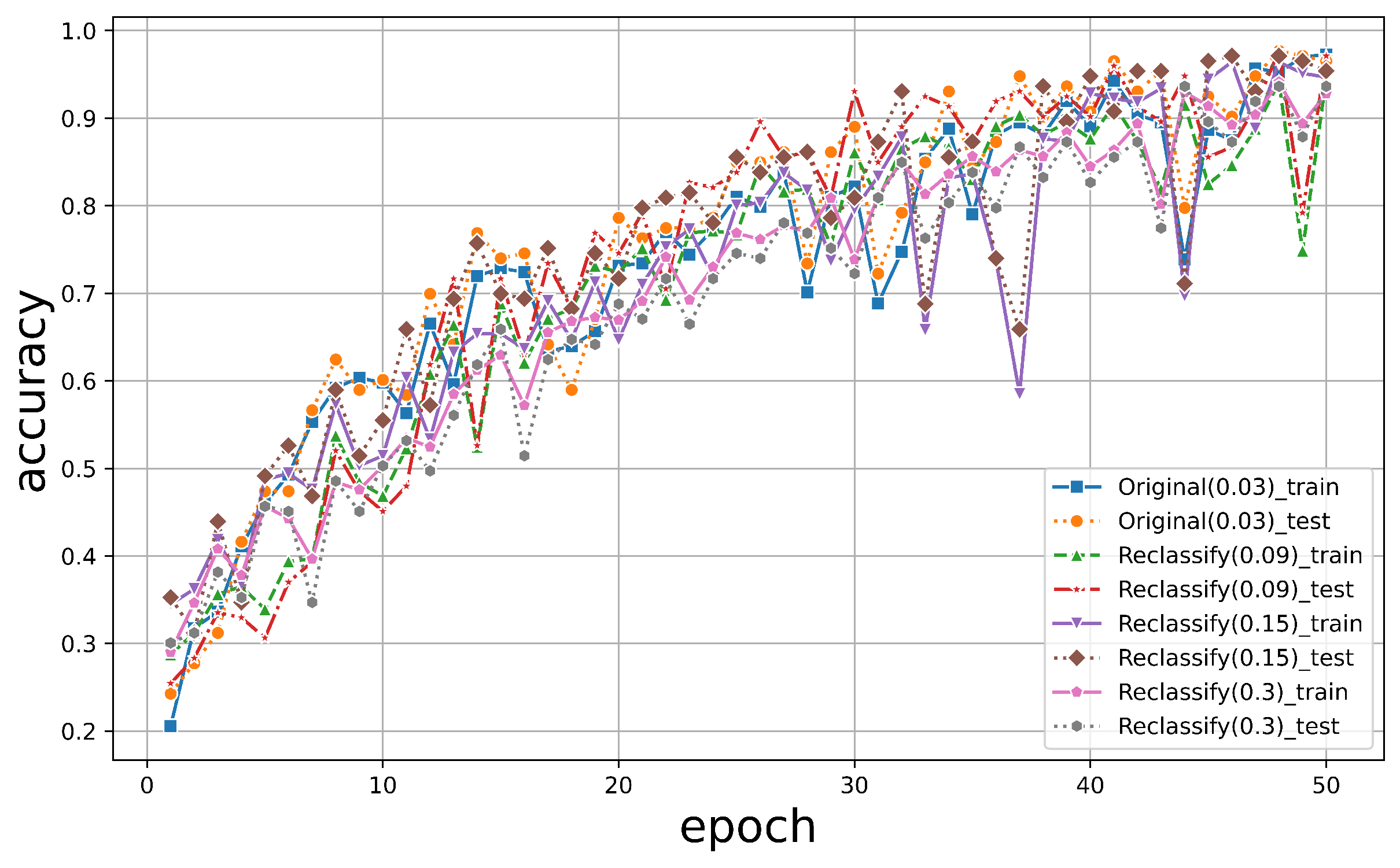
Under 50 epochs, YOLOv5 showed the most volatile performance but yielded an excellent final convergence. In contrast, TF2 showed consistent stability but relatively lower accuracy. The results are illustrated in Figure 8. It was observed that the accuracy of all models exceeded 90% around 40 epochs, with models trained using the PyTorch framework achieving over 95%. While both PyTorch and TF2 achieved a training set accuracy of over 95%, the test set accuracy of TF2 was only around 80%, whereas PyTorch maintained an accuracy of over 95%, maintaining a good performance. For YOLOv5, it was noticed that when the classification threshold was set to 0.5, the training accuracy was generally higher than when the threshold was between 0.5 and 0.95. However, its highest classification accuracy only reached 92%, never exceeding 95%. Moreover, when all three models underwent 50 epochs, the training time for PyTorch was 45 min, while TF2 and YOLOv5 took 76 min and 102 min, respectively (Table 3). The PyTorch classifier demonstrated robust stability, accuracy, and efficiency in tree species classification.
