Machine Learning Algorithms for the Retrieval of Canopy Chlorophyll Content and Leaf Area Index of Crops Using the PROSAIL-D Model with the Adjusted Average Leaf Angle
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Canopy Reflectance Measurements
2.3. Measurements of the CCC and LAI
2.4. Spectra Simulation Datasets
2.5. Leaf Inclination Angle Optimization Datasets
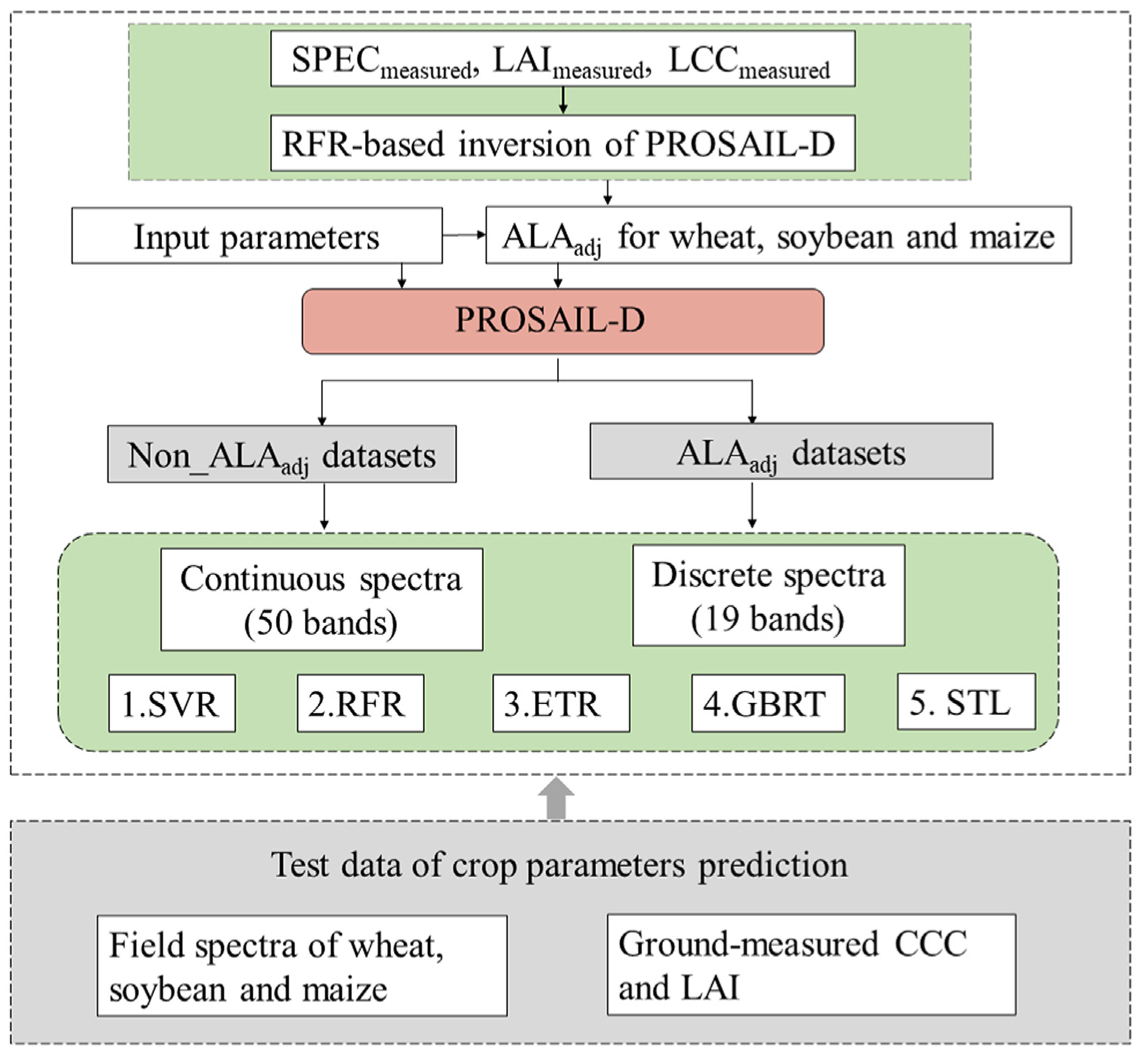
2.6. Modeling and Validating Method
2.6.1. Machine Learning Retrieval Model
2.6.2. Performance Assessment
3. Results
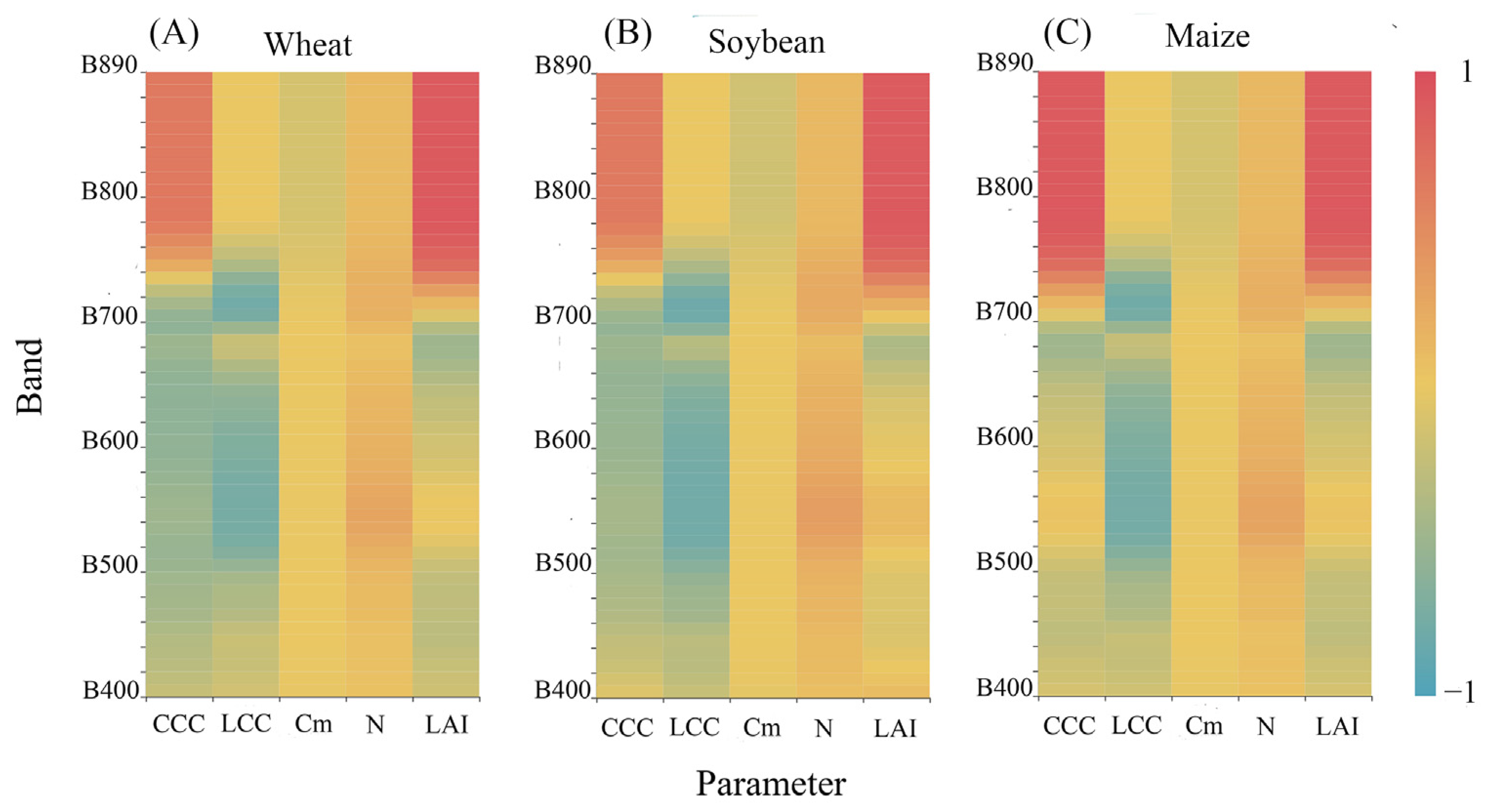
3.1. The Sensitivity of Canopy Reflectance Spectra to Crop Parameters
3.2. ALAadj Parameterization of Wheat, Soybean, and Maize from PROSAIL Models
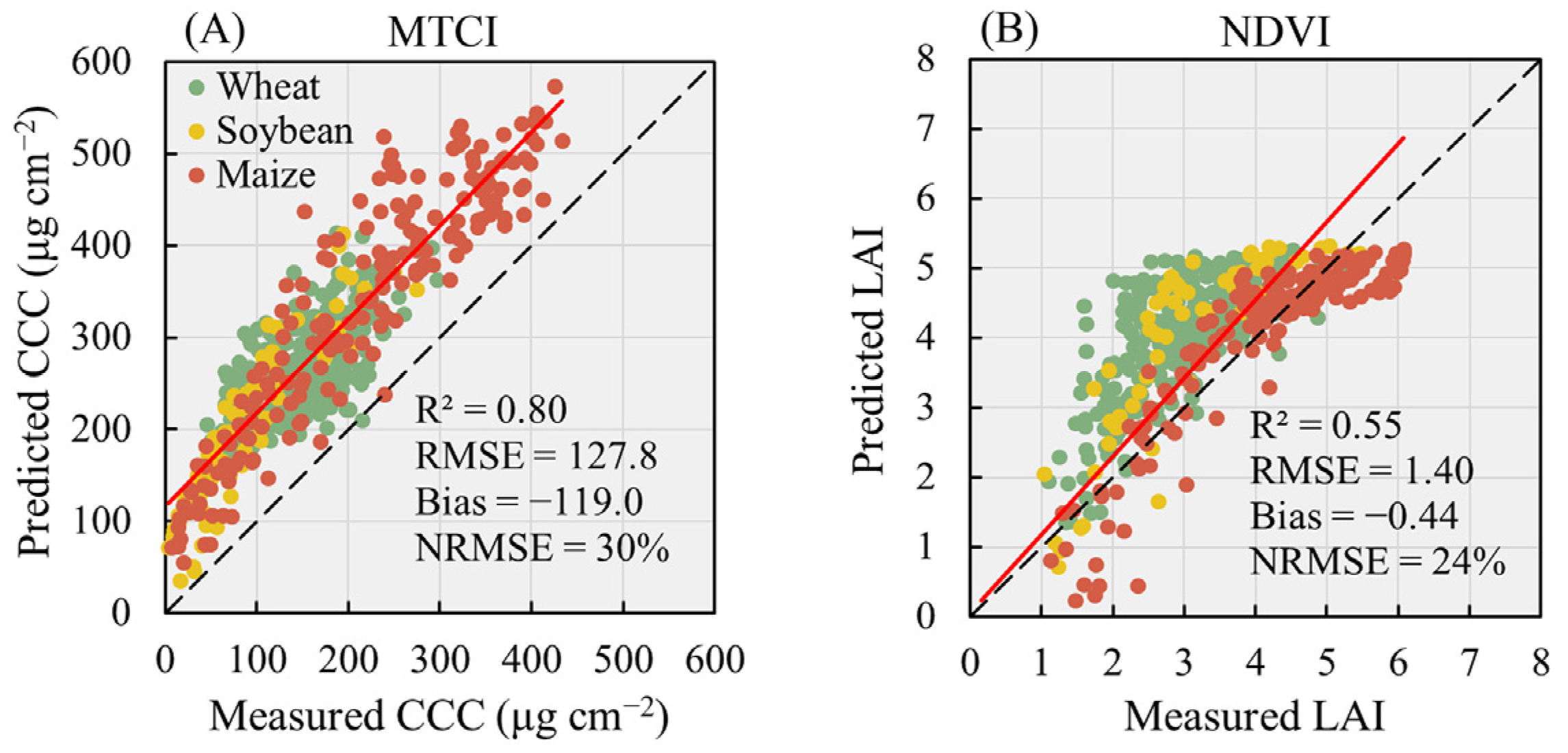
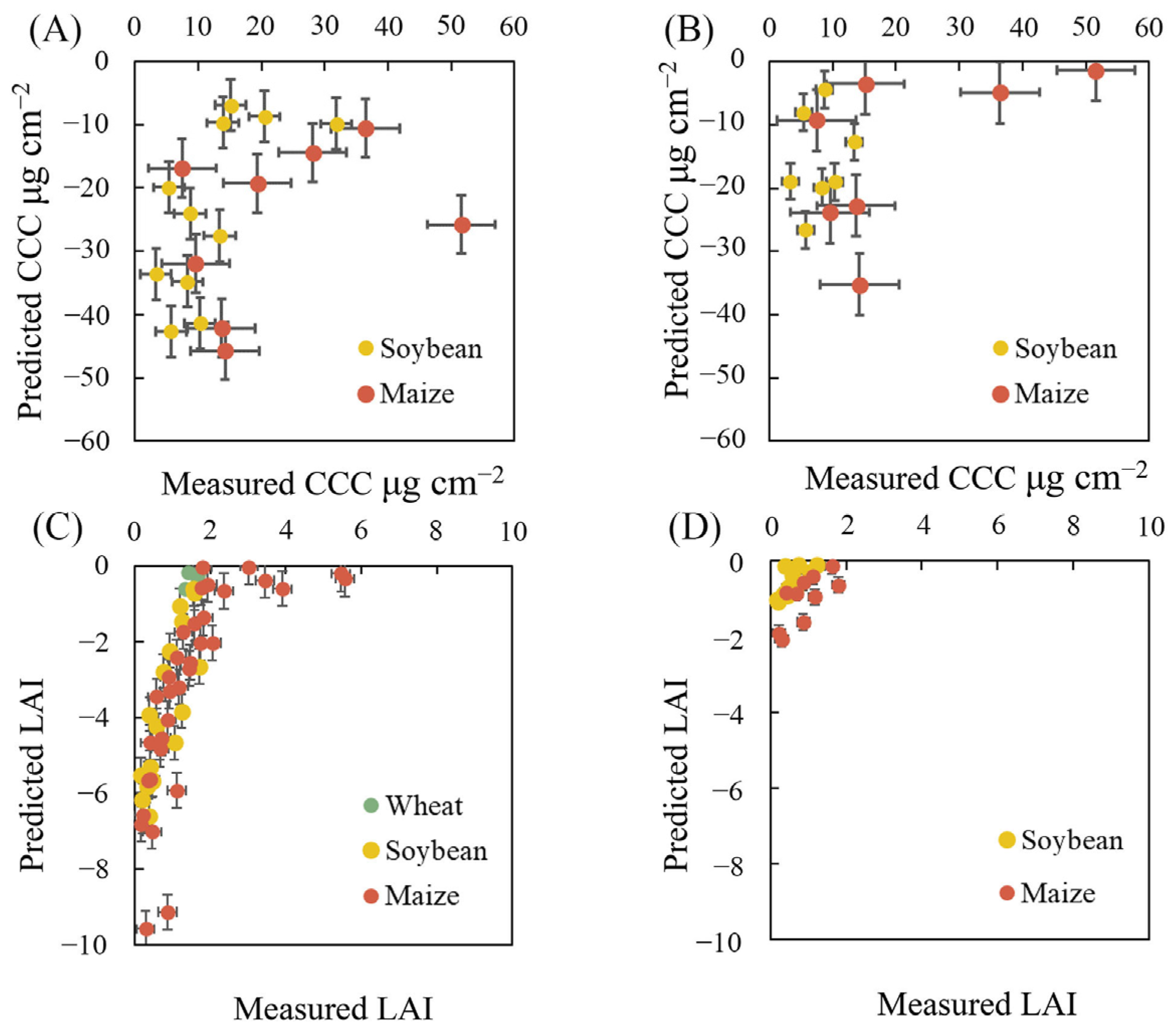
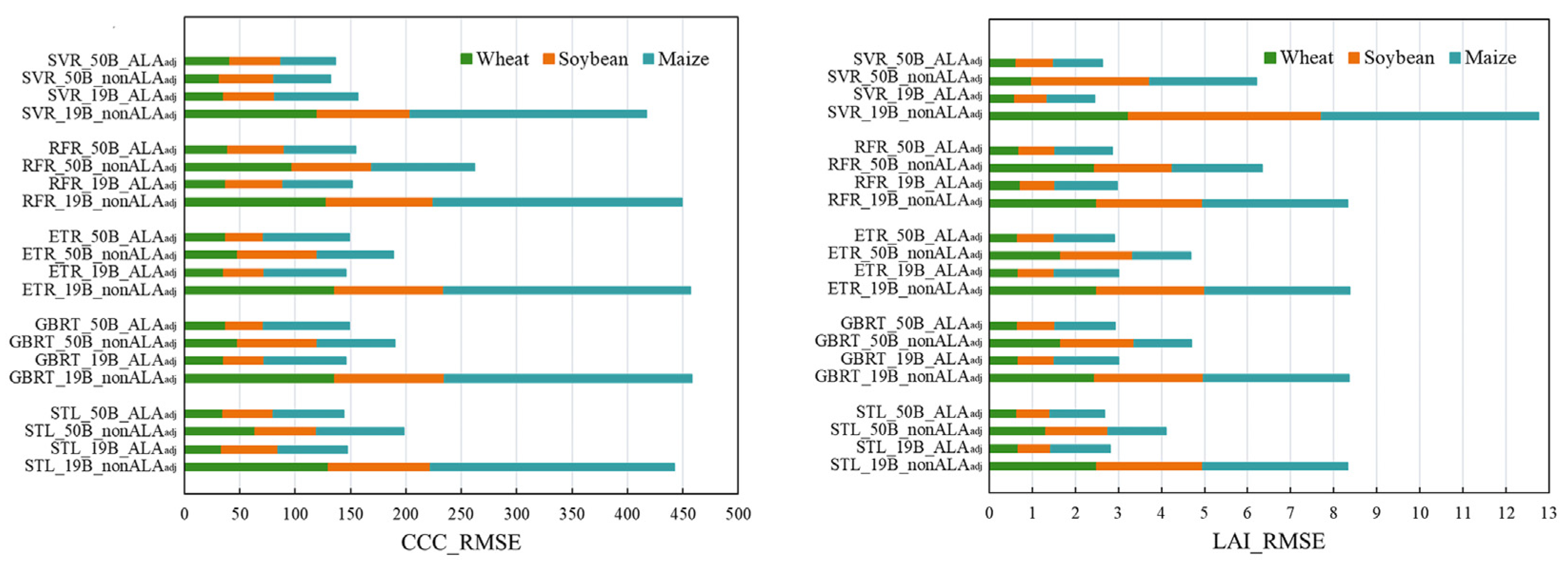
3.3. Performance of Machine Learning Algorithms for CCC Retrieval
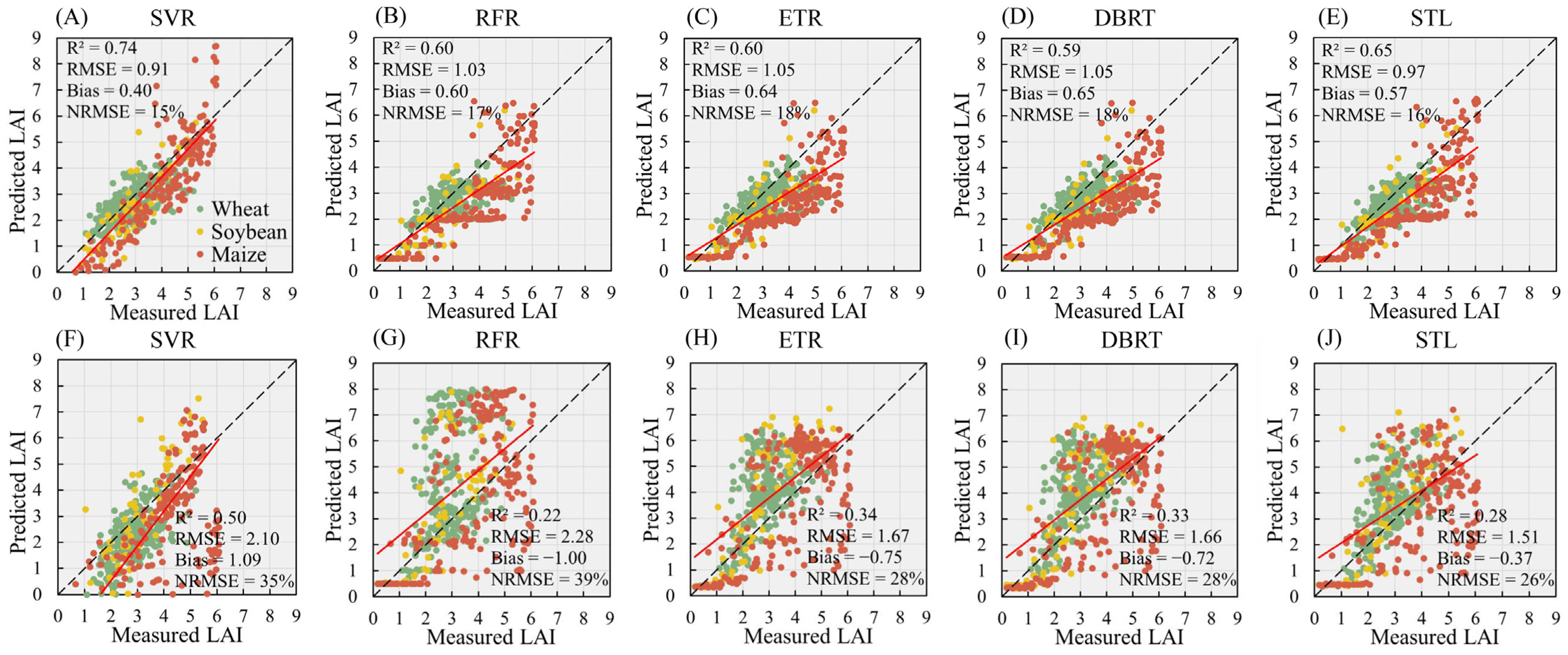
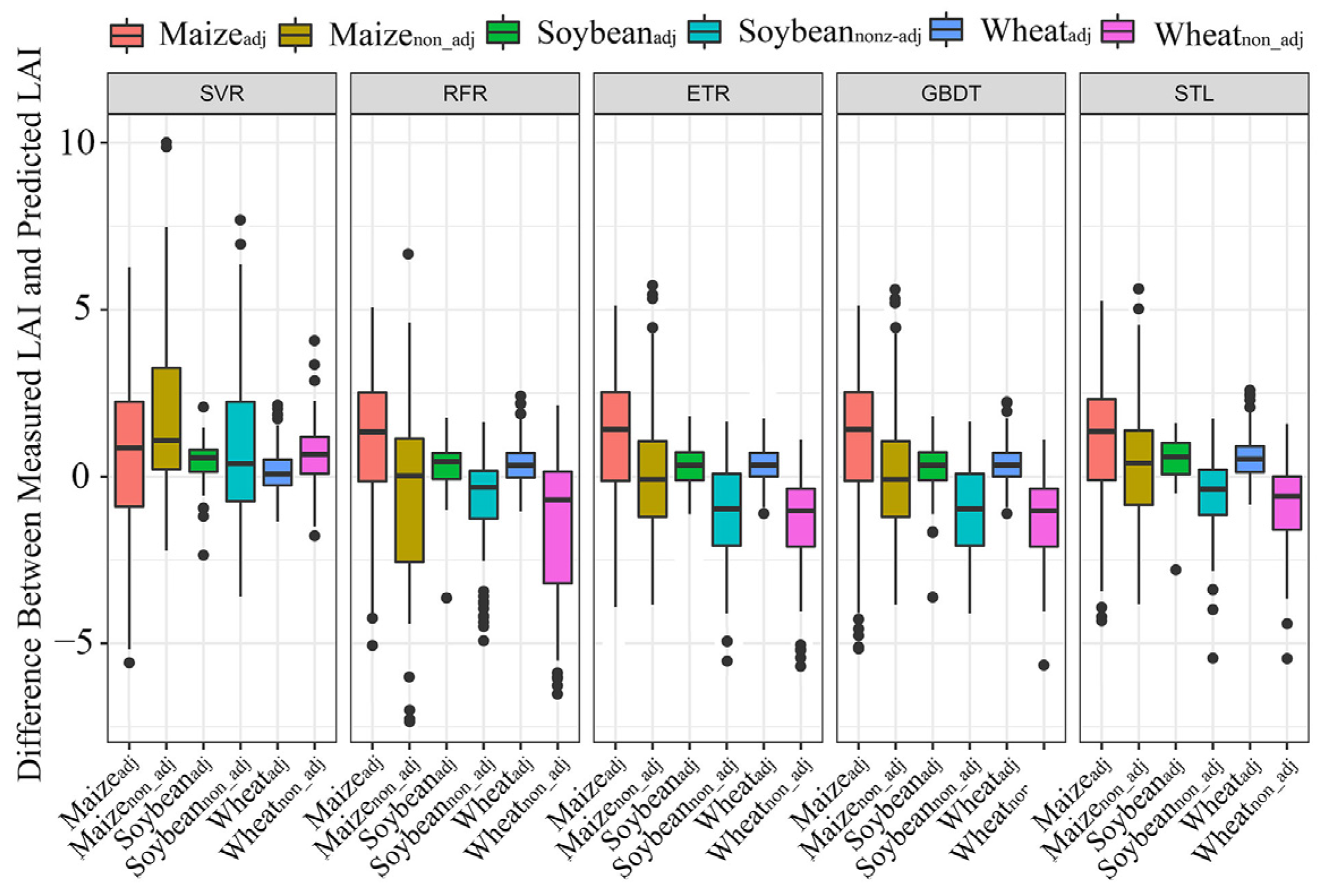
3.4. Performance of Machine Learning Algorithms for LAI Retrieval
4. Discussion
4.1. Application of the ALAadj Parameterization of the PROSAIL Model to Crop CCC and LAI Retrieval
4.2. Performance of the Machine Learning Regression Algorithms for CCC and LAI Retrieval
4.3. Effects of the Uncertainties of the Selected Spectra on the Retrieval Models
4.4. Challenges and Limitations of the Proposed Retrieval Approach
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darvishzadeh, R.; Skidmore, A.; Atzberger, C.; van Wieren, S. Estimation of vegetation LAI from hyperspectral reflectance data: Effects of soil type and plant architecture. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 358–373. [Google Scholar] [CrossRef]
- Qiu, R.; Li, X.; Han, G.; Xiao, J.; Ma, X. Monitoring drought impacts on crop productivity of the U.S. Midwest with solar-induced fluorescence: GOSIF outperforms GOME-2 SIF and MODIS NDVI, EVI, and NIRv. Agric. For. Meteorol. 2022, 323, 109038. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Luo, X.; Bartlett, P.; Chen, B.; Staebler, R.M. Leaf chlorophyll content as a proxy for leaf photosynthetic capacity. Glob. Chang. Biol. 2017, 23, 3513–3524. [Google Scholar] [CrossRef] [PubMed]
- Croft, H.; Chen, J.M.; Zhang, Y. The applicability of empirical vegetation indices for determining leaf chlorophyll content over different leaf and canopy structures. Ecol. Complex. 2014, 17, 119–130. [Google Scholar] [CrossRef]
- Cui, S.; Zhou, K. A comparison of the predictive potential of various vegetation indices for leaf chlorophyll content. Earth Sci. Inform. 2016, 10, 169–181. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C. Inversion of a radiative transfer model for estimating vegetation LAI and chlorophyll in a heterogeneous grassland. Remote Sens. Environ. 2008, 112, 2592–2604. [Google Scholar] [CrossRef]
