Abstract
The Oostvaardersplassen nature reserve in the Netherlands is grazed by large herbivores. Due to their increasing numbers, the area became dominated by short grazed grasslands and biodiversity decreased. From 2018, the numbers are controlled to create a diverse landscape. Fine-scale mapping and monitoring of the aboveground biomass is a tool to evaluate management efforts to restore a heterogeneous and biodiverse area. We developed a random forest model that describes the correlation between field-based samples of aboveground biomass and fifteen height-related vegetation metrics that were calculated from high-density point clouds collected with a handheld LiDAR. We found that two height-related metrics (maximum and 75th percentile of all height points) produced the best correlation with an R2 of 0.79 and a root-mean-square error of 0.073 kg/m2. Grassland segments were mapped by applying a segmentation routine on the normalized grassland’s digital surface model. For each grassland segment, the aboveground biomass was mapped using the point cloud and the random forest AGB model. Visual inspection of video recordings of the scanned trajectories and field observations of grassland patterns suggest that drift and stretch effects of the point cloud influence the map. We recommend optimizing data collection using looped trajectories during scanning to avoid point cloud drift and stretch, test horizontal vegetation metrics in the model development and include seasonal influence of the vegetation status. We conclude that handheld LiDAR is a promising technique to retrieve detailed height-related metrics in grasslands that can be used as input for semi-automated spatio-temporal modelling of grassland aboveground biomass for supporting management decisions in nature reserves.
Keywords:
HMLS; aboveground biomass; OBIA; grassland; random forest; segmentation; Oostvaardersplassen 1. Introduction
Aboveground biomass (AGB) is one the most important ecosystem service measures of grasslands [1,2], to evaluate carbon storage, net primary productivity, and biodiversity [3]. Quantification of the AGB in grasslands improves efficient monitoring of biophysical and ecological processes. The Dutch nature reserve Oostvaardersplassen (Figure 1), located in a polder, and reclaimed from the inland fresh water lake IJsselmeer in 1968 [3], initially developed into a diverse wetland characterized by a heterogeneous mosaic of grasslands, reedbeds, shrubs, and open forests. However, after the introduction of large herbivores in the 1980s and 1990s, and due to their increasing numbers, parts of the Oostvaardersplassen transformed into a short grazed, homogeneous grassland-dominated area [3,4], and as a result bird diversity decreased [5]. Recently, the management policy of the ‘Staatsbosbeheer’, the Dutch national forestry department, aims to transform the area into a more diverse landscape with heterogeneous grasslands, by reducing the numbers of large herbivores [5]. Heterogeneous grasslands are an important breeding and foraging habitat for several wetland related bird species that forage on insects, amphibians and small mammals. To support the transformation into heterogeneous grasslands, detailed mapping of the AGB and vegetation structure in wetlands is essential [6] to provide a baseline for future monitoring.
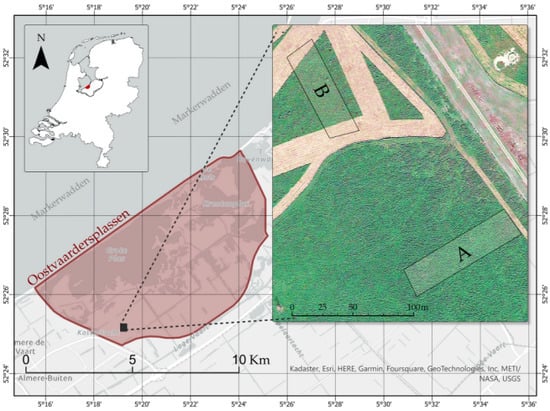
Figure 1.
The two areas A and B in the nature reserve Oostvaardersplassen were selected for handheld LiDAR surveys and vegetation sampling. The aerial backdrop image is from Google Earth. Area B was mowed in the summer of 2021 as a management measure for planting shrubs and trees.
Mapping and monitoring of grassland AGB traditionally rely on ground-based destructive sampling and visual assessment, which is a laborious and costly process [3,7,8,9,10]. An alternative method to map AGB is by using Light Detection and Ranging (LiDAR), a remote sensing technology where the pulses emitted from the sensor can penetrate the vegetation canopy and capture the three-dimensional (3D) physical structure of terrestrial vegetation, detailed topography, and habitats. The vegetation cover, its horizontal variability, height, and vertical variability are four categories that describe how the AGB is distributed in vegetation [10,11]. In many studies, only the height and vertical variability-derived vegetation metrics have been applied in regression models to estimate the AGB [2,8,12,13,14]. These models are based on correlations between AGB values derived from the ground-based, destructive harvested samples and LiDAR-derived structural vegetation metrics. Twenty-six studies [2] explored such regression models for grassland AGB estimation. For instance, Xu et al. [8] compared the simple regression model, stepwise multiple regression, Random Forest (RF) model and artificial neural network model for estimating the AGB of grassland. Stepwise multiple regression produced the highest prediction accuracy (R2 = 0.84, Root Mean Square Error (RMSE) = 48.89 g/m2), followed by RF (R2 = 0.78, RMSE = 68.72 g/m2), simple regression (R2 = 0.80, RMSE = 86.4 g/m2), and artificial neural network (R2 = 0.73, RMSE = 101.40 g/m2) [8]. Li et al. [13] estimated AGB in shrubland and grassland, in which the stepwise multiple regression slightly outperformed the RF model. In our research, we used fifteen input metrics derived from the LiDAR data and only a small sample size. We opted for using the RF model, because it has the advantage that only a relatively small amount of training data is required to support numerous predictors [13].
