Integrating Active and Passive Remote Sensing Data for Mapping Soil Salinity Using Machine Learning and Feature Selection Approaches in Arid Regions
Abstract
1. Introduction
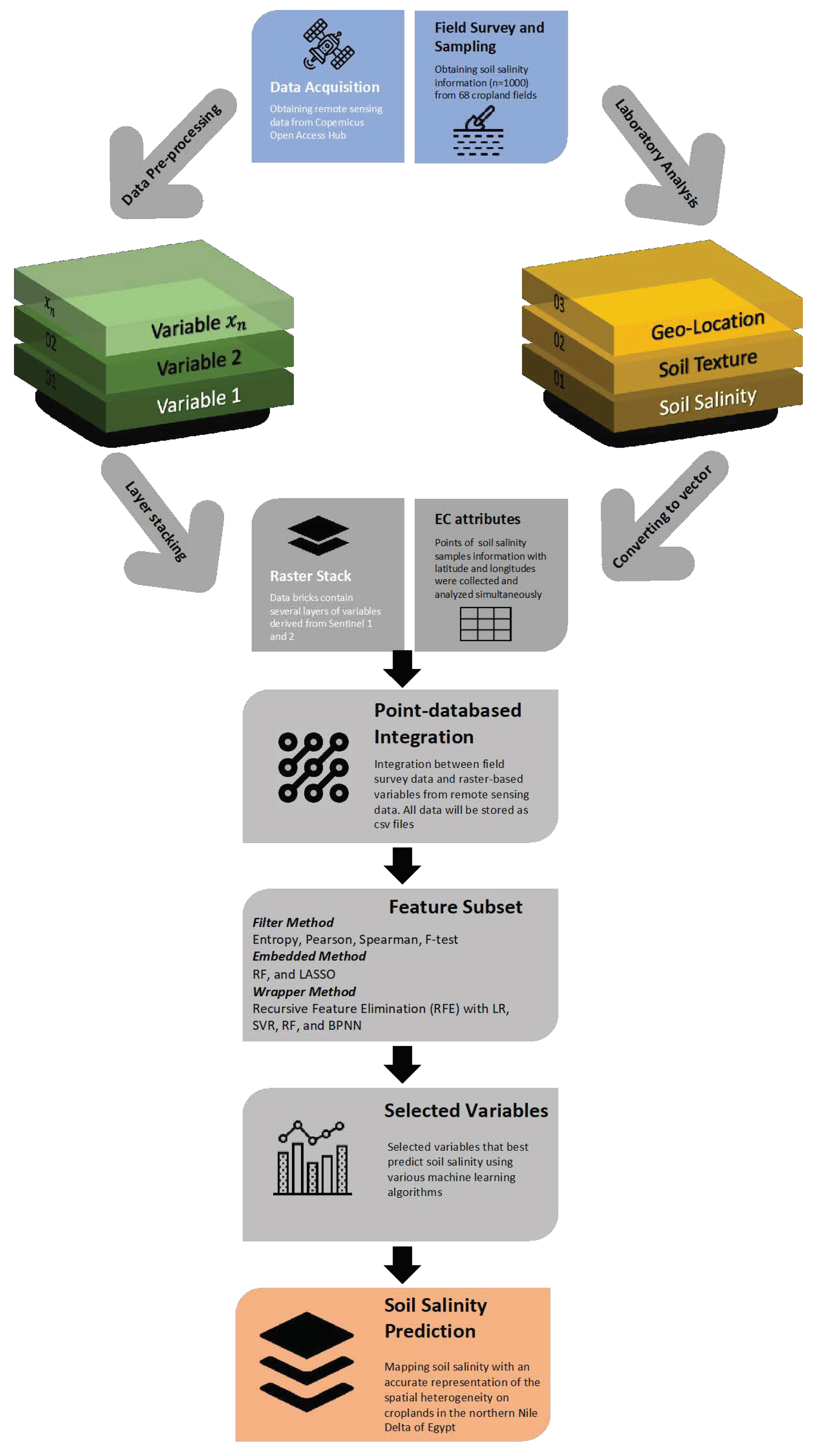
2. Materials and Method
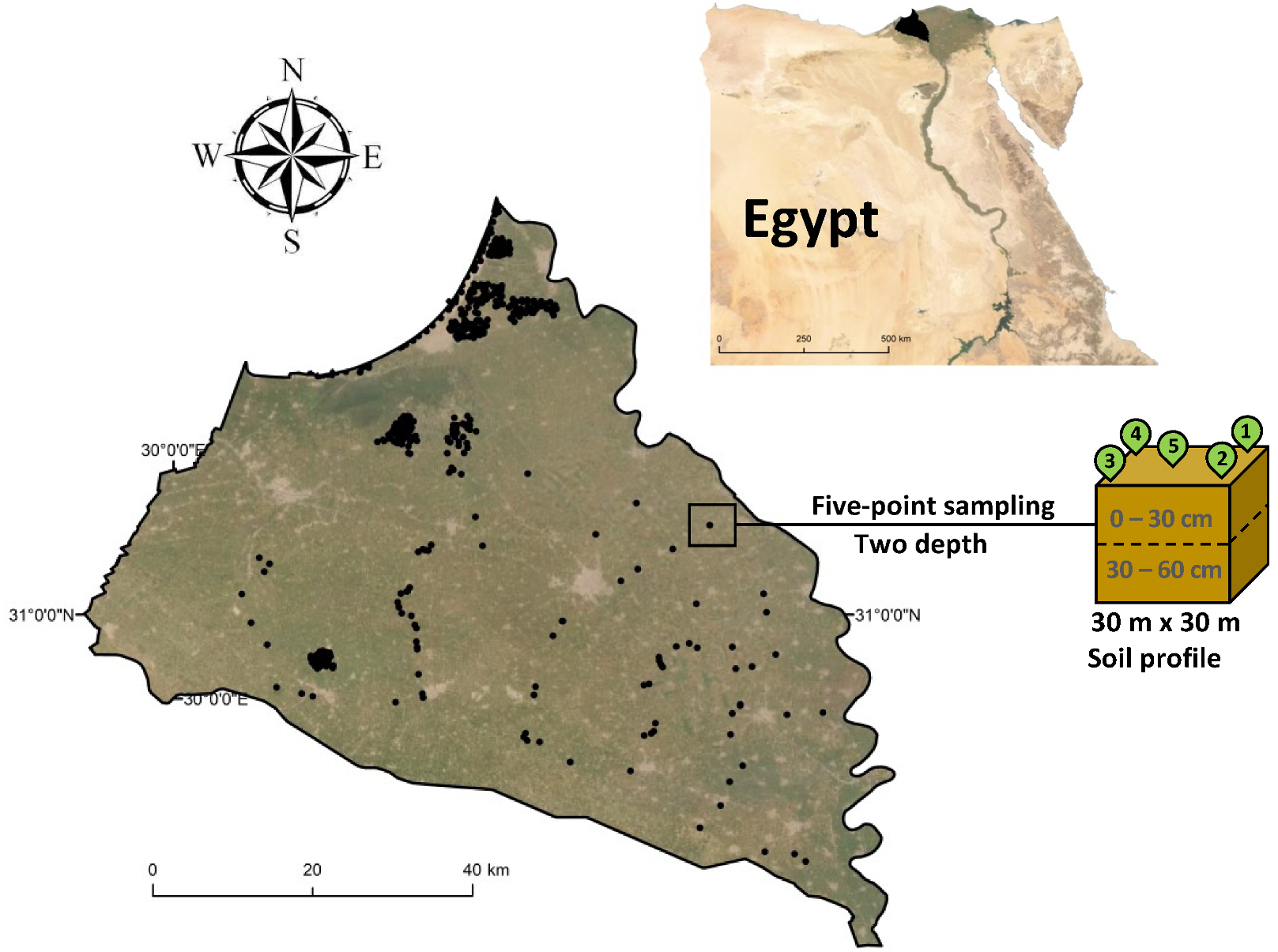
2.1. Study Area Description and Field Survey
2.2. Remote Sensing Data Acquisition and Pre-Processing
2.3. Feature Selection Techniques
2.4. Filter Methods
2.5. Wrapper Methods
2.6. Embedded Methods
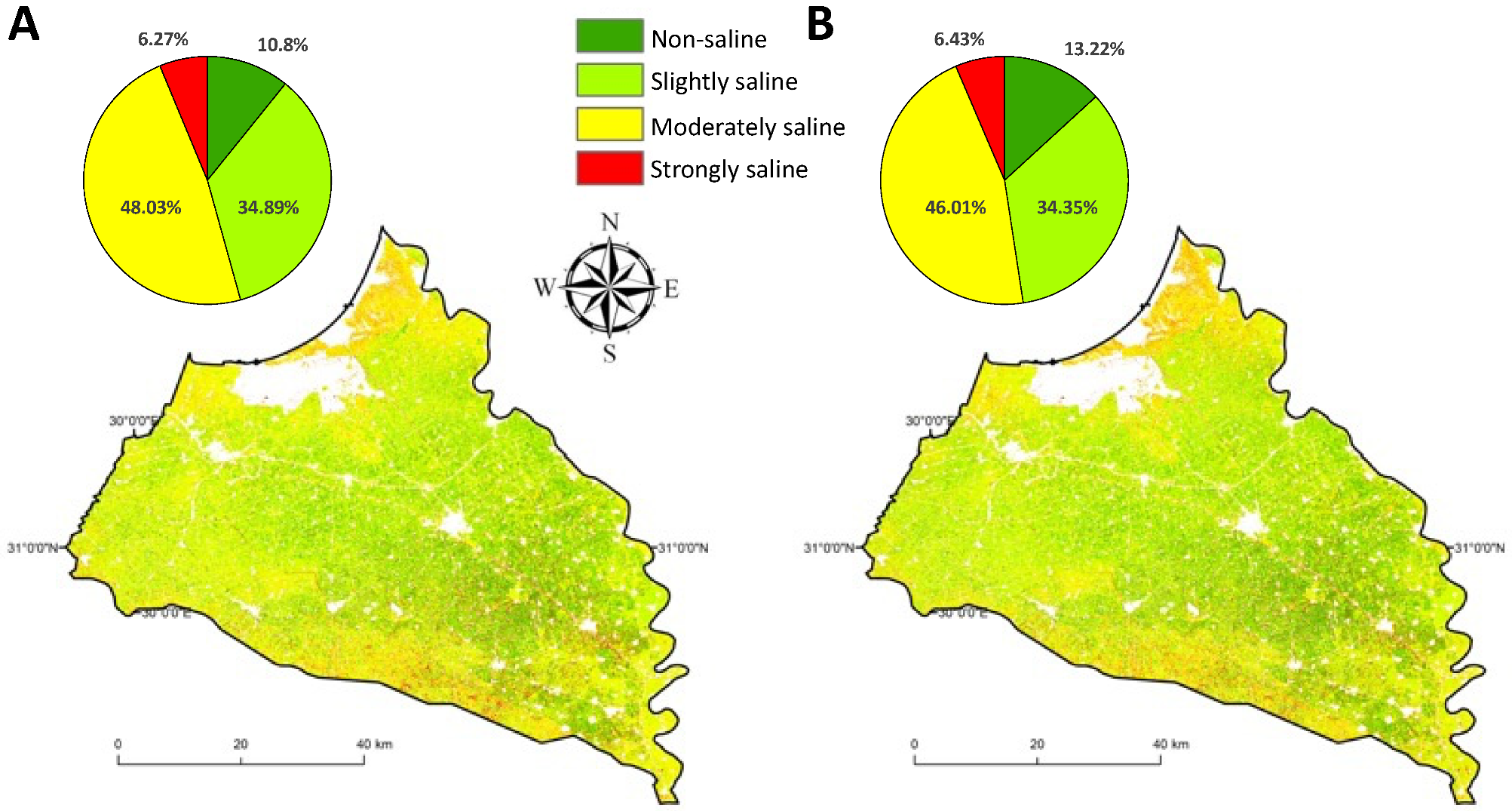
3. Results
3.1. Modeling Assessment
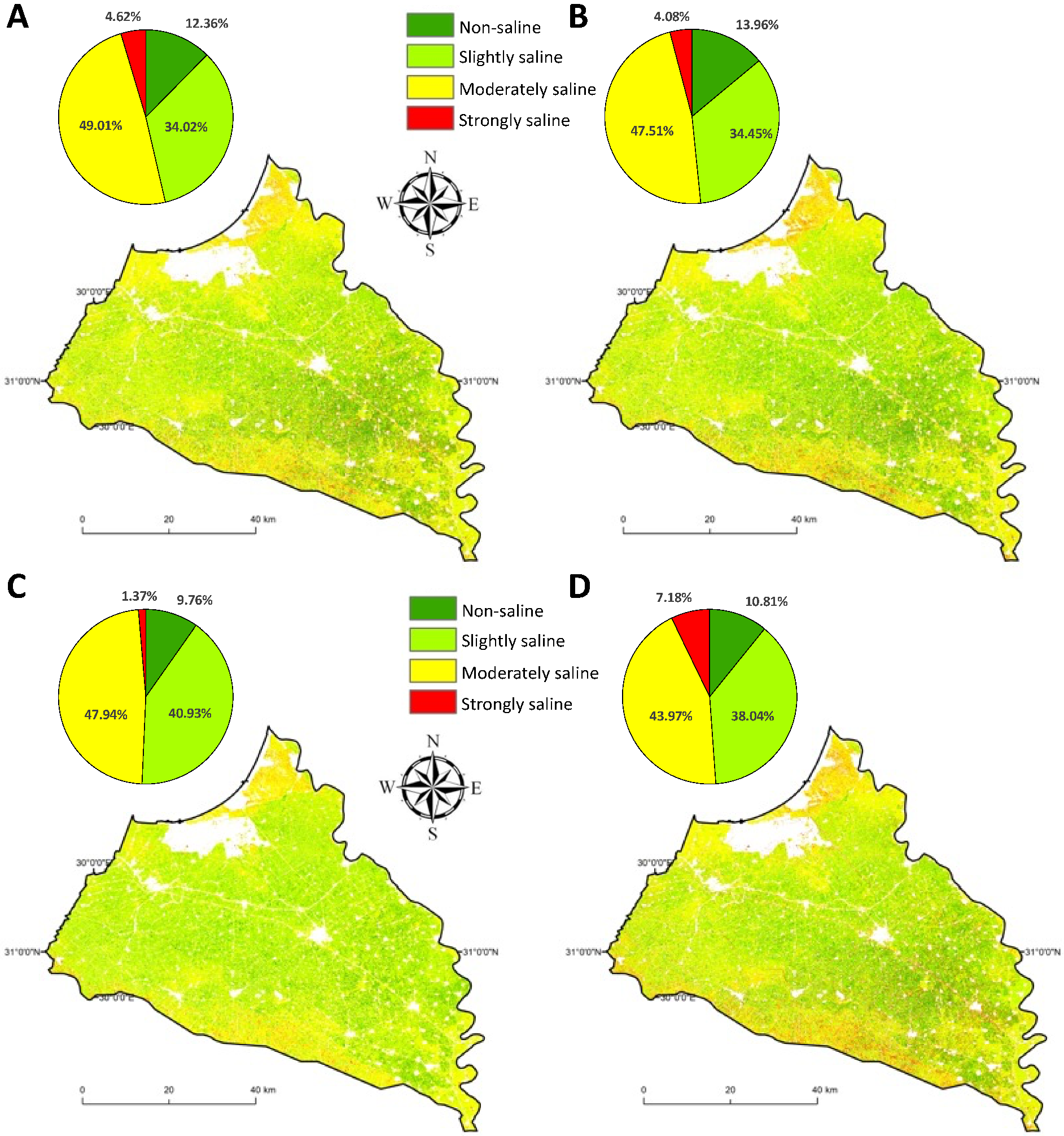
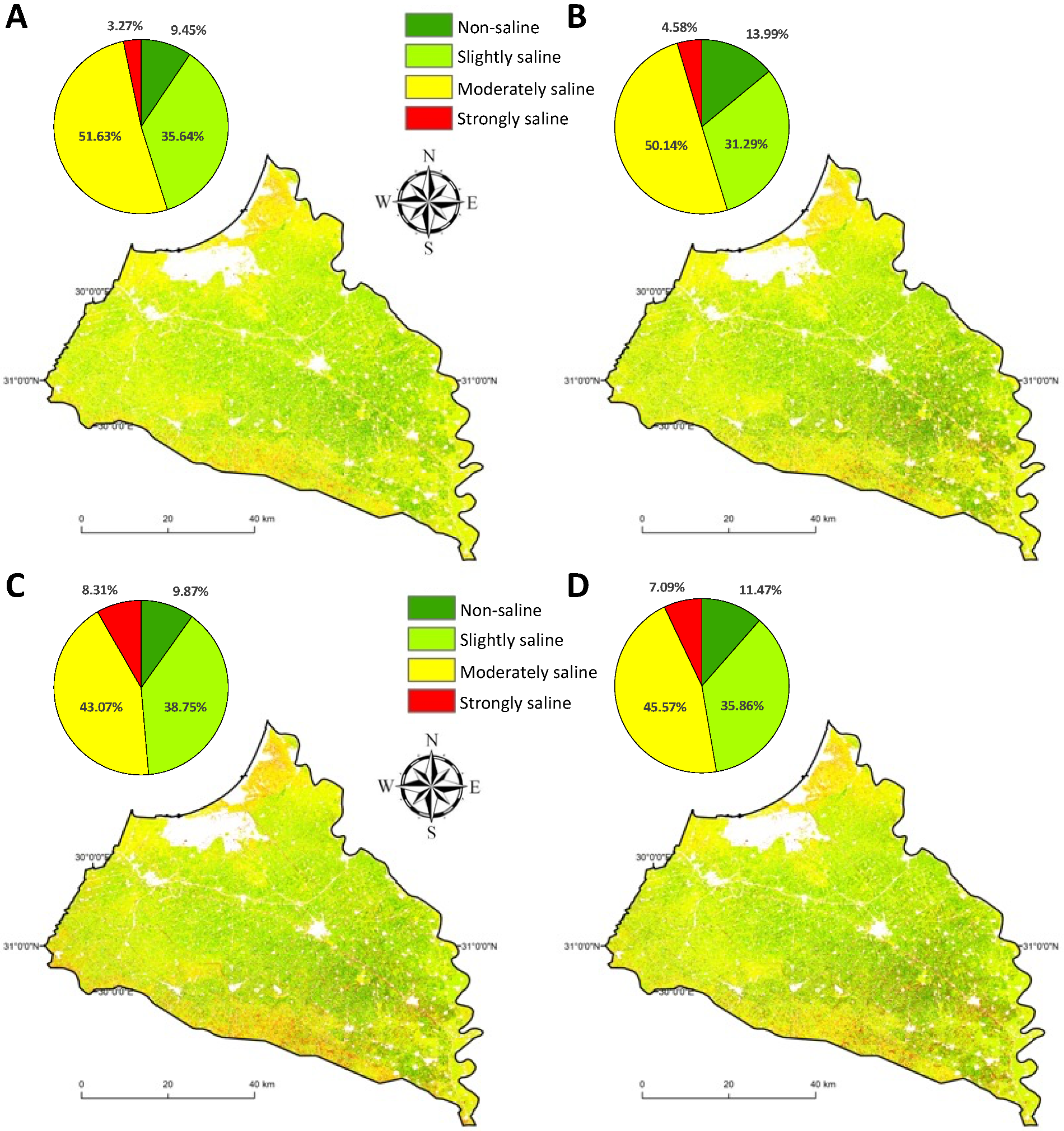
3.2. Feature Selection Approaches for Digital Soil Salinity Mapping
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- AbdelRahman, M.A.E.; Metwaly, M.M.; Afifi, A.A.; D’Antonio, P.; Scopa, A. Assessment of Soil Fertility Status under Soil Degradation Rate Using Geomatics in West Nile Delta. Land 2022, 11, 1256. [Google Scholar] [CrossRef]
- AbdelRahman, M.A.E.; Engel, B.; Eid, M.S.M.; Aboelsoud, H.M. A new index to assess soil sustainability based on temporal changes of soil measurements using geomatics—An example from El-Sharkia, Egypt. All Earth 2022, 34, 147–166. [Google Scholar] [CrossRef]
- AbdelRahman, M.A.E.; Farg, E.; Saleh, A.M.; Sayed, M.; Abutaleb, K.; Arafat, S.M.; Elsharkawy, M.M. Mapping of soils and land-related environmental attributes in modern agriculture systems using geomatics. Sustain. Water Resour. Manag. 2022, 8, 116. [Google Scholar] [CrossRef]
- Aboelsoud, H.M.; AbdelRahman, M.A.E.; Kheir, A.M.S.; Eid, M.S.M.; Ammar, K.A.; Khalifa, T.H.; Scopa, A. Quantitative Estimation of Saline-Soil Amelioration Using Remote-Sensing Indices in Arid Land for Better Management. Land 2022, 11, 1041. [Google Scholar] [CrossRef]
- AbdelRahman, M.A.E.; Afifi, A.A.; D’Antonio, P.; Gabr, S.S.; Scopa, A. Detecting and Mapping Salt-Affected Soil with Arid Integrated Indices in Feature Space Using Multi-Temporal Landsat Imagery. Remote Sens. 2022, 14, 2599. [Google Scholar] [CrossRef]
- Ibrahim, M. Modeling Soil Salinity and Mapping Using Spectral Remote Sensing Data in the Arid and Semi-arid Region. Int. J. Remote Sens. Appl. 2016, 6, 76. [Google Scholar] [CrossRef]
- Mohamed, N.N. Management of salt-affected soils in the Nile Delta. In The Nile Delta; Negm, A., Ed.; The Handbook of Environmental Chemistry; Springer: Cham, Switzerland, 2016; Volume 55, pp. 265–295. [Google Scholar] [CrossRef]
- Hammam, A.; Mohamed, E. Mapping soil salinity in the East Nile Delta using several methodological approaches of salinity assessment. Egypt J. Remote Sens. Space Sci. 2020, 23, 125–131. [Google Scholar] [CrossRef]