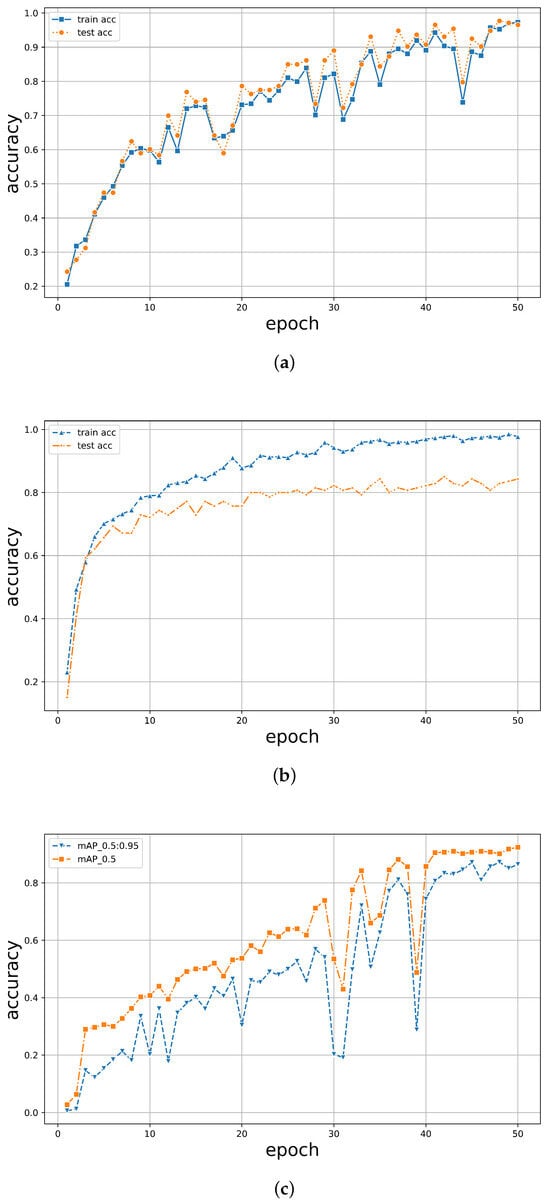
Figure 8.
The accuracy of training and validation of the dataset for (a) PyTorch, (b) TF2, and (c) YOLOv5 as indicated.
4.2. Classification Accuracy Assessment
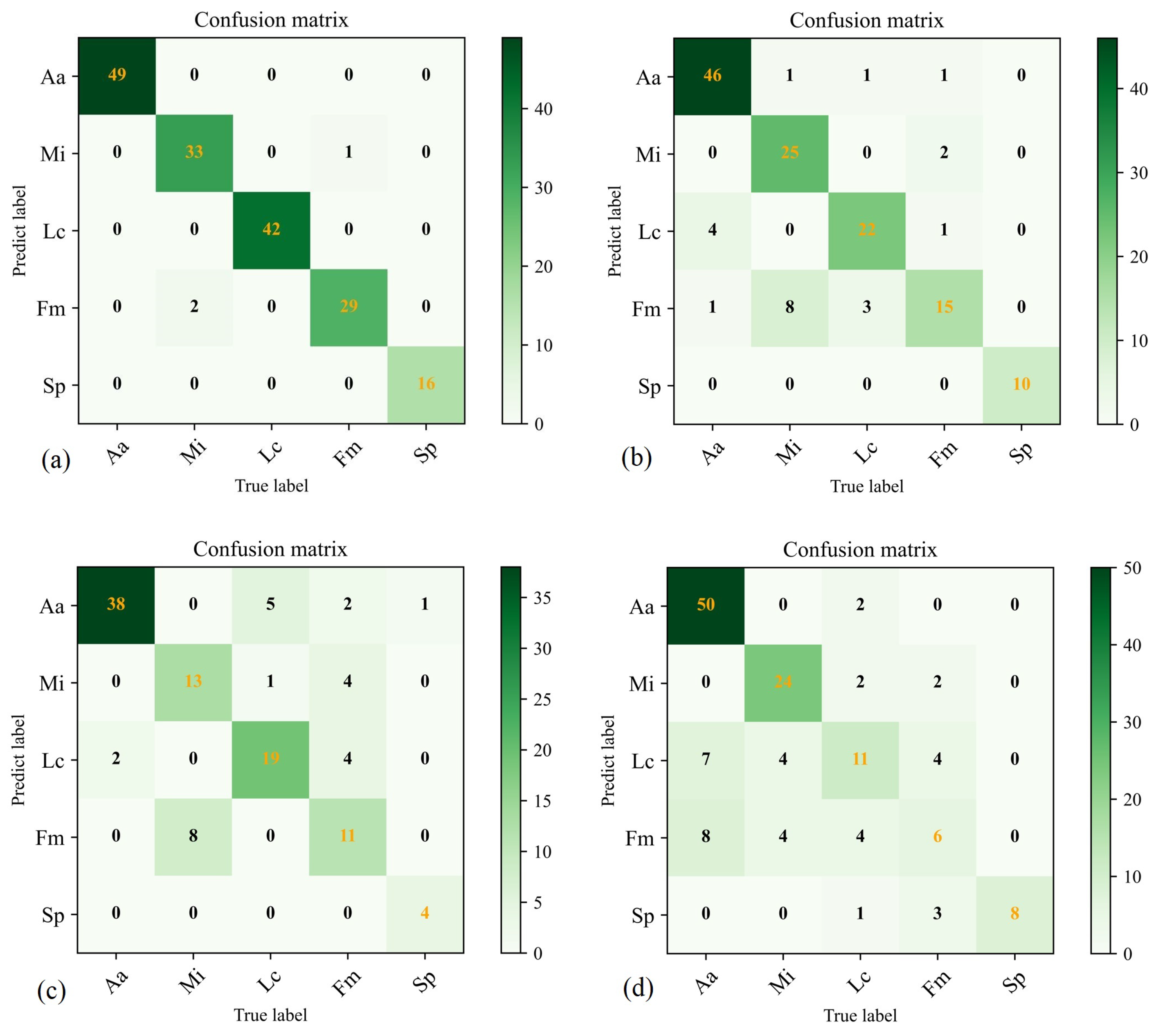
We evaluated each model’s overall classification accuracy and confusion matrices to corroborate our results further, as shown in Figure 9. Precision, recall, specificity, receiver operating characteristic area under the curve (ROC-AUC), and F1-score for each tree class are detailed in Table 4, Table 5, Table 6, Table 7 and Table 8, providing quantitative metrics for the models’ performance in tree species classification tasks. By analysing these results, we can gain a more precise and comprehensive understanding of each model’s performance in tree species classification.