A mosaic of fine-scaled patches with comparable vegetation height and patches with very low vegetation and bare patches occur in the grasslands of the Oostvaardersplassen. This fine-scaled mosaic developed through variations in grazing intensity, trampling by large grazers along movement routes, or variation in growing conditions. To map how these patterns develop over time, we decided to use handheld LiDAR data and object-based image analysis (OBIA). OBIA has previously been used for monitoring, mapping and managing semi-natural grassland habitats [14]. In addition, handheld LiDAR inventories produce high resolution data, and are easily repeatable to support fine scale monitoring of vegetation structure and AGB across seasons. Moreover, they are relatively cost effective, and operation in the terrain is adjustable to changing terrain conditions, an advantage over terrestrial laser scanning campaigns and multispectral imagery.
We applied OBIA to segment the normalized LiDAR-derived canopy height model into structurally homogeneous objects [10] for which the AGB is estimated per object from the metrics used to develop our RF model.
While trees and arable crops show easily detectable physical structures or uniform growth habits, short grazed grassland does not show such distinct growth features [15]. Therefore, such grassland areas are relatively feature-poor environments and require higher point densities to capture the vegetation structure for AGB predictions [10]. Morais et al. [2] conclude from a review of 26 remote sensing grassland AGB studies, that the accuracy increases with the proximity of the sensor to the ground. A terrestrial laser scanner or Handheld Mobile Laser Scanner (HMLS) produce sufficiently high density point clouds, but the HMLS has a far lower acquisition time, enabling quick area coverage [16] and the freedom to adapt, for example, walking routes to complex surface conditions. Consequently, an HMLS device was selected as a novel LiDAR platform for capturing 3D point cloud data in combination with OBIA to estimate aboveground biomass for grassland patches in the Oostvaardersplassen.
2. Study Area
The research was conducted in the Oostvaardersplassen, a 56 km2 Dutch nature reserve managed by the ‘Staatsbosbeheer’—the State Forestry Service. The Oostvaardersplassen is recognized as a Ramsar site (no. 427) and is a Natura 2000 site (site code NL9802054), because it is a key area for breeding and migratory birds. The reserve is a mosaic of novel forest, grasslands, and wetland ecosystems, as it is located in a polder reclaimed from the freshwater IJsselmeer lake in 1968 [17]. The local vegetation consists now of inundated and non-inundated grasslands with more than 95% grasses and low herbs, and less than 25% tall herbs such as thistle [5]. These grasslands have been under heavy grazing pressure by Konik horses (Equus caballus var. konik), red deer (Cervus elaphus), and Heck cattle (Bos taurus var. heck), since their introduction into the Oostvaardersplassen in 1983 and 1992 [3].
Two grassland areas, A (100 by 25 m) and B (80 by 25 m), were selected for AGB prediction (Figure 1) and mapping vegetation patches. In area A (measured: 18 October 2021) the vegetation is composed predominantly of grasses, reeds, nettle, and thistles. In area B (measured: 24 October 2021) similar vegetation types occur in an exclosure which was periodically open and/or closed for the large herbivores and in which strips of grassland were mowed for planting shrubs and trees (Figure 1). After planting in 2022, the exclosures were closed.
3. Materials and Methods
3.1. General Workflow
Grassland AGB is predicted and mapped in the Oostvaardersplassen following three routines (Figure 2). In the first routine, LiDAR data are collected in areas A and B using the HMLS. The vegetation is harvested in both areas to collect samples for AGB lab analyses. In the second routine, an RF model is constructed to predict the AGB based on the lab analysis and testing 15 selected vegetation metrics derived from the LiDAR data. In the third routine, the point cloud is segmented into coherent objects that form the basis for constructing the final AGB map using the best explaining vegetation metrics from the RF model. These routines will be further detailed, including an estimation of potential errors. The data used in this study (R code, input data, and an ArcGIS Pro project) is available via Figshare; see the Data Availability Statement).
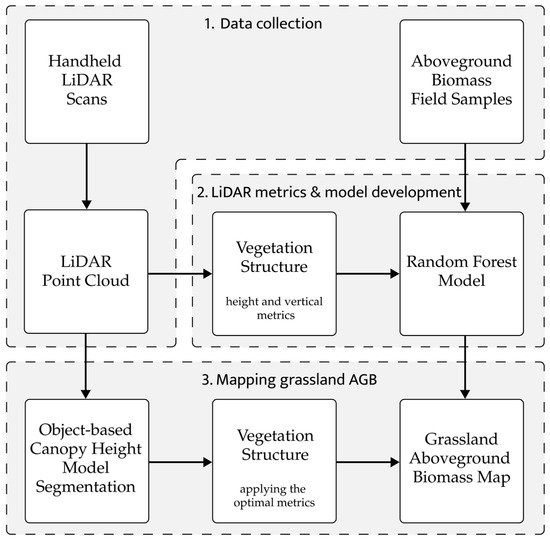
Figure 2.
Workflow with three routines for predicting and mapping grassland AGB in the Oostvaardersplassen. 1. Data collection; 2. LiDAR metrics and model development; and 3. mapping grassland AGB.