- Li, D.; Chen, J.M.; Zhang, X.; Yan, Y.; Zhu, J.; Zheng, H.; Zhou, K.; Yao, X.; Tian, Y.; Zhu, Y.; et al. Improved estimation of leaf chlorophyll content of row crops from canopy reflectance spectra through minimizing canopy structural effects and optimizing off-noon observation time. Remote Sens. Environ. 2020, 248, 111985. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. IEEE J. Sel. Top. Appl. Earth Observ. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H., Jr.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. In NASA SP-351 Third ERTS-1 Symposium; Fraden, S.C., Marcanti, E.P., Becker, M.A., Eds.; Scientific and Technical Information Office, National Aeronautics and Space Administration: Washington, DC, USA, 1974; pp. 309–317. [Google Scholar]
- Atzberger, C.; Darvishzadeh, R.; Immitzer, M.; Schlerf, M.; Skidmore, A.; Le Maire, G. Comparative analysis of different retrieval methods for mapping grassland leaf area index using airborne imaging spectroscopy. Int. J. Appl. Earth Obs. 2015, 43, 19–31. [Google Scholar] [CrossRef]
- Atzberger, C.; Guérif, M.; Baret, F.; Werner, W. Comparative analysis of three chemometric techniques for the spectroradiometric assessment of canopy chlorophyll content in winter wheat. Comput. Electron. Agric. 2010, 73, 165–173. [Google Scholar] [CrossRef]
- Jay, S.; Gorretta, N.; Morel, J.; Maupas, F.; Bendoula, R.; Rabatel, G.; Dutartre, D.; Comar, A.; Baret, F. Estimating leaf chlorophyll content in sugar beet canopies using millimeter- to centimeter-scale reflectance imagery. Remote Sens. Environ. 2017, 198, 173–186. [Google Scholar] [CrossRef]
- Goel, N.S. Inversion of canopy reflectance models for estimation of biophysical parameters from reflectance data. In Theory and Applications of Optical Remote Sensing; Asrar, G., Ed.; Wiley: New York, NY, USA, 1989; pp. 205–251. [Google Scholar]
- Baret, F.; Buis, S. Estimating Canopy Characteristics from Remote Sensing Observations: Review of Methods and Associated Problems. In Advances in Land Remote Sensing: System, Modeling, Inversion and Application; Liang, S., Ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 173–201. [Google Scholar]
- Verhoef, W. Light scattering by leaf layers with application to canopy reflflectance modeling: The SAIL model. Remote Sens. Environ. 1984, 16, 125–141. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for pre-diction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Dhakar, R.; Sehgal, V.K.; Chakraborty, D.; Sahoo, R.N.; Mukherjee, J. Field scale wheat LAI retrieval from multispectral Sentinel 2A-MSI and LandSat 8-OLI imagery: Effect of atmospheric correction, image resolutions and inversion techniques. Geocarto Int. 2021, 36, 2044–2064. [Google Scholar] [CrossRef]