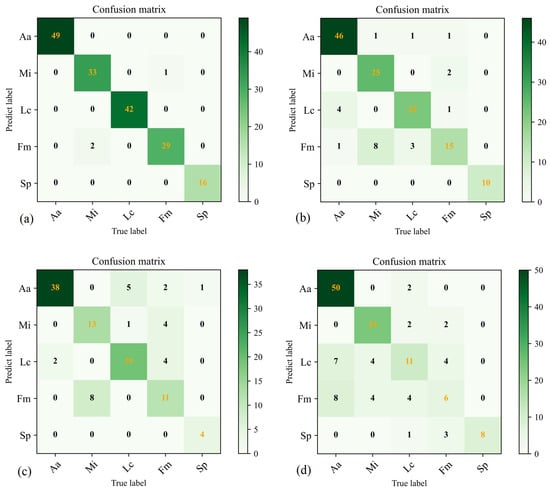
Figure 9.
The confusion matrices for training on the dataset in PyTorch, TF2, YOLOv5, and RF are presented in (a–d), respectively. The color indicates the matching number.

Table 4.
Precision (PyTorch, TF2.0, YOLOv5, and RF).

Table 5.
Recall (PyTorch, TF2.0, YOLOv5, and RF).

Table 6.
Specificity (PyTorch, TF2.0, YOLOv5, and RF).

Table 7.
ROC-AUC (PyTorch, TF2.0, YOLOv5, and RF).

Table 8.
F1-score (PyTorch, TF2.0, YOLOv5 and RF).
The PyTorch classifier achieved the highest overall classification accuracy of 98.26%, followed by TF2 with 84.29%, YOLOv5 with 75.89%, and the traditional random Forest (RF), with an overall classification accuracy of 70.71%. As indicated in Table 4, Table 5, Table 6, Table 7 and Table 8, in PyTorch, the precision, recall, specificity, ROC-AUC, and F1-score for Aa, Lc, and Sp were all 1.0, while Mi maintained these metrics above 0.95. The precision for Fm was slightly lower at 0.935, but the recall and specificity remained above 0.96. In TF2, Sp’s performance was excellent, achieving 1.0 for all three metrics. Aa also maintained results above 0.9. Mi had a lower precision of 0.735, with recall and specificity above 0.92. The precision and recall for Lc ranged between 0.8 and 0.85, but specificity reached 0.965. Although the precision for Fm in TF2 was 0.79, the recall was only 0.556, and the specificity was 0.965. YOLOv5 and RF had less favourable classification results compared to PyTorch and TF2. Despite generally lower performance, YOLOv5 achieved a recall of 1.0 for Sp. RF attained a precision and specificity of 1.0 for Sp, indicating that our dataset performed best in identifying Sp regardless of the classification method.
In summary, our research results suggest that due to the influence of factors such as the blurred edges and interweaving crowns of canopy images and poor texture effects in the original data, the PyTorch classifier performance is superior to that of TF2, YOLOv5, and RF. This demonstrates that PyTorch has significant potential for multi-classification tasks using PTC from high-resolution remote-sensing images. However, in the evaluation of the efficiency of a classification model, there are still many evaluation metrics to consider, along with the need to integrate various features of the dataset and the specific requirements of the classification task. Further research and refinement of these models’ performance using different datasets in various environments are necessary.
4.3. Comparison of the Conventional Nadir View 2D RGB Image and PTC with the Four Classifiers
Additionally, we compared the PTC with the conventional nadir view images using the four classifiers. To keep the model’s performance training consistent in the PTC and conventional images, we also extracted the green band of the 2D nadir view images as the input dataset. The accuracy of the conventional nadir view dataset and PTC are shown in Table 9. Clearly, all ML classifiers were enhanced by 12–14% and about 9% in RF.

Table 9.
The classification results of the conventional nadir view image and PTC using four different classifiers.
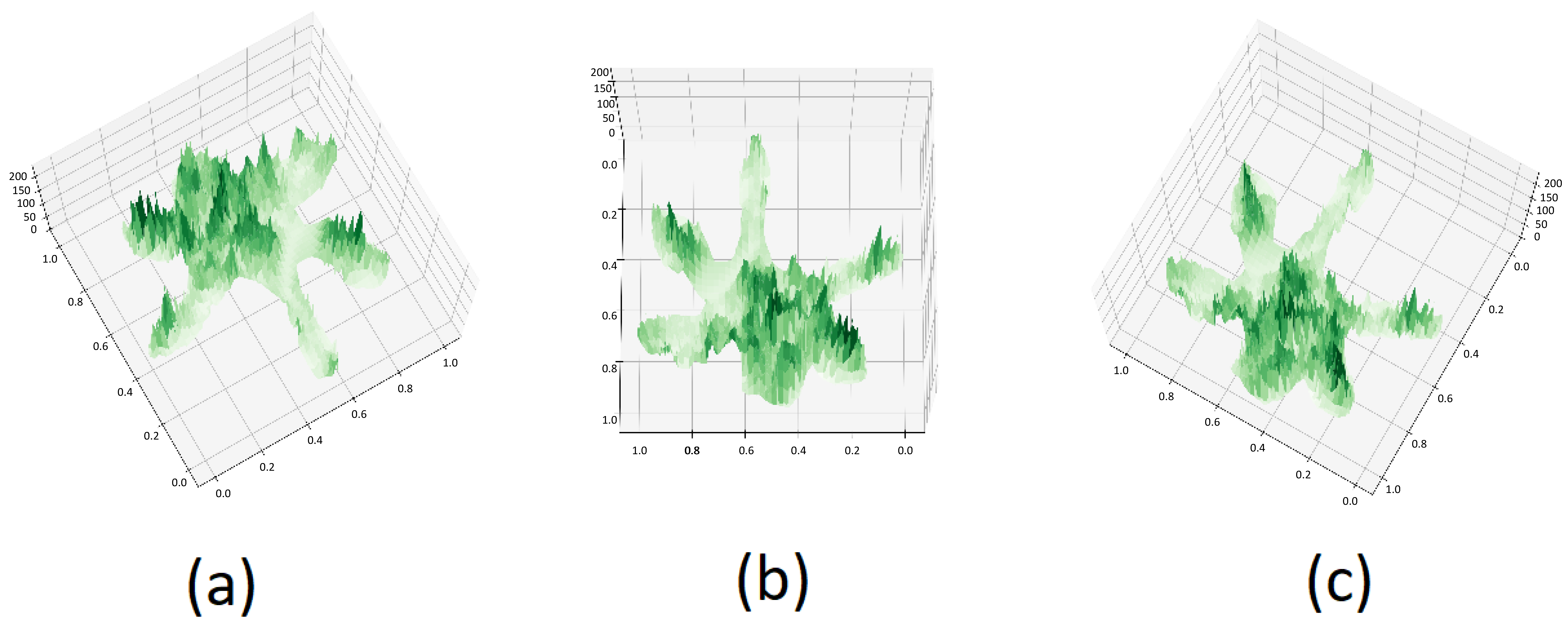
4.4. PTC Azimuth and Elevation Angle Impact Study
To explore the influence of varying azimuth and elevation angles on tree species classification outcomes using PTC, we selected three sets of angles, as illustrated in Figure 10: −120° and 75°, 90° and 75°, and 120° and 75°. PTCs were generated using these different angles and input into our PyTorch-based model for classification. The resultant classification outcomes are shown in Figure 11.
