Evaluation and Comparison of Semantic Segmentation Networks for Rice Identification Based on Sentinel-2 Imagery
Abstract
1. Introduction
2. Materials
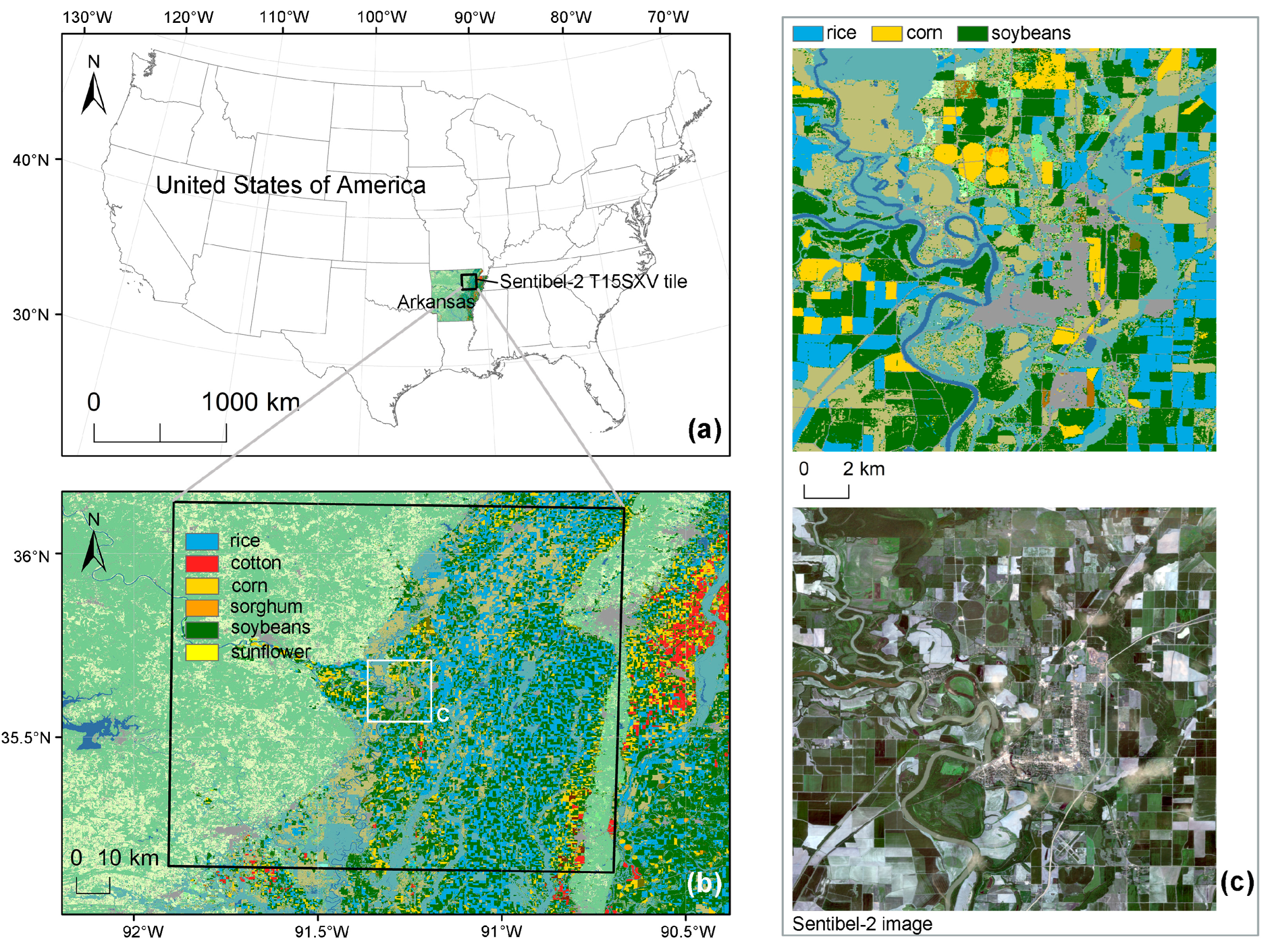
2.1. Study Area
2.2. Datasets
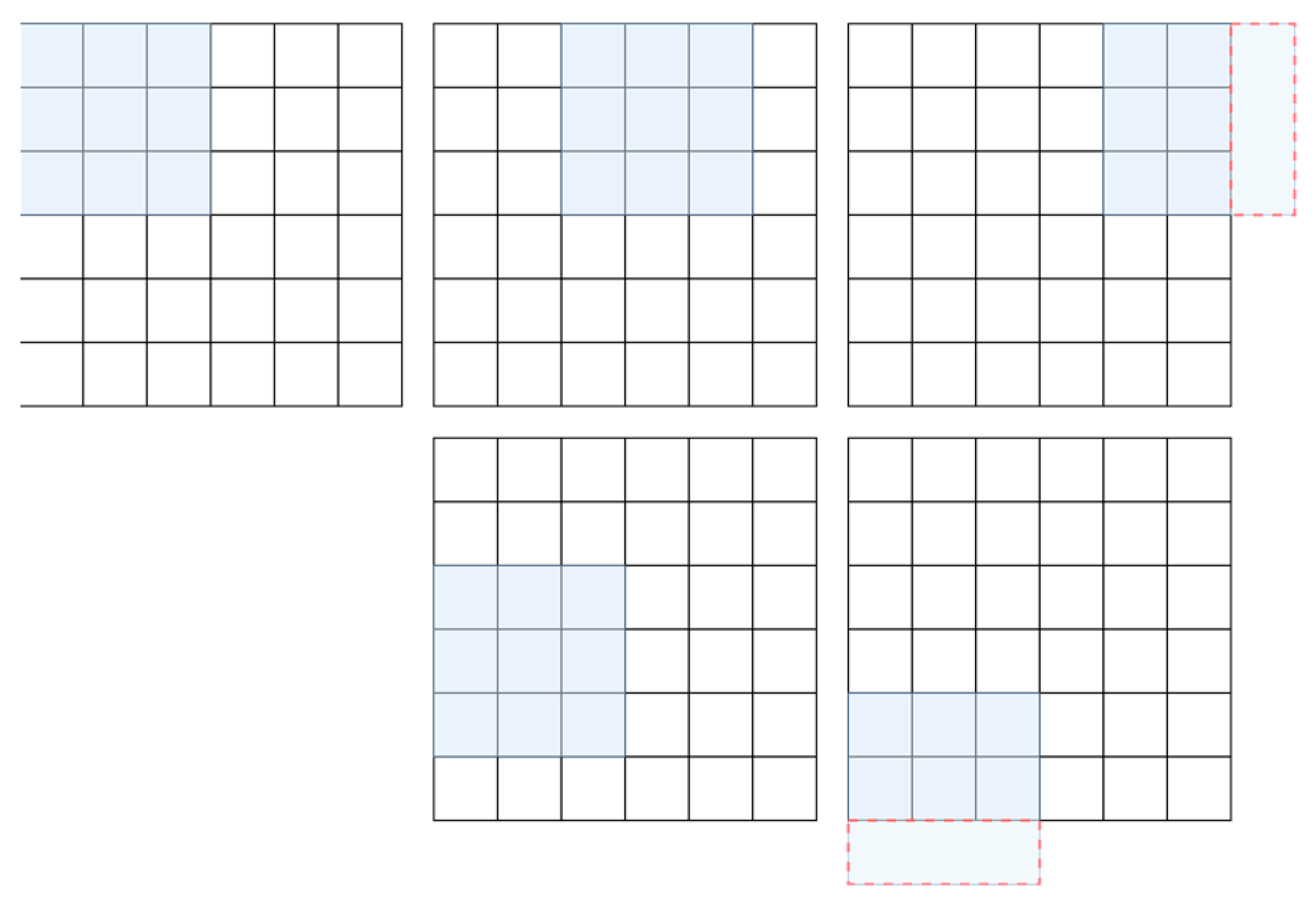
2.2.1. Sentinel-2 and Preprocessing
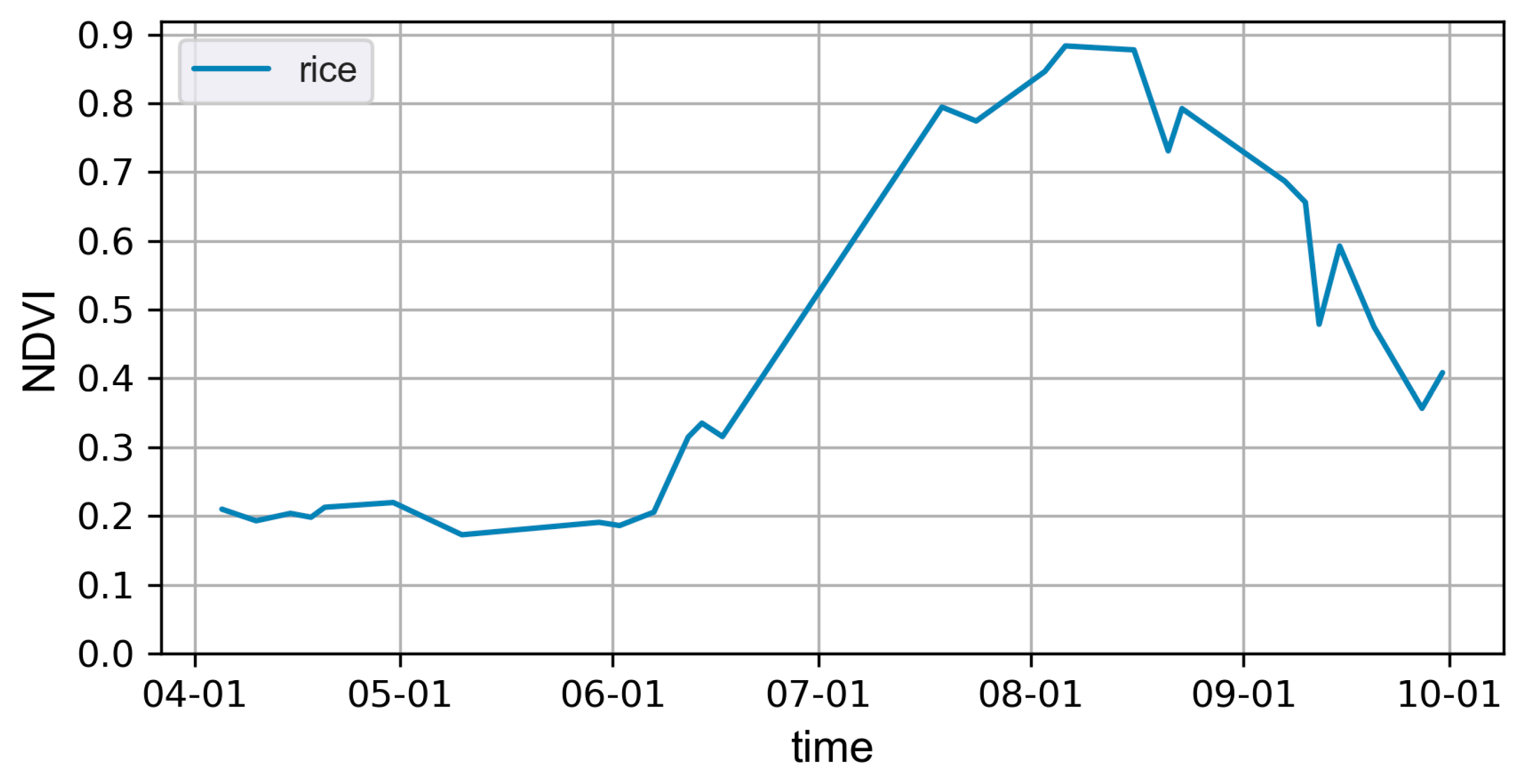
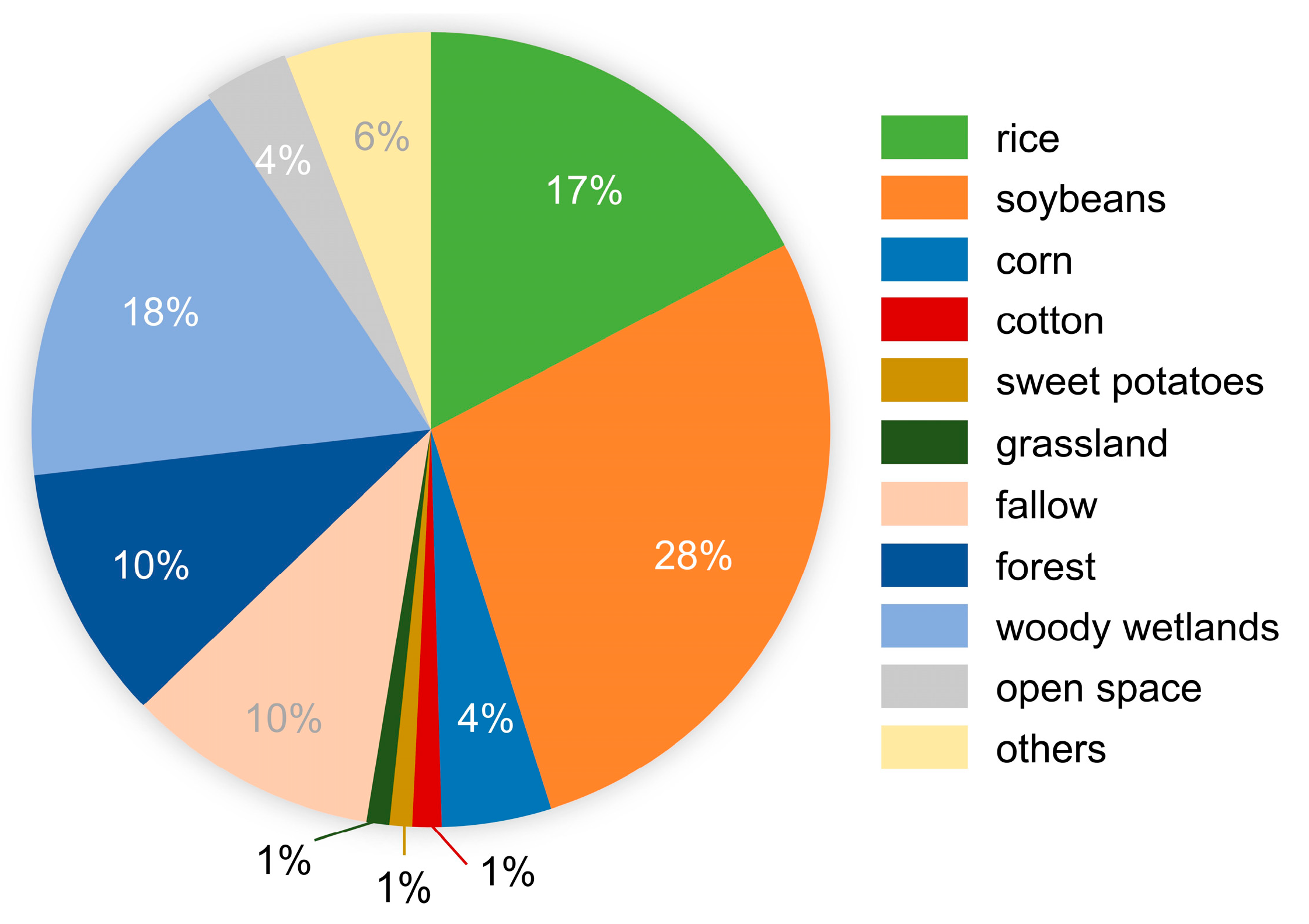
2.2.2. Reference Data and Preprocessing
3. Methodology
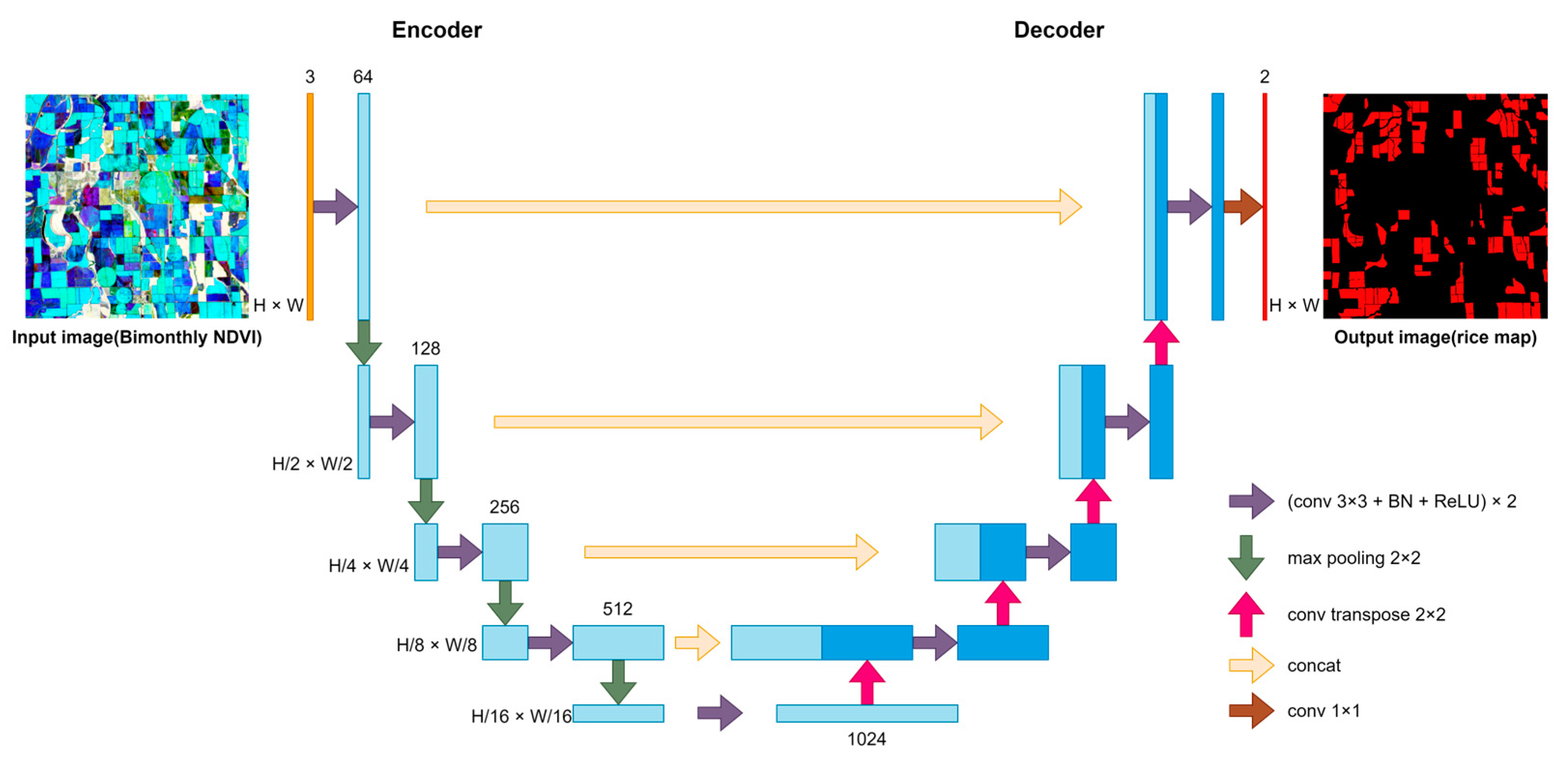
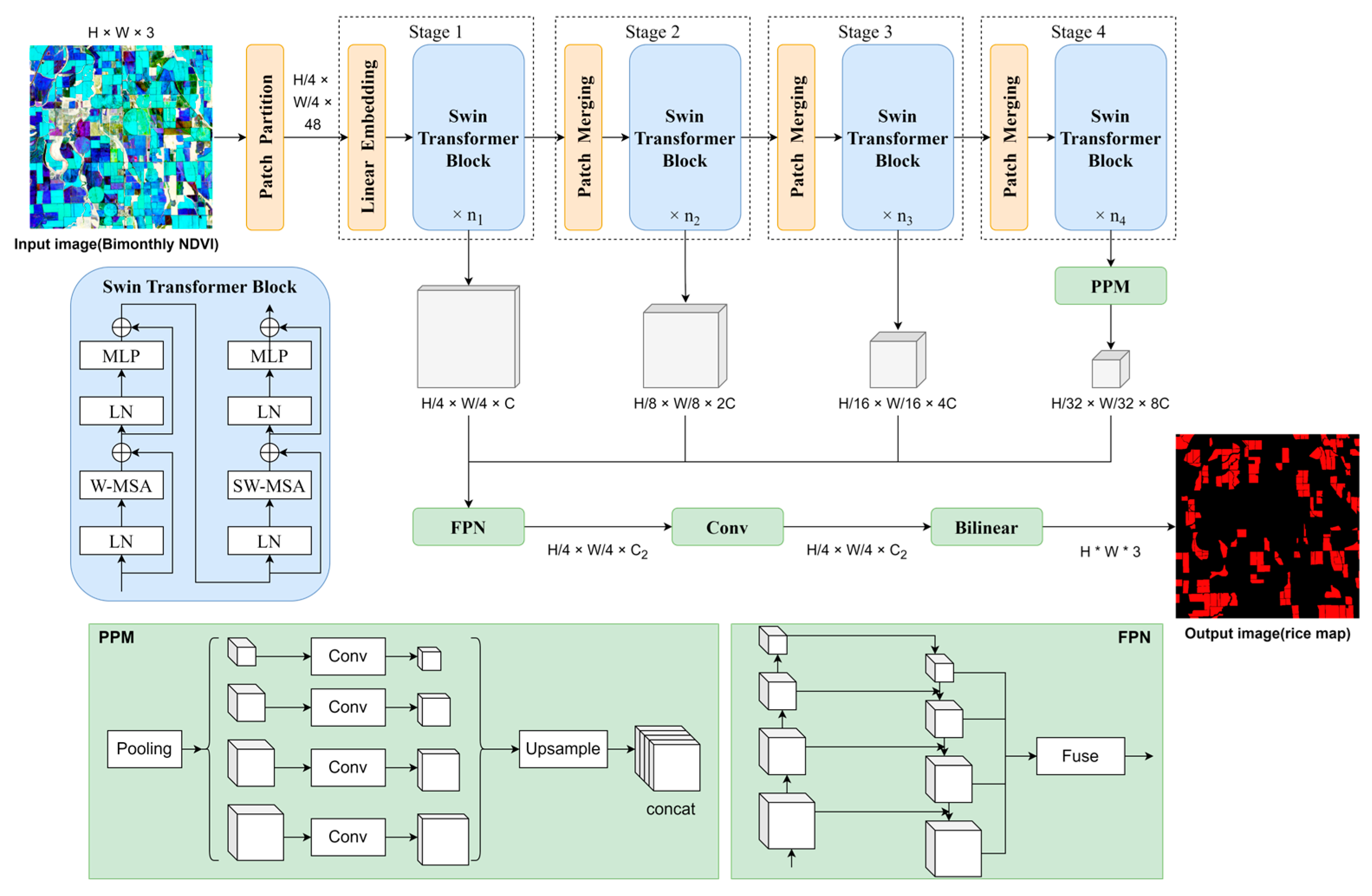
3.1. Three Semantic Segmentation Networks for Rice Identification
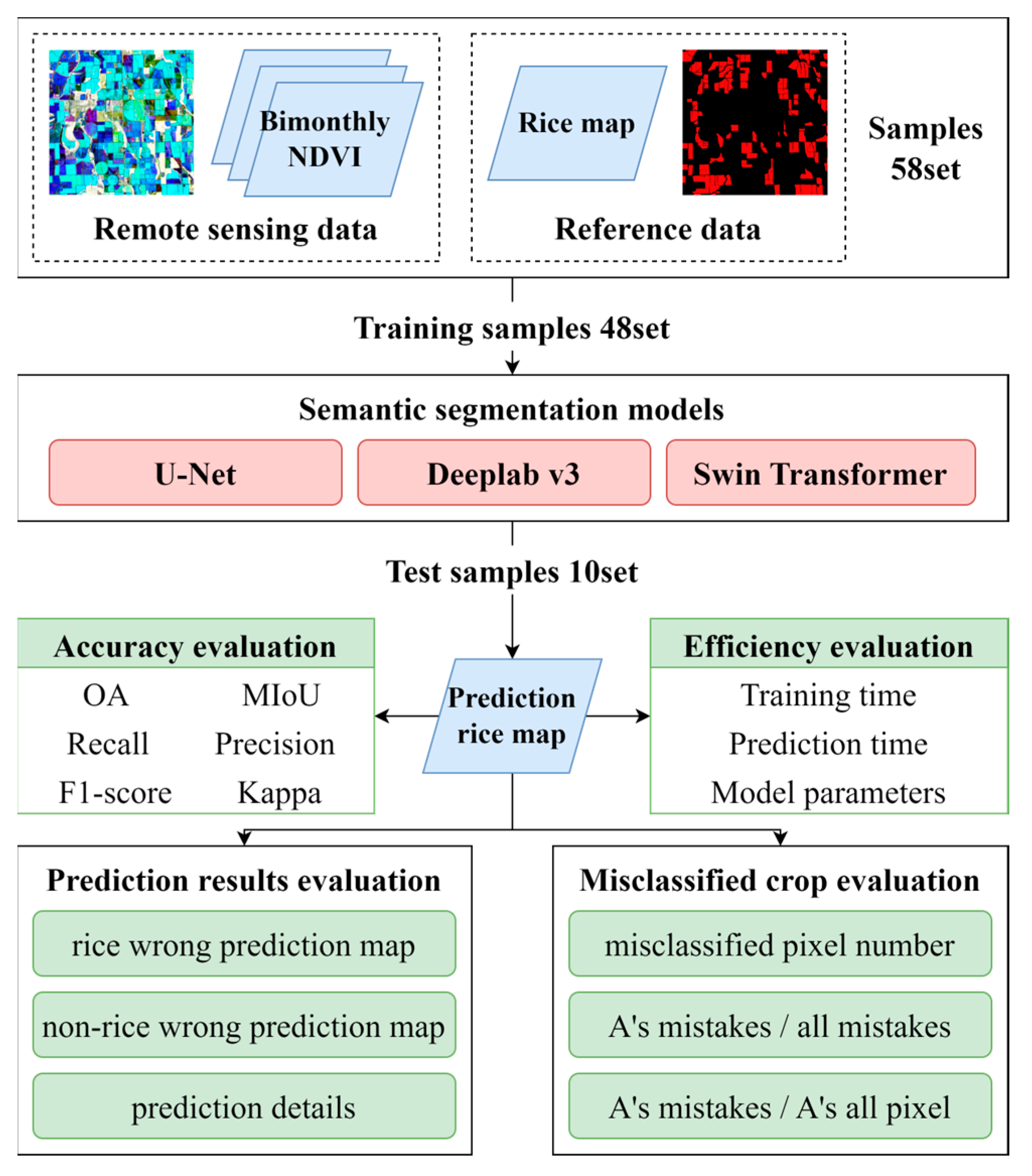
3.2. Evaluation Metrics
3.3. Hardware and Training Settings
4. Results
4.1. Rice Identification Accuracy
4.2. Model Efficiency
4.3. Prediction Results
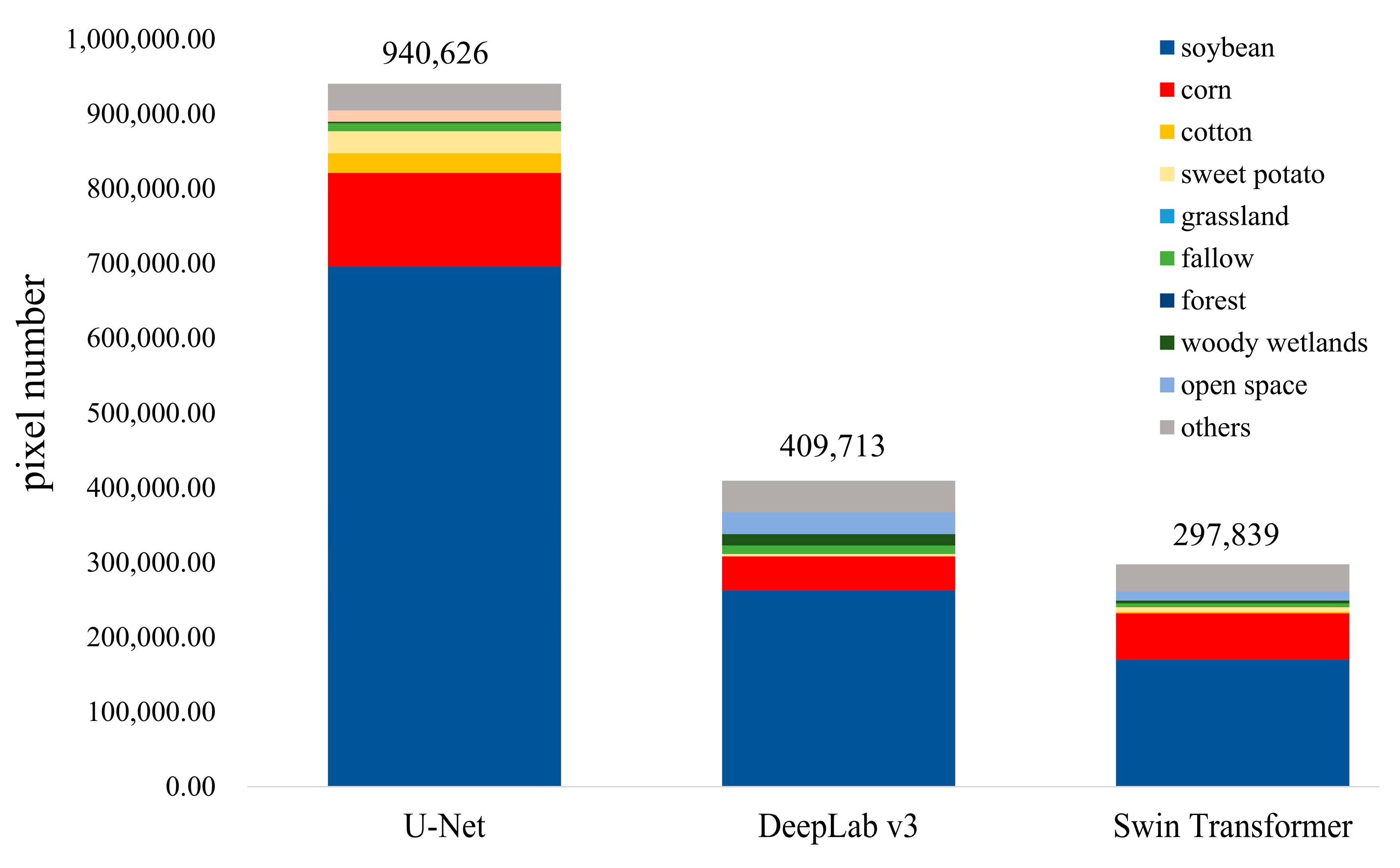
4.4. Misclassified Crops
5. Discussion
5.1. Analysis of Rice Identification Results
5.2. Analysis of Model Efficiency
5.3. Analysis of Misidentification
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elert, E. Rice by the numbers: A good grain. Nature 2014, 514, S50–S51. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef]
- Benayas, J.M.R.; Bullock, J.M. Restoration of Biodiversity and Ecosystem Services on Agricultural Land. Ecosystems 2012, 15, 883–899. [Google Scholar] [CrossRef]