- Jiao, Q.; Sun, Q.; Zhang, B.; Huang, W.; Ye, H.; Zhang, Z.; Zhang, X.; Qian, B. A Random forest algorithm for retrieving canopy chlorophyll content of wheat and soybean trained with PROSAIL simulations using adjusted average leaf angle. Remote Sens. 2021, 14, 98. [Google Scholar] [CrossRef]
- Wang, J.; Lopez-Lozano, R.; Weiss, M.; Buis, S.; Li, W.; Liu, S.; Baret, F.; Zhang, J. Crop specific inversion of PROSAIL to retrieve green area index (GAI) from several decametric satellites using a Bayesian framework. Remote Sens. Environ. 2022, 278, 113085. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Morales, A.; Berjon, A.; Aguera, J. Hyperspectral indices and model simuluation for chlorophyll estimation in open-canopy tree crops. Remote Sens. Environ. 2004, 90, 463–476. [Google Scholar] [CrossRef]
- Kimes, D.S.; Nelson, R.F.; Manry, M.T.; Fung, A.K. Attributes of neural networks for extracting continuous vegetation variables from optical and radar measurements. Int. J. Remote Sens. 1998, 19, 2639–2663. [Google Scholar] [CrossRef]
- An, G.; Xing, M.; He, B.; Liao, C.; Huang, X.; Shang, J.; Kang, H. Using machine learning for estimating rice chlorophyll content from in situ hyperspectral data. Remote Sens. 2020, 12, 3104. [Google Scholar] [CrossRef]
- Danner, M.; Berger, K.; Wocher, M.; Mauser, W.; Hank, T. Efficient RTM-based training of machine learning regression algorithms to quantify biophysical & biochemical traits of agricultural crops. ISPRS J. Photogramm. Remote Sens. 2021, 173, 278–296. [Google Scholar] [CrossRef]
- Kuusk, A. Determination of vegetation canopy parameters from optical measurements. Remote Sens. Environ. 1991, 37, 207–218. [Google Scholar] [CrossRef]
- Asrar, G.; Myeni, R.B.; Kanemasu, E.T. Estimation of Plant Canopy Attributes from Spectral Reflectance Measurements. Theory and Application of Optical Remote Sensing; Wiley: Indianapolis, IN, USA, 1989; pp. 252–296. [Google Scholar]
- Kimes, D.S.; Knyazikhin, Y.; Privette, J.L.; Abuelgasim, A.A.; Gao, F. Inversion methods for physically-based models. Remote Sens. Rev. 2000, 18, 381–439. [Google Scholar] [CrossRef]
- Le Maire, G.; François, C.; Soudani, K.; Berveiller, D.; Pontailler, J.Y.; Bréda, N.; Genet, H.; Davi, H.; Dufrêne, E. Calibration and validation of hyperspectral indices for the estimation of broadleaved forest leaf chlorophyll content, leaf mass per area, leaf area index and leaf canopy biomass. Remote Sens. Environ. 2008, 112, 3846–3864. [Google Scholar] [CrossRef]