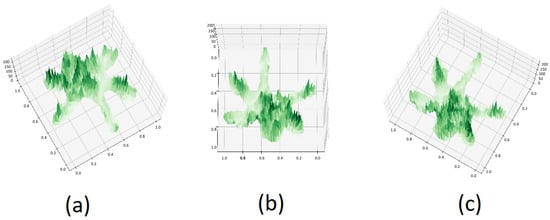
Figure 10.
PTC of Aa with different azimuth and elevation: (a) −120, 75 (b) 90, 75 (c) 120, 75.
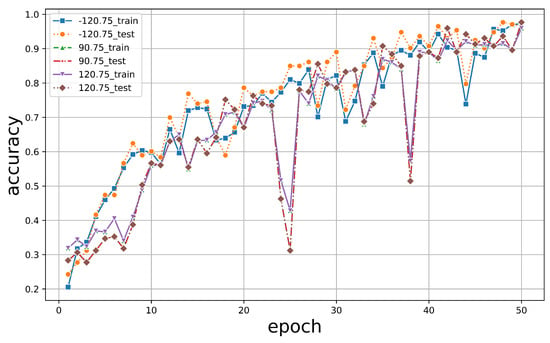
Figure 11.
The PTC classification accuracy by different azimuth and elevation angles.
The graph illustrates that across all angle configurations, the final accuracy of both the training and test datasets consistently exceeded 95%. This suggests that changes in azimuth and elevation angles have minimal impact on the overall classification accuracy. This resilience is logical given PyTorch’s application in training on human images, which does not necessitate specific viewing angles.
4.5. Analysis of the Impact of Different Spatial Resolutions on PTC
We also examined the influence of spatial resolution on PTC. Given that PTC derives height information from greyscale values and partially obscures tree crown details due to viewing angles, it inherently masks out many spatial intricacies, suggesting a natural resilience to changes in spatial resolution. However, we were interested in determining the scale at which classification accuracy would be affected.
To investigate this, we downsampled the original RGB images into lower-resolution versions, which reduced the resolution to a factor of 3, 5, and 10 of the original resolution. We extracted the green band again, cropped out patches of individual trees for each species, and established a resampled dataset, as illustrated in Figure 12 (reduced by a factor of 10 from 0.03 m to 0.3 m). PTCs of different spatial resolutions were created after that. All datasets maintained an accuracy above 90%, while the highest resolution, 0.03 m, achieved a classification accuracy of over 95%, which is shown in Figure 13. Remarkably, we observed no significant decrease in accuracy until the scale was reduced by over ten times. This indicates that as the resolution of the original data decreases, the features displayed by PTC are, to some extent, unfamiliar to the PyTorch classifier, leading to a slight decrease in accuracy.
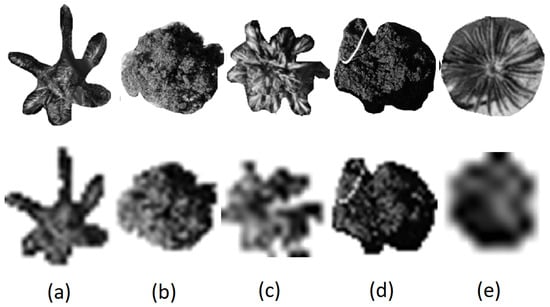
Figure 12.
Resampled images of (a) Aa (b) Mi (c) Lc (d) Fm (e) Sp.
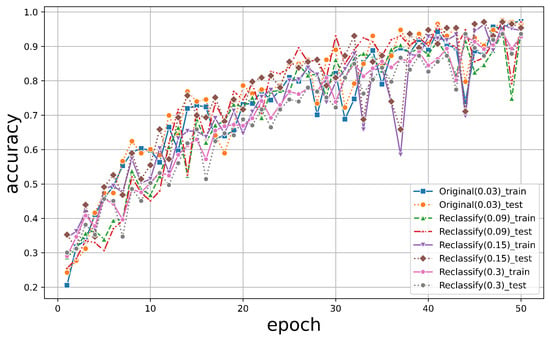
Figure 13.
The classification results of PTCs with different spatial resolutions.