- Alqasemi, A.S.; Ibrahim, M.; Al-Quraishi, A.M.F.; Saibi, H.; Al-Fugara, A.; Kaplan, G. Detection and modeling of soil salinity variations in arid lands using remote sensing data. Open Geosci. 2021, 13, 443–453. [Google Scholar] [CrossRef]
- Ma, G.; Ding, J.; Han, L.; Zhang, Z.; Ran, S. Digital mapping of soil salinization based on Sentinel-1 and Sentinel-2 data combined with machine learning algorithms. Reg. Sustain. 2021, 2, 177–188. [Google Scholar] [CrossRef]
- Mulder, V.; de Bruin, S.; Schaepman, M.; Mayr, T. The use of remote sensing in soil and terrain mapping—A review. Geoderma 2011, 162, 1–19. [Google Scholar] [CrossRef]
- Allbed, A.; Kumar, L.; Aldakheel, Y.Y. Assessing soil salinity using soil salinity and vegetation indices derived from IKONOS high-spatial resolution imageries: Applications in a date palm dominated region. Geoderma 2014, 230–231, 1–8. [Google Scholar] [CrossRef]
- Nguyen, T.G.; Tran, N.A.; Vu, P.L.; Nguyen, Q.-H.; Nguyen, H.D.; Bui, Q.-T. Salinity intrusion prediction using remote sensing and machine learning in data-limited regions: A case study in Vietnam’s Mekong Delta. Geoderma Reg. 2021, 27, e00424. [Google Scholar] [CrossRef]
- Elnaggar, A.A.; Noller, J.S. Application of Remote-sensing Data and Decision-Tree Analysis to Mapping Salt-Affected Soils over Large Areas. Remote Sens. 2009, 2, 151–165. [Google Scholar] [CrossRef]
- Davis, E.; Wang, C.; Dow, K. Comparing Sentinel-2 MSI and Landsat 8 OLI in soil salinity detection: A case study of agricultural lands in coastal North Carolina. Int. J. Remote Sens. 2019, 40, 6134–6153. [Google Scholar] [CrossRef]
- Delavar, M.A.; Naderi, A.; Ghorbani, Y.; Mehrpouyan, A.; Bakhshi, A. Soil salinity mapping by remote sensing south of Urmia Lake, Iran. Geoderma Reg. 2020, 22, e00317. [Google Scholar] [CrossRef]
- Erkin, N.; Zhu, L.; Gu, H.; Tusiyiti, A. Method for predicting soil salinity concentrations in croplands based on machine learning and remote sensing techniques. J. Appl. Remote Sens. 2019, 13, 034520. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, F.; Ma, F.; Chai, X.; Zhu, Z.; Shi, J.; Zhang, S. Spatiotemporal changes in soil salinity in a drip-irrigated field. Geoderma 2009, 149, 243–248. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Žižala, D.; Saberioon, M.; Borůvka, L. Soil organic carbon and texture retrieving and mapping using proximal, airborne and Sentinel-2 spectral imaging. Remote Sens. Environ. 2018, 218, 89–103. [Google Scholar] [CrossRef]