- Herzog, F.; Prasuhn, V.; Spiess, E.; Richner, W. Environmental cross-compliance mitigates nitrogen and phosphorus pollution from Swiss agriculture. Environ. Sci. Policy 2008, 11, 655–668. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Knox, J.W.; Ozdogan, M.; Gumma, M.K.; Congalton, R.G.; Wu, Z.T.; Milesi, C.; Finkral, A.; Marshall, M.; Mariotto, I.; et al. Assessing future risks to agricultural productivity, water resources and food security: How can remote sensing help? Photogramm. Eng. Remote Sens. 2012, 78, 773–782. [Google Scholar]
- Anderegg, J.; Yu, K.; Aasen, H.; Walter, A.; Liebisch, F.; Hund, A. Spectral Vegetation Indices to Track Senescence Dynamics in Diverse Wheat Germplasm. Front. Plant Sci. 2020, 10, 1749. [Google Scholar] [CrossRef]
- Dong, J.W.; Xiao, X.M.; Menarguez, M.A.; Zhang, G.L.; Qin, Y.W.; Thau, D.; Biradar, C.; Moore, B. Mapping paddy rice planting area in northeastern Asia with Landsat 8 images, phenology-based algorithm and Google Earth Engine. Remote Sens. Environ. 2016, 185, 142–154. [Google Scholar] [CrossRef]
- Mosleh, M.K.; Hassan, Q.K.; Chowdhury, E.H. Application of Remote Sensors in Mapping Rice Area and Forecasting Its Production: A Review. Sensors 2015, 15, 769–791. [Google Scholar] [CrossRef]