- Eitel, J.U.H.; Long, D.S.; Gessler, P.E.; Hunt, E.R.; Brown, D.J. Sensitivity of ground-based remote sensing estimates of wheat chlorophyll content to variation in soil reflectance. Soil Sci. Soc. Am. J. 2009, 73, 1715–1723. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Durbha, S.S.; King, R.L.; Younan, N.H. Support vector machines regression for retrieval of leaf area index from multiangle imaging spectroradiometer. Remote Sens. Environ. 2007, 107, 348–361. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Cavallo, D.P.; Cefola, M.; Pace, B.; Logrieco, A.F.; Attolico, G. Contactless and non-destructive chlorophyll content prediction by random forest regression: A case study on fresh-cut rocket leaves. Comput. Electron. Agric. 2017, 140, 303–310. [Google Scholar] [CrossRef]
- Merentitis, A.; Debes, C. Many hands make light work-on ensemble learning techniques for data fusion in remote sensing. IEEE Geosci. Remote Sens. Mag. 2015, 3, 86–99. [Google Scholar] [CrossRef]
- Wei, L.; Yuan, Z.; Zhong, Y.; Yang, L.; Hu, X.; Zhang, Y. An improved gradient boosting regression tree estimation model for soil heavy metal (arsenic) pollution monitoring using hyperspectral remote sensing. Appl. Sci. 2019, 9, 1943. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.; Wang, M.; Li, J. Performance Comparison of Machine learning algorithms for estimating the soil salinity of salt-affected soil using field spectral data. Remote Sens. 2019, 11, 2605. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2000, 29, 1189–1232. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Breiman, L. Stacked regressions. Mach. Learn. 1996, 24, 49–64. [Google Scholar] [CrossRef]
- Schwenker, F. Ensemble methods: Foundations and algorithms. IEEE Comput. Intell. Mag. 2013, 8, 77–79. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, L.; Yu, D.; Feng, K.; Wang, X.; Song, J. Improving leaf area index retrieval using multi-sensor images and stacking learning in subtropical forests of China. Remote Sens. 2021, 14, 148. [Google Scholar] [CrossRef]
- Li, W.; Fu, H.; Yu, L.; Gong, P.; Feng, D.; Li, C.; Clinton, N. Stacked Autoencoder-based deep learning for remote-sensing image classification: A case study of African land-cover mapping. Int. J. Remote Sens. 2016, 37, 5632–5646. [Google Scholar] [CrossRef]
- Long, X.; Li, X.; Lin, H.; Zhang, M. Mapping the vegetation distribution and dynamics of a wetland using adaptive-stacking and Google Earth Engine based on multi-source remote sensing data. Int. J. Appl. Earth Obs. 2021, 102, 102453. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Liang, S.; Li, X.; Liu, J. A stacking ensemble algorithm for improving the biases of forest aboveground biomass estimations from multiple remotely sensed datasets. GIS. Remote Sens. 2022, 59, 234–249. [Google Scholar] [CrossRef]
- Aboneh, T.; Rorissa, A.; Srinivasagan, R. Stacking-Based Ensemble Learning Method for Multi-Spectral Image Classification. Technologies 2022, 10, 17. [Google Scholar] [CrossRef]