3.2. Data Collection
The 3D point cloud data is collected in areas A and B (Figure 1) using a GeoSLAM ZEB-REVO RT Handheld Mobile Laser Scanner [18]. The HMLS potentially records approximately 43,000 points per second, has a relative accuracy of approximately 6 mm, an indoor optimal range of 30 m, and a camera attached during scanning [18]. In feature-poor environments, such as grasslands, point cloud drift, stretch, or discarded areas may occur, because the built-in Simultaneous Localization and Mapping (SLAM) algorithm can experience difficulties in determining forward motion [18]. These difficulties may cause distortions in the scan direction (stretch), or between adjacent scanlines (drift). Discarded areas are areas without data points as the result of incomplete scanning. We therefore kept the scanning range at approximately 10 m to ensure sufficient point densities. We kept the walking speed as constant as possible during scanning, and the scanner oriented in a horizontal position while following the two planned walking trajectories (Figure 3) to reduce potential effects of scanner noise, drift, and stretch [19,20]. Furthermore, we utilized eight AeroPointsTM (Propeller Aero, Surry Hills, NSW, Australia) locators with built-in high precision GPS locators in the field (<10 mm horizontal and vertical accuracy) [21] to define accurate ground control points for georeferencing the generated 3D point cloud. While scanning, the scanner is placed in the center of each AeroPointTM for at least 10 s in order to create exact HMLS coordinates at the AeroPointTM locations. Both coordinates are then used to georeference the point clouds. Once scanned, the raw LiDAR data is preprocessed [22,23] converted into a 3D point cloud, stored in .las format, georeferenced to the RD New coordinate system of the Netherlands, and clipped to the extent of areas A and B (Figure 1) in the CloudCompareTM software, version 2.11.3 [24]. Before the point clouds were ready to be used for analyses, ghost points (3D points resulting from erroneously scanned moving objects) were manually removed in CloudCompareTM. In our study, ghost points were clearly recognized as the HMLS operator’s silhouette, who accidentally produced self-scanned data point returns.
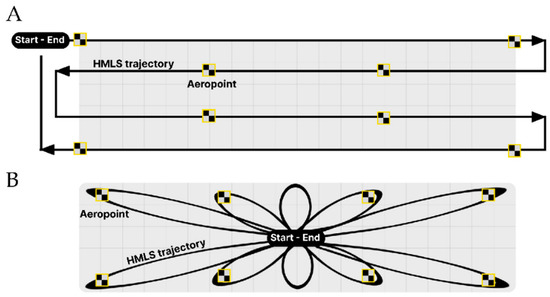
Figure 3.
Two HMLS scan trajectories were tested for point cloud data collection: (A) zigzag trajectory, and (B) looped trajectory. The eight AeroPointsTM (in yellow/black squares) are used to generate fixed GPS points for georeferencing to the Dutch coordinate system (RD New).
In total, 32 vegetation samples were collected and equally divided over areas A and B, according to the design shown in Figure 4A. Two grassland AGB samples were collected 100 cm east and 100 cm west of an AeroPointTM, using a 30 cm circular metal sampler (Figure 4C) and stored in plastic bags. Before harvesting, the maximum vegetation height within the 30 cm diameter sampling locations was measured for error estimation of the maximum height derived from the normalized LiDAR point cloud. Similarly, digital cylindrical samples of the corresponding 3D point cloud were entered in the CloudCompareTM software, which is schematically shown in Figure 4B. All grassland samples were processed using standardized methods (e.g., weighing, oven-drying at 75 °C for 168 h) in the laboratory [25] of the Institute for Biodiversity and Ecosystem Dynamics (IBED) of the University of Amsterdam (UvA) to retrieve the fresh and dry weights of AGB in kg/m2.
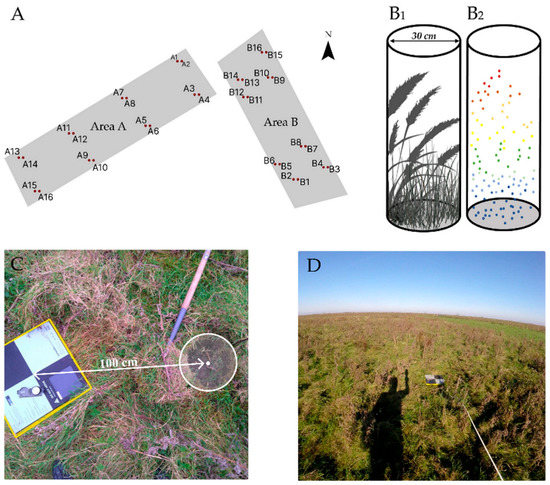
Figure 4.
(A) Locations of the thirty-two grassland samples in areas A and B. The cylinder in (B1) depicts a field-based AGB situation, and (B2) depicts the corresponding LiDAR point cloud. (C) Example of circular vegetation sampling 100 cm west of an AeroPointTM. (D) Video recording example during scan trajectory.
3.3. LiDAR Metrics and Model Development
In the second routine of the workflow (Figure 2), we selected fifteen LiDAR-derived metrics relevant to AGB prediction in grasslands [1,8,10,12,13,14]. In Table 1, nine metrics are categorized according to their height, and six metrics related to vertical variability are listed along with descriptions in terms of their ecological relevance. The height of each LiDAR-point is used to calculate the fifteen metrics. These metrics were calculated for all sampling locations and used to build the RF AGB prediction model. We transformed all sample coordinates east and west of the AeroPointTM GPS locations (Figure 4C), created 30 cm diameter circular polygons, and retrieved the 3D point data within a 30 cm cylinder in ArcGIS Pro [26].

Table 1.
LiDAR-derived metrics and their potential relevance for vegetation structure. Metrics that capture the height and vertical variability-related vegetation structure are calculated from the LiDAR point cloud for all sampling sites. Ecological relevance adapted from Bakx et al. [10].
We rasterized and normalized the classified point cloud by subtracting the digital surface model from the digital terrain model raster to estimate the absolute heights of vegetation in the point cloud. The selected cell size (10 cm) can affect the conservation of information [27], which is especially the case for OBIA-based mapping and monitoring of vegetation patches [28]. Therefore, we tested cell sizes of 1, 5, 10, and 50 cm and suggested that 10 cm cells contain optimal information and detail, which is lost in 50 cm cells, whereas 1 cm cells can contain erroneous heights. In the software R [29], the fifteen metrics were calculated for all the point cloud samples (see Figshare link in the Data Availability Statement).