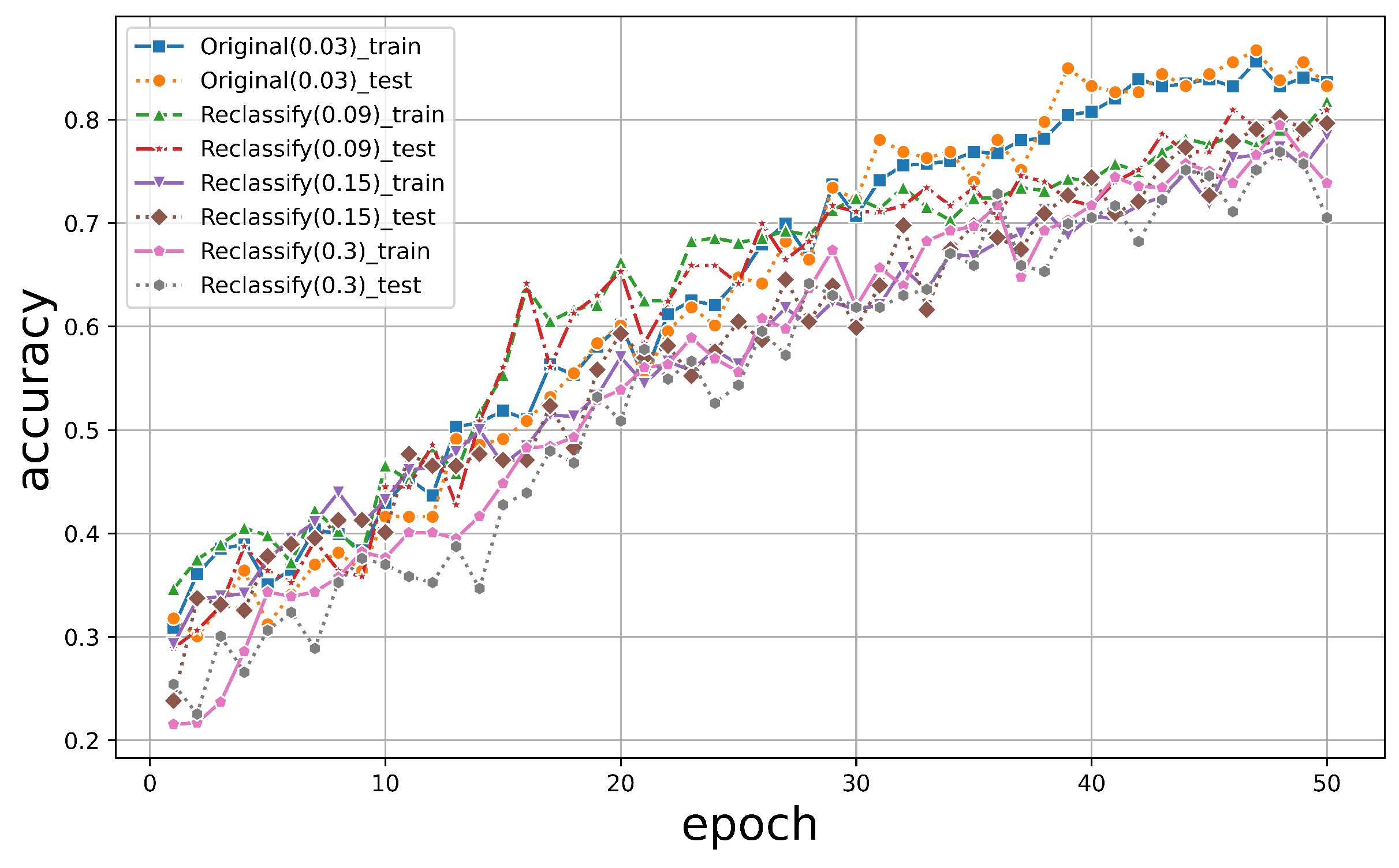
We further explored the resistance of the resolution change for the original images. We did the same resample for the original nadir images. All of these datasets were then input into PyTorch for training, and the accuracy trends on the training and test sets are shown in Figure 14.
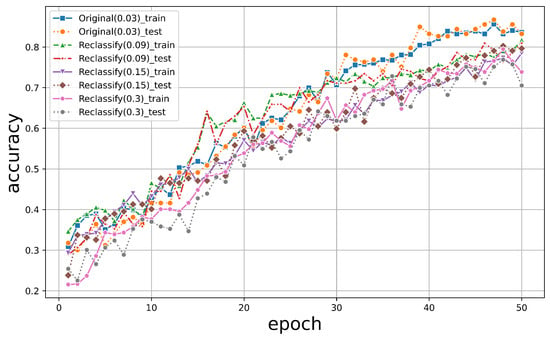
Figure 14.
The graphical representation depicting the variation in accuracy for the two-dimensional dataset.
In contrast, due to reduced resolution, the accuracy of the reclassified original images dropped to between 70% and 80% for both the training and test sets. This was much more significant than was the PTC, which remained approximately at 90% even at a factor of 10.
Therefore, we can conclude that the PTC is much more resistant to reducing spatial resolution, which implies that it has more flexible application areas and robust classification results.
4.6. Other Findings
It is worth mentioning that the accuracy for the tree species Aa was the highest in each classification method. This indicates that the classification model more easily recognizes the PTC, which is favourable to the species that have more spatial features. This could be attributed to its robust trunk and sparse, broad leaves, resulting in higher pixel distinguishability than other tree species. On the other hand, the accuracy for the Fm was relatively low, possibly due to its large crown, complex crown structure, and the intertwining growth of each tree. This leads to unclear boundaries of the crown layer, making it more challenging for the model to accurately recognize its features, thus affecting the classification of the banyan tree. In the future, we aim to improve our classification model algorithm further to achieve comprehensive and higher classification accuracy.