- Thorp, K.R.; Wang, G.; West, A.L.; Moran, M.S.; Bronson, K.F.; White, J.W.; Mon, J. Estimating crop biophysical properties from remote sensing data by inverting linked radiative transfer and ecophysiological models. Remote Sens. Environ. 2012, 124, 224–233. [Google Scholar] [CrossRef]
- Zou, X.; Zhu, S.; Mõttus, M. Estimation of canopy structure of field crops using sentinel-2 bands with vegetation indices and machine learning algorithms. Remote Sens. 2022, 14, 2849. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; van der Tol, C.; Magnani, F.; Mohammed, G.; Moreno, J. Global sensitivity analysis of the SCOPE model: What drives simulated canopy-leaving sun-induced fluorescence? Remote Sens. Environ. 2015, 166, 8–21. [Google Scholar] [CrossRef]
- Berger, K.; Atzberger, C.; Danner, M.; D’Urso, G.; Mauser, W.; Vuolo, F.; Hank, T. Evaluation of the PROSAIL model capabilities for future hyperspectral model environments: A review study. Remote Sens. 2018, 10, 85. [Google Scholar] [CrossRef]
- Ma, X.; Wang, T.; Lu, L. A Refined four-stream radiative transfer model for row-planted crops. Remote Sens. 2020, 12, 1290. [Google Scholar] [CrossRef]
- Gao, L.; Darvishzadeh, R.; Somers, B.; Johnson, B.A.; Wang, Y.; Verrelst, J.; Wang, X.; Atzberger, C. Hyperspectral response of agronomic variables to background optical variability: Results of a numerical experiment. Agric. For. Meteorol. 2002, 326, 109178. [Google Scholar] [CrossRef]
- Combal, B.; Baret, F.; Weiss, M.; Trubuil, A.; Mace, D.; Pragnere, A.; Myneni, R.; Knyazikhin, Y.; Wang, L. Retrieval of canopy biophysical variables from bidirectional reflectance: Using prior information to solve the ill-posed inverse problem. Remote Sens. Environ. 2003, 84, 1–15. [Google Scholar] [CrossRef]
- Atzberger, C. Object-based retrieval of biophysical canopy variables using artificial neural nets and radiative transfer models. Remote Sens. Environ. 2004, 93, 53–67. [Google Scholar] [CrossRef]
- Atzberger, C.; Richter, K. Spatially constrained inversion of radiative transfer models for improved LAI mapping from future Sentinel-2 imagery. Remote Sens. Environ. 2012, 120, 208–218. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT+SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Danner, M.; Berger, K.; Wocher, M.; Mauser, W.; Hank, T. Fitted PROSAIL parameterization of leaf inclinations, water content and brown pigment content for winter wheat and maize canopies. Remote Sens. 2019, 11, 1150. [Google Scholar] [CrossRef]
- Cui, B.; Zhao, Q.; Huang, W.; Song, X.; Ye, H.; Zhou, X. A new integrated vegetation index for the estimation of winter wheat leaf chlorophyll content. Remote Sens. 2019, 11, 974. [Google Scholar] [CrossRef]