We developed a Random Forest (RF) regression model [30] in R using the randomForest package to predict grassland AGB by using the field and lab information to determine the best performing LiDAR-derived metrics. Random Forest regression is an ensemble-learning algorithm that can rank and select important variables for biomass prediction. By bootstrapping the samples, it constructs decision trees, each with a randomized subset of predictors [14]. Important hyperparameters in the model that can be tuned are Ntree and Mtry. Ntree selects the amount of decision trees and Mtry determines the number of features that are randomly selected at each node [31]. The RF model was grown using a randomized subset of the 15 LiDAR-derived metrics (Table 1). In the RF model reruns, predictors are one-by-one removed based on the lowest %IncMSE values, until the corresponding R2 and RMSE values stop improving.
3.4. Mapping Grassland AGB
In the third routine (Figure 2), we segmented the rasterized and normalized canopy height model into objects and then applied the RF model to assign AGB values to each grassland object. We decided to use objects instead of pixels for several technical and practical reasons. OBIA is known to successfully handle high resolution data [31], while potential salt-and-pepper effects are avoided. In practice, the OBIA-based results can be evaluated by consulting the video stream to inspect homogeneous vegetation objects and low vegetation, elongated animal movement path in the grasslands, for example. In the eCognitionTM software, version 10.1.1 [32], the normalized canopy height model 10 cm raster was segmented by applying the multiresolution image segmentation algorithm to create objects [33], by using the optimal combination of the scale, shape, and compactness parameter settings, which were guided by the ESP2 tool [34]. The ESP2 tool selects the best fitting scale parameter by increasing the scale parameters with constant increments until the local variance does not further improve. Once optimal parameters have been derived, objects are created, for which the 15 metrics are calculated. The RF model is run to compute the predicted AGB for each object.
3.5. Validation
Many opportunities exist for introducing errors and error propagation, either related to the data collection protocol, the model development, or the mapping routine. We quantified or described the major error sources and how they might influence the results. For example, point cloud drift is dependent on GeoSLAM’s algorithm’s capability to detect fixed features in feature-poor grassland environments. Since this is a black box procedure, the effect of point cloud drift is only visible after initial point cloud preprocessing. The distance between locations in the georeferenced point clouds and the AeroPointTM-derived GPS locations can be expressed as RMS values. We quantified the RF model performance by calculating R2 and RMSE values, but for the segmentation validation, we used visual inspection of the terrain situation using video recordings of the LiDAR scan trajectories. In the discussion, we provide recommendations on how the procedures and technical steps in our three workflow routines can be optimized to reduce potential errors as much as possible.
4. Results
4.1. Data Collection: Zigzag and Looped Trajectory
Field experiences with an HMLS for collecting point cloud data in grasslands are limited and only assessed without a fixed trajectory [35]. Here, we tested both a zigzag and a looped trajectory for scanning areas A and B. In Figure 5, the relative point densities are displayed in relation to the planned scan trajectories. Although drift occurs in all point clouds, the looped trajectory is less affected than the point cloud in the zigzag trajectory. Some parts of the looped trajectory of area A have relatively low point densities, which are unrelated to mowing activity. In general, however, the effect of drift and fewer data points in the raw point cloud is lower in the looped trajectories in both areas, compared to the zigzag trajectory. Despite the lower RMS errors for the zigzag trajectories (Figure 5), we opted to use the zigzag trajectory for area A and the looped trajectory for area B during further AGB model development and mapping.
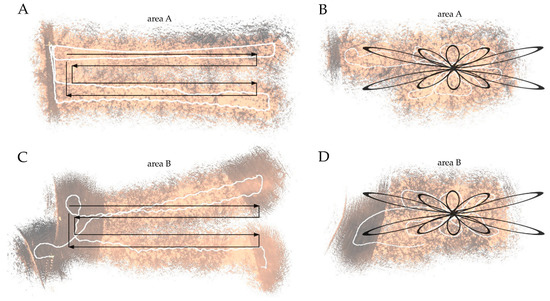
Figure 5.
Results of the zigzag (A,C) and looped (B,D) walking trajectories (in black: planned trajectories, in white: recorded trajectories). The point cloud backdrop images show variation in low to high point densities (grey to colored) and the influence of point cloud drift. The corresponding RMS are 1.58 (A), 1.48 (B), 3.12 (C), 2.89 (D).
Table 2 presents the results of the laboratory analyses of 32 grassland AGB samples. The values range between 0.40 and 1.01 kg/m2 in areas A and B, which suggests that on a fine spatial scale, variation in grassland AGB exists. The maximum vegetation height (Hmax) results from terrain measurements and corresponding point cloud retrieval indicate that mean differences between observation and prediction are 26 cm.

Table 2.
Laboratory results for the AGB in kg/m2. For location of samples see Figure 4.
4.2. LiDAR Metrics and Model Development
After scanning and preprocessing, sampling locations B15 and B16 (Figure 4) appeared to contain too few data points (Figure 5) and were omitted from further analysis. For the 30 remaining locations, the observed versus the predicted AGB is plotted in Figure 6. Including all metrics, and using an Ntree of 500 and a Mtry of two, an R2 of 0.76 was obtained with an RMSE of 0.078 kg/m2 (Table 3). The RF model with four metrics (i.e., Hmax, Hmean, Hmedian, 75th), an Ntree of nine, and a Mtry of four, predicts the AGB with an R2 of 0.78 and a RMSE of 0.075 kg/m2. The highest R2 (0.79) and lowest RMSE (0.073) were obtained with an Ntree of 25, a Mtry of two, and using Hmax and 75th as predictors.