- Hengl, T.; De Jesus, J.M.; Heuvelink, G.B.M.; Gonzalez, M.R.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; de Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- Zhang, Z.; Ding, J.; Zhu, C.; Wang, J.; Ma, G.; Ge, X.; Li, Z.; Han, L. Strategies for the efficient estimation of soil organic matter in salt-affected soils through Vis-NIR spectroscopy: Optimal band combination algorithm and spectral degradation. Geoderma 2020, 382, 114729. [Google Scholar] [CrossRef]
- Kalambukattu, J.G.; Kumar, S.; Raj, R.A. Digital soil mapping in a Himalayan watershed using remote sensing and terrain parameters employing artificial neural network model. Environ. Earth Sci. 2018, 77, 203. [Google Scholar] [CrossRef]
- Zhou, T.; Geng, Y.; Chen, J.; Pan, J.; Haase, D.; Lausch, A. High-resolution digital mapping of soil organic carbon and soil total nitrogen using DEM derivatives, Sentinel-1 and Sentinel-2 data based on machine learning algorithms. Sci. Total Environ. 2020, 729, 138244. [Google Scholar] [CrossRef]
- Roelofsen, H.D.; van Bodegom, P.M.; Kooistra, L.; van Amerongen, J.J.; Witte, J.-P.M. An evaluation of remote sensing derived soil pH and average spring groundwater table for ecological assessments. Int. J. Appl. Earth Obs. Geoinform. 2015, 43, 149–159. [Google Scholar] [CrossRef]
- Dong, W.; Wu, T.; Luo, J.; Sun, Y.; Xia, L. Land parcel-based digital soil mapping of soil nutrient properties in an alluvial-diluvia plain agricultural area in China. Geoderma 2019, 340, 234–248. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, Y.; Wang, X.; Liu, X.; Zhang, X.; Zhang, S. Comparing interpolation methods to predict soil total phosphorus in the Mollisol area of Northeast China. Catena 2018, 174, 59–72. [Google Scholar] [CrossRef]
- Elhag, M. Evaluation of Different Soil Salinity Mapping Using Remote Sensing Techniques in Arid Ecosystems, Saudi Arabia. J. Sens. 2016, 2016, 7596175. [Google Scholar] [CrossRef]
- Bian, L.; Wang, J.; Liu, J.; Han, B. Spatiotemporal Changes of Soil Salinization in the Yellow River Delta of China from 2015 to 2019. Sustainability 2021, 13, 822. [Google Scholar] [CrossRef]
- Wu, W.; Mhaimeed, A.S.; Al-Shafie, W.M.; Ziadat, F.; Dhehibi, B.; Nangia, V.; De Pauw, E. Mapping soil salinity changes using remote sensing in Central Iraq. Geoderma Reg. 2014, 2–3, 21–31. [Google Scholar] [CrossRef]
- Arnous, M.O.; El-Rayes, A.; Green, D.R. Hydrosalinity and environmental land degradation assessment of the East Nile Delta region, Egypt. J. Coast. Conserv. 2015, 19, 491–513. [Google Scholar] [CrossRef]
- Elhaddad, A.; Garcia, L. Detecting soil salinity levels in agricultural lands using satellite imagery. In Proceedings of the Ameri-can Society for Photogrammetry and Remote Sensing Annual Conference, Reno, NV, USA, 1–5 May 2006. [Google Scholar]