- Prasad, A.K.; Chai, L.; Singh, R.P.; Kafatos, M. Crop yield estimation model for Iowa using remote sensing and surface parameters. Int. J. Appl. Earth Obs. Geoinf. 2006, 8, 26–33. [Google Scholar] [CrossRef]
- Wardlow, B.D.; Egbert, S.L.; Kastens, J.H. Analysis of time-series MODIS 250 m vegetation index data for crop classification in the US Central Great Plains. Remote Sens. Environ. 2007, 108, 290–310. [Google Scholar] [CrossRef]
- Ramteke, I.K.; Rajankar, P.B.; Reddy, G.O.; Kolte, D.M.; Sen, T.K.J.I.J.o.E.; Sciences, E. Optical remote sensing applications in crop mapping and acreage estimation: A review. Int. J. Ecol. Environ. Sci. 2020, 2, 696–703. [Google Scholar]
- Zhao, H.; Chen, Z.; Liu, J. Deep Learning for Crop Classifiction of Remote Sensing Data: Applications and Challenges. J. China Agric. Resour. Reg. Plan. 2020, 41, 35–49. [Google Scholar]
- Xiao, X.M.; Boles, S.; Frolking, S.; Li, C.S.; Babu, J.Y.; Salas, W.; Moore, B. Mapping paddy rice agriculture in South and Southeast Asia using multi-temporal MODIS images. Remote Sens. Environ. 2006, 100, 95–113. [Google Scholar] [CrossRef]
- Zhong, L.H.; Hu, L.N.; Yu, L.; Gong, P.; Biging, G.S. Automated mapping of soybean and corn using phenology. ISPRS J. Photogramm. Remote Sens. 2016, 119, 151–164. [Google Scholar] [CrossRef]