- Verma, S.B.; Dobermann, A.; Cassman, K.G.; Walters, D.T.; Knops, J.M.; Arkebauer, T.J.; Suyker, A.E.; Burba, G.G.; Amos, B.; Yang, H.; et al. Annual carbon dioxide exchange in irrigated and rainfed maize-based agroecosystems. Agric. For. Meteorol. 2005, 131, 77–96. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Huang, W.; Zhao, C. Detection of leaf and canopy EWT by calculating REWT from reflectance spectra. Int. J. Remote Sens. 2010, 31, 2681–2695. [Google Scholar] [CrossRef]
- Rundquist, D.; Perk, R.; Leavitt, B.; Keydan, G.; Gitelson, A. Collecting spectral data over cropland vegetation using machine-positioning versus hand-positioning of the sensor. Comput. Electron. Agric. 2004, 43, 173–178. [Google Scholar] [CrossRef]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Liu, L.; Huang, W.; Zhao, C.; Lu, Y. Estimation of nitrogen status in middle and bottom layers of winter wheat canopy by using ground-measured canopy reflectance. Commun. Soil Sci. Plant Anal. 2007, 36, 2289–2302. [Google Scholar] [CrossRef]
- Viña, A.; Henebry, G.M.; Gitelson, A.A. Satellite monitoring of vegetation dynamics: Sensitivity enhancement by the Wide Dynamic Range Vegetation Index. Geophys. Res. Lett. 2004, 31, 373–394. [Google Scholar] [CrossRef]
- Féret, J.B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]
- Verhoef, W.; Jia, L.; Xiao, Q.; Su, Z. Unified optical-thermal four-stream radiative transfer theory for homogeneous vegetation canopies. IEEE Trans. Geosci. Remote Sens. 2007, 45, 1808–1822. [Google Scholar] [CrossRef]
- Qian, X.; Liu, L. Retrieving crop leaf chlorophyll content using an improved look-up-table approach by combining multiple canopy structures and soil backgrounds. Remote Sens. 2020, 12, 2139. [Google Scholar] [CrossRef]
- Verhoef, W. Theory of Radiative Transfer Models Applied in Optical Remote Sensing of Vegetation Canopies. Ph.D. Thesis, Wageningen Agricultural University, Wageningen, The Netherlands, 1998. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Sun, J.; Yang, J.; Shi, S.; Chen, B.; Du, L.; Gong, W.; Song, S. Estimating rice leaf nitrogen concentration: Influence of regression algorithms based on passive and active leaf reflectance. Remote Sens. 2017, 9, 951. [Google Scholar] [CrossRef]
- Chen, J.M.; Black, T.A. Foliage area and architecture of plant canopies from sunfleck size distributions. Agric. For. Meteorol. 1992, 60, 249–266. [Google Scholar] [CrossRef]
- Garrigues, S.; Lacaze, R.; Baret, F.; Morisette, J.T.; Weiss, M.; Nickeson, J.E.; Fernandes, R.; Plummer, S.; Shabanov, N.V.; Myneni, R.; et al. Validation and intercomparison of global Leaf Area Index products derived from remote sensing data. J. Geophys. Res. Space Phys. 2008, 113, 02028. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.; Hornero, A.; Beck, P.; Kattenborn, T.; Kempeneers, P.; Hernández-Clemente, R. Chlorophyll content estimation in an open-canopy conifer forest with Sentinel-2A and hyperspectral imagery in the context of forest decline. Remote Sens. Environ. 2019, 223, 320–335. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Wang, R.; Mo, G.; Luo, S.; Luo, X.; He, L.; Gonsamo, A.; Arabian, J.; Zhang, Y.; et al. The global distribution of leaf chlorophyll content. Remote Sens. Environ. 2020, 236, 111479. [Google Scholar] [CrossRef]