- Setia, R.; Lewis, M.; Marschner, P.; Segaran, R.R.; Summers, D.; Chittleborough, D. Severity of salinity accurately detected and classified on a paddock scale with high resolution multispectral satellite imagery. Land Degrad. Dev. 2013, 24, 375–384. [Google Scholar] [CrossRef]
- Chen, L.; Xing, M.; He, B.; Wang, J.; Shang, J.; Huang, X.; Xu, M. Estimating Soil Moisture Over Winter Wheat Fields During Growing Season Using Machine-Learning Methods. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 3706–3718. [Google Scholar] [CrossRef]
- Wang, J.; Peng, J.; Li, H.; Yin, C.; Liu, W.; Wang, T.; Zhang, H. Soil Salinity Mapping Using Machine Learning Algorithms with the Sentinel-2 MSI in Arid Areas, China. Remote Sens. 2021, 13, 305. [Google Scholar] [CrossRef]
- Aubert, M.; Baghdadi, N.; Zribi, M.; Douaoui, A.; Loumagne, C.; Baup, F.; El Hajj, M.; Garrigues, S. Analysis of TerraSAR-X data sensitivity to bare soil moisture, roughness, composition and soil crust. Remote Sens. Environ. 2011, 115, 1801–1810. [Google Scholar] [CrossRef]
- Mohamed, E.S.; Ali, A.; El-Shirbeny, M.; Abutaleb, K.; Shaddad, S.M. Mapping soil moisture and their correlation with crop pattern using remotely sensed data in arid region. Egypt J. Remote Sens. Space Sci. 2020, 23, 347–353. [Google Scholar] [CrossRef]
- Zhang, Z.; Ding, J.; Wang, J.; Ge, X. Prediction of soil organic matter in northwestern China using fractional-order derivative spectroscopy and modified normalized difference indices. Catena 2019, 185, 104257. [Google Scholar] [CrossRef]
- Periasamy, S.; Ravi, K.P. A novel approach to quantify soil salinity by simulating the dielectric loss of SAR in three-dimensional density space. Remote Sens. Environ. 2020, 251, 112059. [Google Scholar] [CrossRef]
- Wang, N.; Xue, J.; Peng, J.; Biswas, A.; He, Y.; Shi, Z. Integrating Remote Sensing and Landscape Characteristics to Estimate Soil Salinity Using Machine Learning Methods: A Case Study from Southern Xinjiang, China. Remote Sens. 2020, 12, 4118. [Google Scholar] [CrossRef]
- De Bernardis, C.; Vicente-Guijalba, F.; Martinez-Marin, T.; Lopez-Sanchez, J.M. Contribution to Real-Time Estimation of Crop Phenological States in a Dynamical Framework Based on NDVI Time Series: Data Fusion with SAR and Temperature. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3512–3523. [Google Scholar] [CrossRef]
- Farahmand, N.; Sadeghi, V. Estimating Soil Salinity in the Dried Lake Bed of Urmia Lake Using Optical Sentinel-2 Images and Nonlinear Regression Models. J. Indian Soc. Remote Sens. 2020, 48, 675–687. [Google Scholar] [CrossRef]
- Sahbeni, G. A PLSR model to predict soil salinity using Sentinel-2 MSI data. Open Geosci. 2021, 13, 977–987. [Google Scholar] [CrossRef]