- Cai, Y.P.; Guan, K.Y.; Peng, J.; Wang, S.W.; Seifert, C.; Wardlow, B.; Li, Z. A high-performance and in-season classification system of field-level crop types using time-series Landsat data and a machine learning approach. Remote Sens. Environ. 2018, 210, 35–47. [Google Scholar] [CrossRef]
- Ustuner, M.; Sanli, F.B.; Abdikan, S.; Esetlili, M.T.; Kurucu, Y. Crop Type Classification Using Vegetation Indices of RapidEye Imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, XL-7, 195–198. [Google Scholar] [CrossRef]
- Mekhalfi, M.L.; Nicolo, C.; Bazi, Y.; Al Rahhal, M.M.; Alsharif, N.A.; Al Maghayreh, E. Contrasting YOLOv5, Transformer, and EfficientDet Detectors for Crop Circle Detection in Desert. IEEE Geosci. Remote Sens. Lett. 2022, 19, 3003205. [Google Scholar] [CrossRef]
- Patil, R.R.; Kumar, S. Rice Transformer: A Novel Integrated Management System for Controlling Rice Diseases. IEEE Access 2022, 10, 87698–87714. [Google Scholar] [CrossRef]
- Chew, R.; Rineer, J.; Beach, R.; O’Neil, M.; Ujeneza, N.; Lapidus, D.; Miano, T.; Hegarty-Craver, M.; Polly, J.; Temple, D.S. Deep Neural Networks and Transfer Learning for Food Crop Identification in UAV Images. Drones 2020, 4, 7. [Google Scholar] [CrossRef]
- Xu, J.F.; Yang, J.; Xiong, X.G.; Li, H.F.; Huang, J.F.; Ting, K.C.; Ying, Y.B.; Lin, T. Towards interpreting multi-temporal deep learning models in crop mapping. Remote Sens. Environ. 2021, 264, 112599. [Google Scholar] [CrossRef]