There is very limited work reported on reforming the features from the top view to the side view. Among the scant studies, Sun et al., 2023 [34] used UAV LiDAR to generate the side view of the tree and achieved a 88.3% accuracy based on the Transformer and 63.3% from RF. In contrast, in our study, by simply outputting the green band of RGB images and creating PTC, we achieved a classification accuracy of over 95% with more straightforward data processing and less workload. This further highlights the sensitivity of DL classification algorithms to the recognition of tree crown three-dimensional models.
5. Conclusions
In this paper, we propose a novel approach to the input data reformation method, wherein pseudo tree crown’s (PTCs) are generated from conventional nadir-view images. This transformation augments feature distinctiveness, thereby potentially enhancing classification outcomes, which is particularly crucial in scenarios with limited datasets, as demonstrated herein. We assessed the efficacy of PTCs across four ML methodologies for classifying tree species within the Cangshan campus of Fujian Normal University, utilising high-resolution aerial imagery obtained through low-altitude UAV flights. Promising results have been obtained by utilising individual tree PTCs derived from these high-spatial-resolution aerial images. Our findings demonstrate that integrating PTC images into the PyTorch classifier consistently yields a classification accuracy exceeding 95%.
We are excited to present the first application of PTC in image classification, yielding a 95% accuracy compared to utilising nadir view images directly with PyTorch-based classification (87%).Furthermore, among the various AI-based classification methodologies, including TF2.0 and YOLOv5, PyTorch demonstrated superior robustness, accuracy, and efficiency.
Future research will incorporate LiDAR data to explore the correlation between physical tree crowns and PTCs. Preliminary findings indicate encouraging results. If successful, this endeavour will directly link 2D nadir images to 3D tree structures, facilitating the integration of additional parameters—such as diameter at breast height (DBH)—into classification models, a feat previously unattainable using solely 2D image data.
Author Contributions
Conceptualization, K.Z.; Methodology, K.Z.; Software, S.M.; Validation, S.M.; Formal analysis, S.M. and K.Z.; Investigation, S.M. and K.Z.; Data curation, H.Z.; Writing—original draft, K.Z.; Writing—review & editing, K.Z. and H.Z.; Visualization, S.M.; Supervision, K.Z., H.Z. and J.L.; Project administration, K.Z. and J.L.; Funding acquisition, H.Z. and J.L. All authors have read and agreed to the published version of the manuscript.
Funding
We would like to thank Fujian Province Forestry Science and Technology Project (2022FKJ03) for the financial support.
Data Availability Statement
All data and Python source codes are available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | artificial intelligence |
| CNN | convolutional neural network |
| DL | deep learning |
| GNN | graph neural network |
| ML | machine learning |
| MLP | multilayer perception |
| PTC | pseudo tree crown |
| SSPCNN | spectral-spatial parallel convolutional neural network |
| SVM | support vector machine |
| UAV | unmanned aerial vehicle |
| ITS | individual tree species |
References
- Pause, M.; Schweitzer, C.; Rosenthal, M.; Keuck, V.; Bumberger, J.; Dietrich, P.; Heurich, M.; Jung, A.; Lausch, A. In Situ/Remote Sensing Integration to Assess Forest Health—A Review. Remote Sens. 2016, 8, 471. [Google Scholar] [CrossRef]
- Lausch, A.; Borg, E.; Bumberger, J.; Dietrich, P.; Heurish, M.; Huth, A.; Jung, A.; Klenke, R.; Knapp, S.; Mollenhauer, H.; et al. Understanding Forest Health with Remote Sensing, Part III: Requirements for a Scalable Multi-Source Forest Health Monitoring Network Based on Data Science Approaches. Remote Sens. 2018, 10, 1120. [Google Scholar] [CrossRef]
- Trisasongko, B.H.; Paull, D. A review of remote sensing applications in tropical forestry with a particular emphasis in the plantation sector. Geocarto Int. 2018, 35, 317–339. [Google Scholar] [CrossRef]
- Gyamfi-Ampadu, E.; Gebreslasie, M. Two Decades Progress on the Application of Remote Sensing for Monitoring Tropical and Sub-Tropical Natural Forests: A Review. Forests 2021, 12, 739. [Google Scholar] [CrossRef]
- Slavik, M.; Kuzelka, K.; Modlinder, R.; Surovy, P. Spatial Analysis of Dense LiDAR Point Clouds for Tree Species Group Classification Using Individual Tree Metrics. Forests 2023, 14, 1581. [Google Scholar] [CrossRef]
- Gao, C.; Zheng, Y.; Li, N.; Li, Y.; Qin, Y.; Piao, J.; Quan, Y.; Chang, J.; Jin, D.; He, X.; et al. A Survey of Graph Neural Networks for Recommender Systems: Challenges, Methods, and Directions. ACM Trans. Recomm. Syst. 2023, 1, 1–51. [Google Scholar] [CrossRef]
- He, T.; Zhou, H.; Xu, C.; Hu, J.; Xue, X.; Xu, L.; Lou, X.; Zeng, K.; Wang, Q. Deep Learning in Forest Tree Species Classification Using Sentinel-2 on Google Earth Engine: A Case Study of Qingyuan County. Sustainability 2023, 15, 2741. [Google Scholar] [CrossRef]