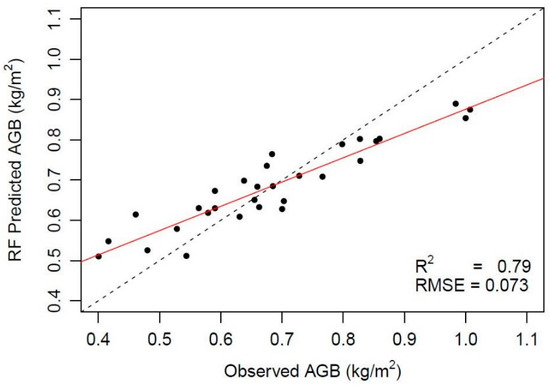
Figure 6.
Relationship between the measured and predicted AGB values.

Table 3.
R2 and RMSE result of various predictors, Ntree, and Mtry combinations.
4.3. Mapping Grassland AGB
We tested various cell sizes (1, 5, 10, and 50 cm) of the digital surface model as input for the segmentation routine. We found that the most realistic cell size was 10 cm, which ensures that sufficient data points are used while keeping most details in the digital surface model. The multiresolution segmentation settings in the eCognitionTM software version 10.1.1 (shape value of 0.1, and a compactness/smoothness value of 0.5) were guided by the ESP2 tool [34], in order to optimize the detection of elongated objects, formed by grazing routes of deer and horses, and of blocky or rounded segments, related to less disturbed (and often higher) vegetation patches. In total, 368 objects were created for area A and 253 for area B (Figure 7); the latter contains a mowed area with little height variation and larger segments, which seem unrelated to variations in the canopy height model. The AGB values were categorized into four RGB classes and assigned to each segment by using the RF model correlation and presented in yellow-to-green legend colors. Area A was characterized by higher mean AGB values (0.74 kg/m2) in comparison to area B (0.69 kg/m2). Field measurements, supported by video observations, also demonstrated that Hmax, and vegetation density in area A were higher than in area B. In area B, distinct NNW to SSW variations in AGB values were mapped (supported by video), as well as an abrupt linear transition in the southern part, which indicates the mowed versus non-mowed area.
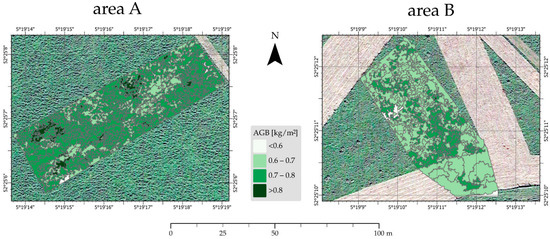
Figure 7.
Segmented areas (A) and (B) with the per object variation of AGB (in kg/m2).
5. Discussion
We created a field-based (semi-)automated workflow for predicting grassland AGB for the Oostvaardersplassen nature reserve in the Netherlands, which consists of three routines: (1) field data collection (handheld laser scanning and vegetation sampling), (2) development of an RF model for AGB prediction, and (3) mapping of the AGB using OBIA. Our results show that AGB mapping provides a relatively fast and accurate inventory of local variation and patterns of AGB in grasslands. However, in the discussion, we consider the steps in the workflow that can be optimized by further work.
5.1. Data Collection
Existing workflows for handheld LiDAR point cloud data collection in relatively featureless and degraded grasslands are not available. We tested a zigzag and a looped walking trajectory (Figure 3) to evaluate the potential effects on scanning accuracy. The looped trajectory, with the base station in the center, produced results with the lowest RMS errors during the georeferencing process. Areas with relatively low point densities were collected in the zigzag trajectory. The looped trajectory facilitates more scan overlap (and thus higher point densities), which promotes more uniformity and quality in the final point clouds, which is in line with the findings of Potter [36] for forested areas. Therefore, we recommend using a predefined looped walking trajectory to optimize the success of the SLAM algorithm, maintain internal positioning [21,37], and place the end and start positions in the center of the scanned area. Terrain obstacles that could hamper walking speed or cause deviation of the predefined route, should be avoided as much as possible by pre-inspecting the desired walking route. From our experience, we recommend that for grassland areas larger than approximately 2500 m2, multiple overlapping scans are preferred, as was also demonstrated in forested areas [37,38]. In featureless grassland areas, scanning of artificial landmarks, such as wooden poles, can increase the internal position and contribute to reduction of drift and stretch in the raw scans.
The vegetation height measurement of Hmax in the field and in the point cloud resulted in an average of 26 cm overestimation of the point cloud vegetation heights and the field observed vegetation heights. This is partly attributed to the ghost point effect of the 3D silhouette of the recording person, which was accidentally scanned and not completely removed in the final 3D point cloud. During field measurements, both the scanning and the measurement of Hmax could potentially be influenced by wind force that affects the position of isolated higher grass plumes, especially when only a few occur in the sampled 30 cm diameter. Acceptable results with slightly larger harvested sample size of 50 × 50 cm quadrats were used by Koma et al. [6] in reedbeds, where Phragmites australis vegetation reached >5 m height. In the degraded grasslands of the Oostvaardersplassen, most AGB is concentrated within approximately 15 cm above terrain level, which is regarded as both seasonal effect and/or the effect of grazing and trampling by the large grazers. Studies in forests report that measurements of tree heights between 5 and 8 m with the ZEB-REVO HMLS contained an average overestimation of 0.65 m [21,39]. Overestimation of woody vegetation has also been reported with the HMLS LiBackpack D50 device in a study by Levick et al. [16]. Overestimation is not necessarily an issue for the RF model development because both training and application are applied to the same point cloud.