- Shang, R.H.; He, J.H.; Wang, J.M.; Xu, K.M.; Jiao, L.C.; Stolkin, R. Dense connection and depthwise separable convolution based CNN for polarimetric SAR image classification. Knowl.-Based Syst. 2020, 194, 105542. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Yang, S.T.; Gu, L.J.; Ren, R.Z.; He, F.C. Research on Crop Classification in Northeast China Based on Deep Learning for Sentinel-2 Data. In Proceedings of the Conference on Earth Observing Systems XXIV, San Diego, CA, USA, 11–15 August 2019. [Google Scholar]
- Dong, J.W.; Xiao, X.M. Evolution of regional to global paddy rice mapping methods: A review. ISPRS J. Photogramm. Remote Sens. 2016, 119, 214–227. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Z.; Jiang, H.; Liu, J. Early growing stage crop species identification in southern China based on sentinel-1A time series imagery and one-dimensional CNN. Trans. Chin. Soc. Agric. Eng. 2020, 36, 169–177. [Google Scholar]
- Zhong, L.H.; Hu, L.; Zhou, H.; Tao, X. Deep learning based winter wheat mapping using statistical data as ground references in Kansas and northern Texas, US. Remote Sens. Environ. 2019, 233, 111411. [Google Scholar] [CrossRef]