- Liu, P.; Ren, C.; Wang, Z.; Jia, M.; Yu, W.; Ren, H.; Xia, C. Evaluating the Potential of Sentinel-2 Time Series Imagery and Machine Learning for Tree Species Classification in a Mountainous Forest. Remote Sens. 2024, 16, 293. [Google Scholar] [CrossRef]
- Liu, H.; Su, X.; Zhang, C.; An, H. Landscape tree species recognition using RedEdge-MX: Suitability analysis of two different texture extraction forms under MLC and RF supervision. Open Geosci. 2022, 14, 985–994. [Google Scholar] [CrossRef]
- Freeman, E.A.; Moisen, G.G.; Frescino, T.S. Evaluating effectiveness of down-sampling for stratified designs and unbalanced prevalence in Random Forest models of tree species distributions in Nevada. Ecol. Model. 2012, 233, 1–10. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Qin, Y.; Chi, M.; Liu, X.; Zhang, Y.; Zeng, Y.; Zhao, Z.; Hinton, G.E. Classification of high resolution urban remote sensing images using deep networks by integration of social media photos. In Proceedings of the IGARSS 2018 (IEEE International Geoscience and Remote Sensing Symposium), Valencia, Spain, 22–27 July 2018; pp. 7243–7446. [Google Scholar]
- Marrs, J.; Ni-Meister, W. Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data. Remote Sens. 2019, 11, 819. [Google Scholar] [CrossRef]
- Egli, S.; Hopke, M. CNN-Based Tree Species Classification Using High Resolution RGB Image Data from Automated UAV Observations. Remote Sens. 2020, 12, 3892. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Li, Q.; Jing, L. CNN-based tree species classification using airborne lidar data and high-resolution satellite image. In Proceedings of the IGARSS 2020 (IEEE International Geoscience and Remote Sensing Symposium), Waikoloa, HI, USA, 26 September–2 October 2020; pp. 2679–2682. [Google Scholar]
- Liang, J.; Li, P.; Zhao, H.; Han, L.; Qu, M. Forest species classification of UAV hyperspectral image using deep learning. In Proceedings of the 2020 Chinese Automation Congress (CAC), Shanghai, China, 6–8 November 2020; pp. 7126–7130. [Google Scholar]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree species classification of drone hyperspectral and RGB imagery with deep learning convolutional neural networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef]
- Plesoiamu, A.; Stupariu, M.; Sandric, I.; Patru-Stupariu, I.; Dragut, L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Shi, Y.; Ma, D.; Lv, J.; Li, J. ACTL: Asymmetric Convolutional Transfer Learning for Tree Species Identification Based on Deep Neural Network. IEEE Access 2021, 9, 13643–13654. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, K.; Zhu, Y.; Wang, X.; Yun, T. Individual tree crown segmentation directly from UAV-borne LiDAR data using the PointNet of deep learning. Forest 2021, 12, 131. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, Y.; Im, J.; Zhao, Y.; Zhen, Z. A deep-learning-based tree species classification for natural secondary forests using unmanned aerial vehicle hyperspectral images and LiDAR. Ecol. Indic. 2024, 159, 111608. [Google Scholar] [CrossRef]
- Hou, C.; Liu, Z.; Chen, Y.; Wang, S.; Liu, A. Tree Species Classification from Airborne Hyperspectral Images Using Spatial–Spectral Network. Remote Sens. 2023, 15, 5679. [Google Scholar] [CrossRef]
- Michałowska, M.; Rapiński, J.; Janicka, J. Tree species classification on images from airborne mobile mapping using ML.NET. Eur. J. Remote Sens. 2023, 56, 2271651. [Google Scholar] [CrossRef]
- Hou, J.; Zhou, H.; Hu, J.; Yu, H.; Hu, H. A Multi-Scale Convolution and Multi-Layer Fusion Network for Remote Sensing Forest Tree Species Recognition. Remote Sens. 2023, 15, 4732. [Google Scholar] [CrossRef]
- Wang, N.; Pu, T.; Zhang, Y.; Liu, Y.; Zhang, Z. More appropriate DenseNetBL classifier for small sample tree species classification using UAV-based RGB imagery. Heliyon 2023, 9, e20467. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.; Lim, J.; Kim, K.; Yim, K.; Lee, W. Deepening the Accuracy of Tree Species Classification: A Deep Learning-Based Methodology. Forests 2023, 14, 1602. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Lian, Z.; Yang, N. Semi-Supervised Tree Species Classification for Multi-Source Remote Sensing Images Based on a Graph Convolutional Neural Network. Forests 2023, 14, 1211. [Google Scholar] [CrossRef]
- Huang, Y.; Wen, X.; Gao, Y.; Zhang, Y.; Lin, G. Tree Species Classification in UAV Remote Sensing Images Based on Super-Resolution Reconstruction and Deep Learning. Remote Sens. 2023, 15, 2942. [Google Scholar] [CrossRef]
- Chen, X.; Shen, X.; Cao, L. Tree Species Classification in Subtropical Natural Forests Using High-Resolution UAV RGB and SuperView-1 Multispectral Imageries Based on Deep Learning Network Approaches: A Case Study within the Baima Snow Mountain National Nature Reserve, China. Remote Sens. 2023, 15, 2697. [Google Scholar] [CrossRef]
- Lee, E.; Baek, W.; Jung, H. Mapping Tree Species Using CNN from Bi-Seasonal High-Resolution Drone Optic and LiDAR Data. Remote Sens. 2023, 15, 2140. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.; Tao, Y.; Sun, J.; Liu, X.; Yu, P.; Wang, T. DGRec: Graph Neural Network for Recommendation with Diversified Embedding Generation. In Proceedings of the Sixteenth ACM International Conference on Web Search and Data Mining (WSDM ’23), Singapore, 27 February–3 March 2023. [Google Scholar] [CrossRef]