- Ravi, K.P.; Periasamy, S. Systematic discrimination of irrigation and upheaval associated salinity using multitemporal SAR data. Sci. Total Environ. 2021, 790, 148148. [Google Scholar] [CrossRef]
- Rudiyanto; Minasny, B.; Setiawan, B.I.; Saptomo, S.K.; McBratney, A.B. Open digital mapping as a cost-effective method for mapping peat thickness and assessing the carbon stock of tropical peatlands. Geoderma 2018, 313, 25–40. [Google Scholar] [CrossRef]
- Tripathi, A.; Tiwari, R.K. Synergetic utilization of sentinel-1 SAR and sentinel-2 optical remote sensing data for surface soil moisture estimation for Rupnagar, Punjab, India. Geocarto Int. 2020, 37, 2215–2236. [Google Scholar] [CrossRef]
- Hoa, P.V.; Giang, N.V.; Binh, N.A.; Hai, L.V.H.; Pham, T.-D.; Hasanlou, M.; Bui, D.T. Soil Salinity Mapping Using SAR Sentinel-1 Data and Advanced Machine Learning Algorithms: A Case Study at Ben Tre Province of the Mekong River Delta (Vietnam). Remote Sens. 2019, 11, 128. [Google Scholar] [CrossRef]
- Pittman, R.C. Improvement of Soil Property Mapping in Northern Ontario’s Great Clay Belt Using Multi-Source Remotely Sensed Data; YorkSpace: Toronto, ON, Canada, 2020. [Google Scholar]
- Song, T.; Wang, Z.; Xie, P.; Han, N.; Jiang, J.; Xu, D. A Novel Dual Path Gated Recurrent Unit Model for Sea Surface Salinity Prediction. J. Atmos. Ocean. Technol. 2020, 37, 317–325. [Google Scholar] [CrossRef]
- Hosseini, M.; Bahrami, H.; Khormali, F.; Khavazi, K.; Mokhtassi-Bidgoli, A. Artificial Intelligence Statistical Analysis of Soil Respiration Improves Predictions Compared to Regression Methods. J. Soil Sci. Plant Nutr. 2021, 21, 2242–2251. [Google Scholar] [CrossRef]
- Schulz, K.; Hänsch, R.; Sörgel, U. Machine learning methods for remote sensing applications: An overview. In Proceedings of the Volume 10790 SPIE Remote Sensing, Berlin, Germany, 10–13 September 2018; Volume 10790, p. 1079002. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an Afromontane landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Achieng, K.O. Modelling of soil moisture retention curve using machine learning techniques: Artificial and deep neural net-works vs support vector regression models. Comput. Geosci. 2019, 133, 104320. [Google Scholar] [CrossRef]
- Sahour, H.; Gholami, V.; Vazifedan, M. A comparative analysis of statistical and machine learning techniques for mapping the spatial distribution of groundwater salinity in a coastal aquifer. J. Hydrol. 2020, 591, 125321. [Google Scholar] [CrossRef]
- Bokde, N.D.; Ali, Z.H.; Al-Hadidi, M.T.; Farooque, A.A.; Jamei, M.; Al Maliki, A.A.; Beyaztas, B.H.; Faris, H.; Yaseen, Z.M. Total Dissolved Salt Prediction Using Neurocomputing Models: Case Study of Gypsum Soil Within Iraq Region. IEEE Access 2021, 9, 53617–53635. [Google Scholar] [CrossRef]