5.2. LiDAR Metrics and Model Development
We selected fifteen height and vertical variability-related LiDAR metrics from the literature to develop an RF model for AGB mapping of grassland ecosystems [1,8,12,13,14]. The model performance was optimized by selecting those combinations of metrics that resulted in the lowest error. The highest R2 (0.79) and lowest RMSE (0.073 kg/m2) used only the Hmax and 75th as predictors, which is comparable to the RF model (R2 = 0.78 and RMSE = 0.075 kg/m2) that used four height metrics (Hmax, Hmean, Hmedian and 75th). These results suggest that in (grazed and degraded) grasslands with limited spatial variation in AGB values, height metrics outweigh the vertical variability metrics. Both increasing the sample size and splitting the dataset into a training and a validation set is expected to increase model stability. Morais et al. [2] reviewed 26 studies that used various machine learning methods to predict AGB in grassland-dominated areas, most of them based on satellite imagery as data source (R2 ranging from 0.22–0.94). In three studies, RF models were used in combination with LiDAR data (R2 values of 0.59, 0.61 and 0.79), which is in line with our findings.
Although RF models seem to perform well in grassland dominated areas, other machine learning methods (e.g., stepwise multiple regression, support vector machine, partial least squares regression) could be suitable for remotely sensed grassland AGB estimates as well [2,13], especially if the number of ABG field samples are increased to improve model stability and for upscaling to larger extents. Fine scale LiDAR measurements are sensitive to local environmental disturbances that might cause under- or over-estimation of the vegetation height. In low and very dense vegetation in the first 15 cm, such as the Oostvaardersplassen grasslands, the Hmin depends on the pulse penetration potential [20]. Full waveform LiDAR, in contrast to ALS data, has been reported to better capture ground points [6], but comparisons with HMLS data are unavailable. Still, small irregularities (rabbit holes, ant hills, grazer’s imprints in the clayey subsoil) can lead to lower Hmin values per segment. Furthermore, the wind’s influence might have impact on the Hmax calculation. Scanning should be performed preferably during windless conditions to reduce this type of sensitivity. Our model provides insights into the vegetation structure and biomass conditions in autumn, and can be extended to spring and summer conditions as well, and to other grasslands with similar vegetation structures. We expect that with increasing variability of vegetation cover over time in the Oostvaardersplassen (e.g., a mosaic of grassland, shrubs and trees), other combinations of LiDAR metrics (including horizontal metrics), will contribute to variations in vegetation structure [11] and the prediction of AGB in the RF model. This would require harmonization of sampling design and field sampling protocols and would promote the use of indicators of ecological change [6].
5.3. Mapping Grassland AGB
OBIA has predominantly been used to map land use and land cover using high resolution imagery [31], such as aerial (drone) imagery [15,32], but has also been applied to sensors with coarser resolutions and in combination with pixel-based hybrid approaches [39,40]. OBIA-based studies in grassland-dominated habitats, however, are scarce [41,42]. Our segmentation is based on a LiDAR-derived canopy height model raster and allowed direct comparison between field-measured Hmax and the Hmax and Hmin calculated from the point cloud. There is no temporal mismatch between AGB sampling and collecting LiDAR data. Field inspection and video recordings during the scanning confirmed that patterns of lower vegetation height (and thus lower AGB) were present and likely the result of grazing activity. Despite the SLAM algorithm’s difficulty to detect forward motion in feature-poor environments and potential error propagation, realistic AGB maps were produced for the autumn situation. To support monitoring and mapping of AGB in grasslands throughout the season, our workflow should be tested for different growing phases across the seasons. Another interesting development is to investigate the synergy with other sensor products [43], such as national-wide LiDAR inventories [44] or Sentinel imagery, in order to upscale our workflow to other grasslands.
6. Concluding Remarks
Handheld LiDAR scanning is a promising technique to capture high density point clouds for the rapid retrieval of detailed height-related metrics in grasslands across small spatial extents. In combination with vegetation samples harvested at the same date, our workflow presents a rapid method for mapping patterns and variation in grassland AGB. For the Oostvaardersplassen nature reserve, the two height metrics (Hmax and the 75th percentile of the grassland vertical vegetation height) proved to be optimal metrics for the AGB prediction model. We recommend capturing the 3D point clouds during windless conditions, applying looped scanning trajectories, and placing artificial markers in featureless grasslands, in order to reduce the potential effects of stretch and drift, and to avoid low point density areas. Future research could focus on further optimization of our three workflow routines, especially by increasing the sample size, training and validation data, increasing the mapping extent, collecting data in other seasons, and testing the synergy with other sensors, such as existing nation-wide LiDAR data or Sentinel imagery.
Author Contributions
Conceptualization, A.C.S.; methodology, J.S.d.N. and A.C.S.; software, J.S.d.N.; validation, A.C.S. and J.S.d.N.; formal analysis, J.S.d.N.; investigation, A.C.S. and J.S.d.N.; data curation, J.S.d.N.; writing—original draft preparation, J.S.d.N.; writing—review and editing, A.C.S., K.F.R. and P.C.; visualization, J.S.d.N.; supervision, A.C.S., K.F.R. and P.C.; project administration, A.C.S.; internal funding acquisition, A.C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was internally funded by the Institute for Biodiversity and Ecosystem Dynamics (IBED) of the Universiteit van Amsterdam.
Data Availability Statement
The data presented in this study are available on Figshare (https://doi.org/10.6084/m9.figshare.22251274.v3, accessed on 3 March 2023).