- Yang, S.T.; Gu, L.J.; Li, X.F.; Gao, F.; Jiang, T. Fully Automated Classification Method for Crops Based on Spatiotemporal Deep-Learning Fusion Technology. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5405016. [Google Scholar] [CrossRef]
- Cai, Y.; Liu, S.; Lin, H.; Zhang, M. Extraction of paddy rice based on convolutional neural network using multi-source remote sensing data. Remote Sens. Land Resour. 2020, 32, 97–104. [Google Scholar]
- Hong, S.L.; Jiang, Z.H.; Liu, L.Z.; Wang, J.; Zhou, L.Y.; Xu, J.P. Improved Mask R-CNN Combined with Otsu Preprocessing for Rice Panicle Detection and Segmentation. Appl. Sci. 2022, 12, 11701. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, S.Y.; Shao, Y.Y. Object-oriented crops classification for remote sensing images based on convolutional neural network. In Proceedings of the Conference on Image and Signal Processing for Remote Sensing XXIV, Berlin, Germany, 10–12 September 2018. [Google Scholar]
- Liu, G.; Jiang, X.; Tang, B. Application of Feature Optimization and Convolutional Neural Network in Crop Classification. J. Geo-Inf. Sci. 2021, 23, 1071–1081. [Google Scholar]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. In The Handbook of Brain Theory Neural Networks; MIT Press: Cambridge, MA, USA, 1995; Volume 3361, p. 1995. [Google Scholar]
- Maggiori, E.; Tarabalka, Y.; Charpiat, G.; Alliez, P. Convolutional Neural Networks for Large-Scale Remote-Sensing Image Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 645–657. [Google Scholar] [CrossRef]