- Muller, S.J.; Van Niekerk, A. An evaluation of supervised classifiers for indirectly detecting salt-affected areas at irrigation scheme level. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 138–150. [Google Scholar] [CrossRef]
- Alamdar, S.; Ghazban, F.; Zarei, A. Efficiency of Machine Learning Algorithms in Soil Salinity Detection Using LAND-SAT-8 Oli Imagery. ISPRS Annals of the Photogrammetry. Remote Sens. Spat. Inf. Sci. 2023, 10, 49–55. [Google Scholar]
- Xiao, C.; Ji, Q.; Chen, J.; Zhang, F.; Li, Y.; Fan, J.; Hou, X.; Yan, F.; Wang, H. Prediction of soil salinity parameters using machine learning models in an arid region of northwest China. Comput. Electron. Agric. 2023, 204, 107512. [Google Scholar] [CrossRef]
- Mzid, N.; Boussadia, O.; Albrizio, R.; Stellacci, A.M.; Braham, M.; Todorovic, M. Salinity Properties Retrieval from Senti-nel-2 Satellite Data and Machine Learning Algorithms. Agronomy 2023, 13, 716. [Google Scholar] [CrossRef]
- Kaya, F.; Schillaci, C.; Keshavarzi, A.; Başayiğit, L. Predictive Mapping of Electrical Conductivity and Assessment of Soil Salinity in a Western Türkiye Alluvial Plain. Land 2022, 11, 2148. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Schmidt, K.; Toomanian, N.; Heung, B.; Behrens, T.; Mosavi, A.; Band, S.S.; Amirian-Chakan, A.; Fathabadi, A.; Scholten, T. Improving the spatial prediction of soil salinity in arid regions using wavelet transformation and support vector regression models. Geoderma 2020, 383, 114793. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, F.; Zhang, X.; Chan, N.W.; Kung, H.-T.; Ariken, M.; Zhou, X.; Wang, Y. Regional suitability prediction of soil salinization based on remote-sensing derivatives and optimal spectral index. Sci. Total Environ. 2021, 775, 145807. [Google Scholar] [CrossRef]
- Rafik, A.; Ibouh, H.; Fels, A.E.A.E.; Eddahby, L.; Mezzane, D.; Bousfoul, M.; Amazirh, A.; Ouhamdouch, S.; Bahir, M.; Gourfi, A.; et al. Soil Salinity Detection and Mapping in an Environment under Water Stress between 1984 and 2018 (Case of the Largest Oasis in Africa-Morocco). Remote Sens. 2022, 14, 1606. [Google Scholar] [CrossRef]
- Jiang, X.; Duan, H.; Liao, J.; Guo, P.; Huang, C.; Xue, X. Estimation of Soil Salinization by Machine Learning Algo-rithms in Different Arid Regions of Northwest China. Remote Sens. 2022, 14, 347. [Google Scholar] [CrossRef]
- Merembayev, T.; Amirgaliyev, Y.; Saurov, S.; Wójcik, W. Soil Salinity Classification Using Machine Learning Algo-rithms and Radar Data in the Case from the South of Kazakhstan. J. Ecol. Eng. 2022, 23, 61–67. [Google Scholar] [CrossRef]