Acknowledgments
We would like to thank the State Forestry department of the Netherlands ‘Staatsbosbeheer’ for their general support and allowing access to the grasslands in the Oostvaardersplassen. Furthermore, we thank the laboratory of IBED for allowing use of their laboratory facilities to analyze the AGB samples. We thank the GIS-Studio of the UvA (www.GIS-Studio.nl, accessed on 10 February 2023) for providing access to software and computational power.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wijesingha, J.; Moeckel, T.; Hensgen, F.; Wachendorf, M. Evaluation of 3D point cloud-based models for the prediction of grassland biomass. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 352–359. [Google Scholar] [CrossRef]
- Morais, T.G.; Teixeira, R.F.; Figueiredo, M.; Domingos, T. The use of machine learning methods to estimate aboveground biomass of grasslands: A review. Ecol. Indic. 2021, 130, 108081. [Google Scholar] [CrossRef]
- Cooper, S.D.; Roy, D.P.; Schaaf, C.B.; Paynter, I. Examination of the potential of terrestrial laser scanning and structure-from-motion photogrammetry for rapid nondestructive field measurement of grass biomass. Remote Sens. 2017, 9, 531. [Google Scholar] [CrossRef]
- Cornelissen, P.; Gresnigt, M.C.; Vermeulen, R.; Bokdam, J.; Smit, R. Transition of a Sambucus nigra L. dominated woody vegetation into grassland by a multi-species herbivore assemblage. J. Nat. Conserv. 2014, 22, 84–92. [Google Scholar] [CrossRef]
- Kuypers, H.E.; Op den Kelder, R.; Cornelissen, P. Jaarrapportage Oostvaardersplassen 2022. In Staatsbosbeheer Report; State Forestry Department of The Netherlands: Amsterdam, The Netherlands, 2023; pp. 1–39. [Google Scholar]
- Koma, Z.; Seijmonsbergen, A.C.; Kissling, W.D. Classifying wetland-related land cover types and habitats using fine-scale lidar metrics derived from country-wide Airborne Laser Scanning. Remote Sens. Ecol. Conserv. 2021, 7, 80–96. [Google Scholar] [CrossRef]
- Wang, D.; Xin, X.; Shao, Q.; Brolly, M.; Zhu, Z.; Chen, J. Modeling aboveground biomass in Hulunber grassland ecosystem by using unmanned aerial vehicle discrete lidar. Sensors 2017, 17, 180. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.T.; Lamb, D.W. A combination of plant NDVI and LiDAR measurements improve the estimation of pasture biomass in tall fescue (Festuca arundinacea var. Fletcher). Remote Sens. 2016, 8, 109. [Google Scholar] [CrossRef]
- Xu, K.; Su, Y.; Liu, J.; Hu, T.; Jin, S.; Ma, Q.; Zhai, Q.; Wang, R.; Zhang, J.; Li, Y.; et al. Estimation of degraded grassland aboveground biomass using machine learning methods from terrestrial laser scanning data. Ecol. Indic. 2020, 108, 105747. [Google Scholar] [CrossRef]
- Bakx, T.R.; Koma, Z.; Seijmonsbergen, A.C.; Kissling, W.D. Use and categorization of light detection and ranging vegetation metrics in avian diversity and species distribution research. Divers. Distrib. 2019, 25, 1045–1059. [Google Scholar] [CrossRef]
- Jansen, V.S.; Kolden, C.A.; Greaves, H.E.; Eitel, J.U.H. Lidar provides novel insights into the effect of pixel size and grazing intensity on measures of spatial heterogeneity in a native bunchgrass ecosystem. Remote Sens. Environ. 2019, 235, 111432. [Google Scholar] [CrossRef]
- Zhang, X.; Bao, Y.; Wang, D.; Xin, X.; Ding, L.; Xu, D.; Hou, L.; Shen, J. Using UAV LiDAR to Extract Vegetation Parameters of Inner Mongolian Grassland. Remote Sens. 2021, 13, 656. [Google Scholar] [CrossRef]
- Li, A.; Dhakal, S.; Glenn, N.F.; Spaete, L.P.; Shinneman, D.J.; Pilliod, D.S.; Arklem, R.S.; McIlroy, S.K. Lidar aboveground vegetation biomass estimates in shrublands: Prediction, uncertainties and application to coarser scales. Remote Sens. 2017, 9, 903. [Google Scholar] [CrossRef]
- Ventura, D.; Napoleone, F.; Cannucci, S.; Alleaume, S.; Valentini, E.; Casoli, E.; Burrascano, S. Integrating low-altitude drone based-imagery and OBIA for mapping and manage semi natural grassland habitats. J. Environ. Manag. 2022, 321, 115723. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Brüninghoff, D.; Hensgen, F.; Wachendorf, M.; Astor, T. Methods for LiDAR-based estimation of extensive grassland biomass. Comput. Electron. Agric. 2019, 156, 693–699. [Google Scholar] [CrossRef]
- Levick, S.R.; Whiteside, T.; Loewensteiner, D.A.; Rudge, M.; Bartolo, R. Leveraging TLS as a calibration and validation tool for MLS and ULS mapping of savanna structure and biomass at landscape-scales. Remote Sens. 2021, 13, 257. [Google Scholar] [CrossRef]
- Feiten en Cijfers van de Oostvaardersplassen. Available online: https://www.staatsbosbeheer.nl/Over-Staatsbosbeheer/Dossiers/oostvaardersplassen-beheer/feiten-en-cijfers (accessed on 16 September 2021).