| Site Name | Country | Latitude/Longitude (°) | Crop Species | Landcover (ha) | Sampling Periods |
|---|---|---|---|---|---|
| XTS | China | 40.18/116.44 | Wheat | 167 | 4–5/2002; 4–5/2004 |
| US-Ne1 | America | 41.17/−96.48 | Maize | 48.7 | (6–9)/(2001–2005) |
| US-Ne2 | America | 41.165/−96.47 | Soybean and maize | 52.4 | 6–9/2002; 6–9/2004 |
| US-NE3 | America | 41.18/−96.44 | Soybean and maize | 65.4 | 6–9/2002 |
| Parameters | Description | Units | Range | |
|---|---|---|---|---|
| Leaf | N | Leaf structure index | - | 1, 1.5, 2 |
| LCC | Leaf chlorophyll content | μg cm−2 | 10~80; interval, 10 | |
| Cm | Leaf dry matter content | g cm−2 | 0.003, 0.004, 0.005, 0.006 | |
| Cb | Leaf brown pigment content | - | 0 | |
| Cw | Equivalent water thickness | cm | 0.02 | |
| Car | Leaf carotenoid content | μg cm−2 | 25% LCC | |
| CAnt | Leaf anthocyanin content | μg cm−2 | 2 | |
| Canopy | LAI | Leaf area index | m2 m−2 | 0.5, 1, 2, 3, 4, 5, 6, 7, 8 |
| αsoil | Soil reflectance | - | Five soil reflectance types | |
| ALA | Average leaf angle | Degrees | 10–80 degrees | |
| hotS | Hot spot parameter | m m−1 | 0.05 | |
| skyl | Fraction of diffuse incoming solar radiation | - | 0.5 | |
| Observed Geometry | θs | Solar zenith angle | Degrees | 0, 10, 20, 30, 40, 50, 60 |
| θv | View zenith angle | Degrees | 0 | |
| φ | Sun-sensor azimuth angle | Degrees | 0 |
| Crop Type | Using SPEC_LAI | Using SPEC_LAI_LCC | This Study | ||
|---|---|---|---|---|---|
| Mean | StDv | Mean | StDv | ||
| Wheat | 61.8 | 5.2 | 62.0 | 5.2 | 62 |
| Soybean | 45.7 | 9.3 | 45.1 | 9.3 | 45 |
| Maize | 59.8 | 8.1 | 60.3 | 8.1 | 60 |
| ML Algorithm | Crop Type | ALAadj Dadaset | Non-ALAadj Dataset | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | Bias | NRMSE | R2 | RMSE | Bias | NRMSE | ||
| SVR | Wheat | 0.59 | 41.2 | −25.5 | 16% | 0.62 | 31.7 | −6.6 | 13% |
| Soybean | 0.84 | 46.1 | −29.3 | 17% | 0.85 | 49.5 | −28.2 | 18% | |
| Maize | 0.86 | 49.3 | 23.4 | 12% | 0.86 | 50.8 | 27.6 | 12% | |
| RFR | Wheat | 0.54 | 39.4 | −17.4 | 16% | 0.2 | 97.2 | −57.7 | 38% |
| Soybean | 0.84 | 50.8 | −25.8 | 19% | 0.75 | 72.4 | −43.8 | 26% | |
| Maize | 0.78 | 64.3 | 19.5 | 15% | 0.53 | 93.8 | −10.4 | 22% | |
| ETR | Wheat | 0.48 | 37.4 | 7.1 | 15% | 0.37 | 48.0 | −4.7 | 19% |
| Soybean | 0.83 | 34.0 | −9.3 | 13% | 0.85 | 72.2 | −44.8 | 27% | |
| Maize | 0.73 | 77.6 | 43.1 | 18% | 0.68 | 69.7 | 18.2 | 16% | |
| GBRT | Wheat | 0.48 | 37.4 | 7.0 | 14% | 0.37 | 48.0 | −4.7 | 19% |