- Estévez, V.; Beucher, A.; Mattbäck, S.; Boman, A.; Auri, J.; Björk, K.-M.; Österholm, P. Machine learning techniques for acid sulfate soil mapping in southeastern Finland. Geoderma 2021, 406, 115446. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Hamzehpour, N.; Hassanzadeh, M.; Heung, B.; Goydaragh, M.G.; Schmidt, K.; Scholten, T. Enhancing the accuracy of machine learning models using the super learner technique in digital soil mapping. Geoderma 2021, 399, 115108. [Google Scholar] [CrossRef]
- Nabiollahi, K.; Taghizadeh-Mehrjardi, R.; Shahabi, A.; Heung, B.; Amirian-Chakan, A.; Davari, M.; Scholten, T. Assessing agricultural salt-affected land using digital soil mapping and hybridized random forests. Geoderma 2020, 385, 114858. [Google Scholar] [CrossRef]
- Gharaibeh, M.A.; Albalasmeh, A.A.; Pratt, C.; El Hanandeh, A. Estimation of exchangeable sodium percentage from sodium adsorption ratio of salt-affected soils using traditional and dilution extracts, saturation percentage, electrical conductivity, and generalized regression neural networks. Catena 2021, 205, 105466. [Google Scholar] [CrossRef]
- Paz, A.M.; Castanheira, N.; Farzamian, M.; Paz, M.C.; Gonçalves, M.C.; Santos, F.A.M.; Triantafilis, J. Prediction of soil sa-linity and sodicity using electromagnetic conductivity imaging. Geoderma 2020, 361, 114086. [Google Scholar] [CrossRef]
- El Bilali, A.; Taleb, A.; Nafii, A.; Alabjah, B.; Mazigh, N. Prediction of sodium adsorption ratio and chloride concentration in a coastal aquifer under seawater intrusion using machine learning models. Environ. Technol. Innov. 2021, 23, 101641. [Google Scholar] [CrossRef]
- El Bilali, A.; Taleb, A.; Brouziyne, Y. Groundwater quality forecasting using machine learning algorithms for irrigation purposes. Agric. Water Manag. 2020, 245, 106625. [Google Scholar] [CrossRef]
- Wang, F.; Yang, S.; Yang, W.; Yang, X.; Jianli, D. Comparison of machine learning algorithms for soil salinity pre-dictions in three dryland oases located in Xinjiang Uyghur Autonomous Region (XJUAR) of China. Eur. J. Remote Sens. 2019, 52, 256–276. [Google Scholar] [CrossRef]
- Wei, Q.; Nurmemet, I.; Gao, M.; Xie, B. Inversion of Soil Salinity Using Multisource Remote Sensing Data and Parti-cle Swarm Machine Learning Models in Keriya Oasis, Northwestern China. Remote Sens. 2022, 14, 512. [Google Scholar] [CrossRef]
- Abdellatif, A.D.; El Ghonamey, Y.K.; Shoman, M.M. The use of geographic information systems for monitoring some soil properties: Case study Damanhur District, El-Beheira Governorate—Egypt. Ann. Agric. Sci. 2017, 55, 979–988. Available online: http://aasj.bu.edu.eg/index.php (accessed on 12 August 2021).