- GeoSLAM Ltd. Zeb Revo RT User’s Manual v1.0.1; GeoSLAM Ltd.: Nottingham, UK, 2017; 80p. [Google Scholar]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest inventory with terrestrial LiDAR: A comparison of static and hand-held mobile laser scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef]
- Balenović, I.; Liang, X.; Jurjević, L.; Hyyppä, J.; Seletković, A.; Kukko, A. Hand-Held Personal Laser Scanning–Current Status and Perspectives for Forest Inventory Application. Croat. J. For. Eng. J. Theory Appl. For. Eng. 2021, 42, 165–183. [Google Scholar]
- Geometius. Propeller AeroPoints-Technical Specifications. Available online: https://www.geometius.nl/wp-content/uploads/2017/11/Brochures-AeroPoints-1.pdf (accessed on 10 March 2023).
- GeoSLAM Hub, Version 6.1.0; GeoSLAM Ltd.: Nottingham, UK, 2020.
- GeoSLAM Draw, Version 3.5; GeoSLAM Ltd.: Nottingham, UK, 2020.
- CloudCompareTM. 3D Point Cloud and Mesh Processing Software, Version 2.11.3. Available online: https://www.danielgm.net/cc/ (accessed on 21 November 2021).
- Soil and Environmental Chemistry Lab—Institute for Biodiversity and Ecosystem Dynamics. Available online: https://ibed.uva.nl/facilities/physical-and-chemical-lab/soil--environmental-chemistry/soil--environmental-chemistry.html (accessed on 11 February 2022).
- Esri Inc. ArcGIS Pro GIS-Software. Version 2.8.3. 2021. Available online: https://www.esri.com/en-us/arcgis/products/arcgis-pro/overview (accessed on 2 November 2021).
- Hengl, T. Finding the right pixel size. Comput. Geosci. 2006, 32, 1283–1298. [Google Scholar] [CrossRef]
- Blaschke, T.; Johansen, K.; Tiede, D. Object-Based Image Analysis for Vegetation Mapping and Monitoring. In Advances in Environmental Remote Sensing: Sensors, Algorithms, and Applications; Qihao, W., Ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 141–266. ISBN 9781138072916. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing, version 4.0.02; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Trimble Germany GmbH. Trimble Documentation eCognition Developer 10.1.1 User Guide; Trimble Germany GmbH: Munich, Germany, 2022. [Google Scholar]
- Baatz, M.; Schäpe, A. Multiresolution Segmentation: An Optimization Approach for High Quality Multi-Scale Image Segmentation. In Angewandte Geographische Informations-Verarbeitung, XII; Strobl, J., Blaschke, T., Griesbner, G., Eds.; Wichmann Verlag: Karlsruhe, Germany, 2000; pp. 12–23. [Google Scholar]
- Drăguţ, L.; Tiede, D.; Levick, S. ESP: A tool to estimate scale parameter for multiresolution image segmentation of remotely sensed data. Int. J. Geogr. Inf. Sci. 2010, 24, 859–871. [Google Scholar] [CrossRef]
- Obanawa, H.; Yoshitoshi, R.; Watanabe, N.; Sakanoue, S. Portable LiDAR-based method for improvement of grass height measurement accuracy: Comparison with SfM methods. Sensors 2020, 20, 4809. [Google Scholar] [CrossRef]
- Potter, T.L. Mobile Laser Scanning in Forests: Mapping beneath the Canopy. Ph.D. Dissertation, University of Leicester, Leicester, UK, 2019. [Google Scholar]
- Haacke, P. HMLS-ALS Synergy and Accuracy for Complex Forest Stands. Master’s Thesis, University of Amsterdam, Amsterdam, The Netherlands, 2019. [Google Scholar]
- Cabo, C.; Del Pozo, S.; Rodríguez-Gonzálvez, P.; Ordóñez, C.; Gonzalez-Aguilera, D. Comparing terrestrial laser scanning (TLS) and wearable laser scanning (WLS) for individual tree modeling at plot level. Remote Sens. 2018, 10, 540. [Google Scholar] [CrossRef]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Beier, C.M.; Johnson, L. A Comparative Analysis of Pixel-Based and Object-Based Approaches for Forest Above-Ground Biomass Estimation Using Random Forest Model. ISPRS-Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2022, 46, 191–196. [Google Scholar] [CrossRef]
- Aguirre-Gutiérrez, J.; Seijmonsbergen, A.C.; Duivenvoorden, J.F. Optimizing land cover classification accuracy for change detection, a combined pixel-based and object-based approach in a mountainous area in Mexico. Appl. Geogr. 2012, 34, 29–37. [Google Scholar] [CrossRef]
- Hellesen, T.; Matikainen, L. An object-based approach for mapping shrub and tree cover on grassland habitats by use of LiDAR and CIR orthoimages. Remote Sens. 2013, 5, 558–583. [Google Scholar] [CrossRef]
- Zlinszky, A.; Schroiff, A.; Kania, A.; Deák, B.; Mücke, W.; Vári, Á.; Szekely, B.; Pfeifer, N. Categorizing grassland vegetation with full-waveform airborne laser scanning: A feasibility study for detecting Natura 2000 habitat types. Remote Sens. 2014, 6, 8056–8087. [Google Scholar] [CrossRef]
- Bazzo, C.O.G.; Kamali, B.; Hütt, C.; Bareth, G.; Gaiser, T. A Review of Estimation Methods for Aboveground Biomass in Grasslands Using UAV. Remote Sens. 2023, 15, 639. [Google Scholar] [CrossRef]
- Kissling, W.D.; Shi, Y.; Koma, Z.; Meijer, C.; Ku, O.; Nattino, F.; Seijmonsbergen, A.C.; Grootes, M.W. Country-wide data of ecosystem structure from the third Dutch airborne laser scanning survey. Data Brief 2023, 46, 108798. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).