| Soybean | 0.83 | 34.0 | −9.3 | 13% | 0.86 | 72.2 | −44.8 | 26% | |
| Maize | 0.74 | 77.6 | 43.1 | 19% | 0.68 | 69.7 | 18.2 | 17% | |
| STL | Wheat | 0.58 | 34.8 | −7.7 | 14% | 0.38 | 63.8 | −39.5 | 25% |
| Soybean | 0.84 | 45.7 | −21.7 | 17% | 0.85 | 55.3 | −35.9 | 20% | |
| Maize | 0.80 | 63.4 | 29.4 | 15% | 0.63 | 79.0 | −16.3 | 19% | |
| ML Algorithm | Crop Type | ALAadj Dadaset | Non-ALAadj Dataset | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | Bias | NRMSE | R2 | RMSE | Bias | NRMSE | ||
| SVR | Wheat | 0.46 | 0.62 | 0.19 | 15% | 0.47 | 0.99 | 0.64 | 24% |
| Soybean | 0.87 | 0.87 | 0.53 | 16% | 0.85 | 2.73 | 1.00 | 48% | |
| Maize | 0.88 | 1.14 | 0.84 | 21% | 0.55 | 2.84 | 1.77 | 46% | |
| RFR | Wheat | 0.43 | 0.69 | 0.34 | 17% | 0.13 | 2.45 | −1.49 | 60% |
| Soybean | 0.75 | 0.83 | 0.43 | 16% | 0.54 | 1.80 | −0.84 | 34% | |
| Maize | 0.70 | 1.33 | 1.02 | 25% | 0.29 | 2.18 | −0.36 | 37% | |
| ETR | Wheat | 0.47 | 0.65 | 0.31 | 16% | 0.27 | 1.66 | −1.20 | 41% |
| Soybean | 0.75 | 0.86 | 0.51 | 16% | 0.77 | 1.67 | −1.03 | 31% | |
| Maize | 0.71 | 1.40 | 1.12 | 26% | 0.37 | 1.67 | 0.01 | 28% | |
| GBRT | Wheat | 0.48 | 0.66 | 0.30 | 15% | 0.27 | 1.67 | −1.21 | 41% |
| Soybean | 0.75 | 0.87 | 0.50 | 16% | 0.77 | 1.68 | −1.02 | 32% | |
| Maize | 0.70 | 1.40 | 1.13 | 16% | 0.38 | 1.67 | 0.01 | 28% | |
| STL | Wheat | 0.51 | 0.64 | 0.33 | 16% | 0.27 | 1.31 | −0.79 | 32% |
| Soybean | 0.79 | 0.78 | 0.43 | 15% | 0.68 | 1.45 | −0.68 | 27% | |
| Maize | 0.74 | 1.26 | 0.97 | 23% | 0.28 | 1.78 | 0.36 | 30% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Q.; Jiao, Q.; Chen, X.; Xing, H.; Huang, W.; Zhang, B. Machine Learning Algorithms for the Retrieval of Canopy Chlorophyll Content and Leaf Area Index of Crops Using the PROSAIL-D Model with the Adjusted Average Leaf Angle. Remote Sens. 2023, 15, 2264. https://doi.org/10.3390/rs15092264
Sun Q, Jiao Q, Chen X, Xing H, Huang W, Zhang B. Machine Learning Algorithms for the Retrieval of Canopy Chlorophyll Content and Leaf Area Index of Crops Using the PROSAIL-D Model with the Adjusted Average Leaf Angle. Remote Sensing. 2023; 15(9):2264. https://doi.org/10.3390/rs15092264
Chicago/Turabian StyleSun, Qi, Quanjun Jiao, Xidong Chen, Huimin Xing, Wenjiang Huang, and Bing Zhang. 2023. "Machine Learning Algorithms for the Retrieval of Canopy Chlorophyll Content and Leaf Area Index of Crops Using the PROSAIL-D Model with the Adjusted Average Leaf Angle" Remote Sensing 15, no. 9: 2264. https://doi.org/10.3390/rs15092264
APA StyleSun, Q., Jiao, Q., Chen, X., Xing, H., Huang, W., & Zhang, B. (2023). Machine Learning Algorithms for the Retrieval of Canopy Chlorophyll Content and Leaf Area Index of Crops Using the PROSAIL-D Model with the Adjusted Average Leaf Angle. Remote Sensing, 15(9), 2264. https://doi.org/10.3390/rs15092264