- Sun, Z.H.; Di, L.P.; Fang, H.; Burgess, A. Deep Learning Classification for Crop Types in North Dakota. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 2200–2213. [Google Scholar] [CrossRef]
- Marcos, D.; Volpi, M.; Kellenberger, B.; Tuia, D. Land cover mapping at very high resolution with rotation equivariant CNNs: Towards small yet accurate models. ISPRS J. Photogramm. Remote Sens. 2018, 145, 96–107. [Google Scholar] [CrossRef]
- Huang, B.; Zhao, B.; Song, Y.M. Urban land-use mapping using a deep convolutional neural network with high spatial resolution multispectral remote sensing imagery. Remote Sens. Environ. 2018, 214, 73–86. [Google Scholar] [CrossRef]
- Yan, Y.L.; Ryu, Y. Exploring Google Street View with deep learning for crop type mapping. ISPRS J. Photogramm. Remote Sens. 2021, 171, 278–296. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, H.; Wang, G.X.; Sun, H.; Fu, J. Mapping Paddy Rice Using a Convolutional Neural Network (CNN) with Landsat 8 Datasets in the Dongting Lake Area, China. Remote Sens. 2018, 10, 1840. [Google Scholar] [CrossRef]
- Yang, S.; Song, Z.; Yin, H.; Zhang, Z.; Ning, J. Crop Classification Method of UVA Multispectral Remote Sensing Based on Deep Semantic Segmentation. Trans. Chin. Soc. Agric. Mach. 2021, 52, 185–192. [Google Scholar]