- Aboukila, E.; Abdelaty, E. Assessment of saturated soil paste salinity from 1:2.5 and 1:5 soil-water extracts for coarse tex-tured soils. Alex. Sci. Exch. J. 2017, 38, 722–732. [Google Scholar]
- Afifi, A.A.; Darwish, K.M. Detection and impact of land encroachment in El-Beheira governorate, Egypt. Model. Earth Syst. Environ. 2018, 4, 517–526. [Google Scholar] [CrossRef]
- Staff, U. Diagnosis and Improvement of Saline and Alkali Soils; Handbook 60; US Department of Agriculture, United States Salinity Laboratory (USSL): Washington, DC, USA, 1954. [Google Scholar]
- Abrol, I.; Yadav, J.S.P.; Massoud, F.I. Salt-Affected Soils and Their Management; Food and Agriculture Organization: Washington, DC, USA, 1988. [Google Scholar]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Spoto, F.; Sy, O.; Laberinti, P.; Martimort, P.; Fernandez, V.; Colin, O.; Hoersch, B.; Meygret, A. Overview of Sentinel-2. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Munich, Germany, 22–27 July 2012; pp. 1707–1710. [Google Scholar] [CrossRef]
- Rossi, M.; Chiarito, E.; Cigna, F.; Cuozzo, G.; Fontanelli, G.; Paloscia, S.; Santi, E.; Tapete, D.; Notarnicola, C. Multisensor SAR and optical estimation of grassland above-ground biomass and LAI: A case study for the Mazia valley in South Tyrol. In Proceedings of the EGU General Assembly 2021, online, 19–30 April 2021; Volume EGU21-11932. [Google Scholar] [CrossRef]
- Haryanto, T.; Pratama, A.; Suhartanto, H.; Murni, A.; Kusmardi, K.; Pidanic, J. Multipatch-GLCM for Texture Feature Extraction on Classification of the Colon Histopathology Images using Deep Neural Network with GPU Acceleration. J. Comput. Sci. 2020, 16, 280–294. [Google Scholar] [CrossRef]
- Corrales, D.C.; Schoving, C.; Raynal, H.; Debaeke, P.; Journet, E.-P.; Constantin, J. A surrogate model based on feature selection techniques and regression learners to improve soybean yield prediction in southern France. Comput. Electron. Agric. 2022, 192, 106578. [Google Scholar] [CrossRef]
- Pearson, K. The fundamental problem of practical statistics. Biometrika 1920, 13, 1–16. [Google Scholar] [CrossRef]
- Spearman, C. “General Intelligence” Objectively Determined and Measured. In Studies in Individual Differences: The Search for Intelligence; Jenkins, J.J., Paterson, D.G., Eds.; Appleton-Century-Crofts: New York, NY, USA, 1961; Volume 15, pp. 59–73. [Google Scholar] [CrossRef]
- Largeron, C.; Christophe, M.; Mathias, G. Entropy based feature selection for text categorization. In Proceedings of the 2011 ACM Symposium on Applied Computing, Taichung, Taiwan, 21–24 March 2011. [Google Scholar]
- Kroehl, M.E. On the Use of Lasso Regression for Mediation Analysis with Application to Microbiota Data; University of Colorado Denver, Anschutz Medical Campus: Denver, CO, USA, 2014. [Google Scholar]
- Tang, J.; Alelyani, S.; Liu, H. Feature selection for classification: A review. In Data Classification: Algorithms and Applications; CRC Press: Boca Raton, FL, USA, 2014; Volume 37, pp. 37–64. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Kuhn, M.; Johnson, K. Feature Engineering and Selection: A Practical Approach for Predictive Models; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Peng, J.; Biswas, A.; Jiang, Q.; Zhao, R.; Hu, J.; Hu, B.; Shi, Z. Estimating soil salinity from remote sensing and terrain data in southern Xinjiang Province, China. Geoderma 2018, 337, 1309–1319. [Google Scholar] [CrossRef]


| Salinity Class | Soil Salinity (dS/m) | Effect on Crop Plants |
|---|---|---|
| Non-saline | <2 | Negligible salinity effects. |
| Slightly saline | 2–4 | Yields of sensitive crops maybe affected. |
| Moderately saline | 4–8 | Yields of many crops are affected. |
| Strongly saline | 8–16 | Only tolerant crops will survive. |
| Very strongly saline | >16 | Only a few tolerant crops will survive. |
| Descriptions | Selected Variable |
|---|---|
| Sentinel-2, Band (b) selection | b2, b3, b4, b5, b6, b7, b8, b8A, b11 and b12. |
| Sentinel-2, Indices | WDVI, TNDVI, SAVI, NDWI, NDVI, MSAVI, MSAVI2, MNDWI, MCARI, IPVI, GNDVI, DVI, CI, BI and BI2. |
| Sentinal-1, sigma nought (δ0) db | VV and VH. |
| V V_ GLCM | Contrast_VV, Dissimilarity_VV, Homogeneity_VV, AngularSecondMoment_VV, Energy_VV, Entropy_VV, MaximumProbability_VV, Correlation_VV, Mean_VV and StandardDviation_VV. |
| VH_ GLCM | Contrast_VH, Dissimilarity_VH, Homogeneity_VH, AngularSecondMoment_VH, Energy_VH, Entropy_VH, MaximumProbability_VH, Correlation_VH, Mean_VH and StandardDeviation_VH. |
| Learner | Subsetted Variables | No. of Subsetted Variables | RMSE |
|---|---|---|---|
| LR |
| 29 | 0.48174263 |
| RF |
| 24 | 0.52825744 |
| SVR |
| 14 | 0.55513133 |
| BPNN |
| 18 | 0.302719033 |
| Learner | Subsetted Variables | Number of Subsetted Variables | RMSE |
|---|---|---|---|
| RF | WDVI, TNDVI, SAVI, NDWI, NDVI, MSAVI, MSAVI2, IPVI, GNDVI, DVI, CI. VV, VH Mean_VV. Mean_VH. | 15 | 0.52825744 |
| LASSO | band3, band6, band8, band8A, band11, band12, TNDVI, MNDWI, CI, BI2, VV. Contrast_VV, Dissimilarity_VV, Homogeneity_VV, Angular_Second_Moment_VV, Correlation_VV, Mean_VV. Contrast_VH, Dissimilarity_VH, Homogeneity_VH, Angular_Second_Moment_VH, Maximum_Probability_VH, Correlation_VH, Angular_Second_Moment_VH, Maximum_Probability_VH, Correlation_VH. | 23 | 0.5093330 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, S.A.; Metwaly, M.M.; Metwalli, M.R.; AbdelRahman, M.A.E.; Badreldin, N. Integrating Active and Passive Remote Sensing Data for Mapping Soil Salinity Using Machine Learning and Feature Selection Approaches in Arid Regions. Remote Sens. 2023, 15, 1751. https://doi.org/10.3390/rs15071751
Mohamed SA, Metwaly MM, Metwalli MR, AbdelRahman MAE, Badreldin N. Integrating Active and Passive Remote Sensing Data for Mapping Soil Salinity Using Machine Learning and Feature Selection Approaches in Arid Regions. Remote Sensing. 2023; 15(7):1751. https://doi.org/10.3390/rs15071751
Chicago/Turabian StyleMohamed, Sayed A., Mohamed M. Metwaly, Mohamed R. Metwalli, Mohamed A. E. AbdelRahman, and Nasem Badreldin. 2023. "Integrating Active and Passive Remote Sensing Data for Mapping Soil Salinity Using Machine Learning and Feature Selection Approaches in Arid Regions" Remote Sensing 15, no. 7: 1751. https://doi.org/10.3390/rs15071751
APA StyleMohamed, S. A., Metwaly, M. M., Metwalli, M. R., AbdelRahman, M. A. E., & Badreldin, N. (2023). Integrating Active and Passive Remote Sensing Data for Mapping Soil Salinity Using Machine Learning and Feature Selection Approaches in Arid Regions. Remote Sensing, 15(7), 1751. https://doi.org/10.3390/rs15071751