- Cini, A.; Marisca, I.; Bianchi, F.; Alippi, C. Scalable Spatiotemporal Graph Neural Networks. In Proceedings of the Thirty-Seventh AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 20–27 February 2024. [Google Scholar]
- Sun, P.; Yuan, X.; Li, D. Classification of Individual Tree Species Using UAV LiDAR Based on Transformer. Forests 2023, 14, 484. [Google Scholar] [CrossRef]
- Lei, Z.; Li, H.; Zhao, J.; Jing, L.; Tang, Y.; Wang, H.J. Individual Tree Species Classification Based on a Hierarchical Convolutional Neural Network and Multitemporal Google Earth Images. Remote Sens. 2023, 14, 5124. [Google Scholar] [CrossRef]
- Allen, M.; Grieve, S.; Owen, H.; Lines, E. Tree species classification from complex laser scanning data in Mediterranean forests using deep learning. Methods Ecol. Evol. 2023, 14, 1657–1667. [Google Scholar] [CrossRef]
- Li, Y.; Chai, G.; Wang, Y.; Lei, L.; Zhang, X. Ace R-CNN: An attention complementary and edge detection-based instance segmentation algorithm for individual tree species identification using UAV RGB images and LiDAR data. Remote Sens. 2022, 14, 3035. [Google Scholar] [CrossRef]
- Li, M.; Zhou, G.; Li, Z. Fast recognition system for Tree images based on dual-task Gabor convolutional neural network. Multimed. Tools Appl. 2022, 81, 28607–28631. [Google Scholar] [CrossRef]
- Adelabu, S.; Mutanga, O.; Adam, E.; Cho, M.A. Exploiting machine learning algorithms for tree species classification in a semiarid woodland using RapidEye image. J. Appl. Remote Sens. 2013, 17, 073480. [Google Scholar] [CrossRef]
- Rochdi, N.; Yang, X.; Staenz, K.; Patterson, S.; Purdy, B. Mapping Tree Species in a Boreal Forest Area using RapidEye and LiDAR Data. In Proceedings of the Earth Resources and Environmental Remote Sensing 2014 SPIE, Quebec City, QC, Canada, 13–18 July 2014. [Google Scholar] [CrossRef]
- Zhao, D.; Pang, Y.; Liu, L.; Li, Z. Individual Tree Classification Using Airborne LiDAR and Hyperspectral Data in a Natural Mixed Forest of Northeast China. Forests 2020, 11, 303. [Google Scholar] [CrossRef]
- Airlangga, G. Comparative Analysis of Machine Learning Models for Tree Species Classification from UAV LiDAR Data. Bul. Ilm. Sarj. Tek. Elektro 2024, 6, 54–62. [Google Scholar] [CrossRef]
- Seeley, M.; Vaughn, N.; Shanks, B.; Martin, R.; König, M.; Asner, P. Classifying a Highly Polymorphic Tree Species across Landscapes Using Airborne Imaging Spectroscopy. Remote Sens. 2023, 15, 4365. [Google Scholar] [CrossRef]
- Rina, S.; Ying, H.; Shan, Y.; Du, W.; Liu, Y.; Li, R.; Deng, D. Application of Machine Learning to Tree Species Classification Using Active and Passive Remote Sensing: A Case Study of the Duraer Forestry Zone. Remote Sens. 2023, 15, 2596. [Google Scholar] [CrossRef]
- Cha, S.; Lim, J.; Kim, K.; Yim, J.; Lee, W. Uncovering the Potential of Multi-Temporally Integrated Satellite Imagery for Accurate Tree Species Classification. Forests 2023, 14, 746. [Google Scholar] [CrossRef]
- Usman, M.; Ejaz, M.; Nichol, J.; Farid, M.; Abbas, S.; Khan, M. A Comparison of Machine Learning Models for Mapping Tree Species Using WorldView-2 Imagery in the Agroforestry Landscape of West Africa. Int. J. Geo-Inf. 2023, 12, 142. [Google Scholar] [CrossRef]
- Wang, N.; Wang, G. Tree species classification using machine learning algorithms with OHS-2 hyperspectral image. Sci. For. 2023, 51, e3991. [Google Scholar] [CrossRef]
- Kluczek, M.; Zagajewski, M.; Zwijacz-Kozica, T. Mountain Tree Species Mapping Using Sentinel-2, PlanetScope, and Airborne HySpex Hyperspectral Imagery. Remote Sens. 2023, 15, 844. [Google Scholar] [CrossRef]
- Fourier, R.A.; Edwards, G.; Eldridge, N.R. A catalogue of potential spatial discriminators for high spatial resolution digital images of individual crowns. Can. J. Remote Sens. 1995, 3, 285–298. [Google Scholar] [CrossRef]
- Zhang, K.; Hu, B. Individual Urban Tree Species Classification Using Very High Spatial Resolution Airborne Multi-Spectral Imagery Using Longitudinal Profiles. Remote Sens. 2012, 4, 1741–1757. [Google Scholar] [CrossRef]
- Balkenhol, L.; Zhang, K. Identifying Individual Tree Species Structure with High-Resolusion Hyperspectral Imagery Using a Linear Interpretation of the Spectral Signature. In Proceedings of the 38th Canadian Symposium on Remote Sensing, Montreal, QC, Canada, 20–22 June 2017. [Google Scholar]
- Miao, S.; Zhang, K.; Liu, J. An AI-based Tree Species Classification Using a 3D Tree Crown Model Derived From UAV Data. In Proceedings of the 44th Canadian Symposium on Remote Sensing, Yellowknife, NWT, Canada, 19–22 June 2023. [Google Scholar]
- Joche, G. YOLOv5 by Ultralytics, License AGPL-3.0, v7.0. 2020. Available online: https://github.com/ultralytics/yolov5 (accessed on 8 May 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NY, USA, 27–30 June 2016. [Google Scholar]
- Wang, C.; Liao, H. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. arXiv 2024, arXiv:2402.13616. [Google Scholar]
- Hussain, M. YOLO-v1 to YOLO-v8, the Rise of YOLO and Its Complementary Nature toward Digital Manufacturing and Industrial Defect Detection. Machines 2023, 11, 677. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).