- Wei, P.L.; Chai, D.F.; Lin, T.; Tang, C.; Du, M.Q.; Huang, J.F. Large-scale rice mapping under different years based on time-series Sentinel-1 images using deep semantic segmentation model. ISPRS J. Photogramm. Remote Sens. 2021, 174, 198–214. [Google Scholar] [CrossRef]
- Duan, Z.; Liu, J.; Lu, M.; Kong, X.; Yang, N. Tile edge effect and elimination scheme of image classification using CNN-ISS remote sensing. Trans. Chin. Soc. Agric. Eng. 2021, 37, 209–217. [Google Scholar]
- He, Y.; Zhang, X.Y.; Zhang, Z.Q.; Fang, H. Automated detection of boundary line in paddy field using MobileV2-UNet and RANSAC. Comput. Electron. Agric. 2022, 194, 106697. [Google Scholar] [CrossRef]
- Sun, K.Q.; Wang, X.; Liu, S.S.; Liu, C.H. Apple, peach, and pear flower detection using semantic segmentation network and shape constraint level set. Comput. Electron. Agric. 2021, 185, 106150. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Annual Conference on Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Sheng, J.J.; Sun, Y.Q.; Huang, H.; Xu, W.Y.; Pei, H.T.; Zhang, W.; Wu, X.W. HBRNet: Boundary Enhancement Segmentation Network for Cropland Extraction in High-Resolution Remote Sensing Images. Agriculture 2022, 12, 1284. [Google Scholar] [CrossRef]
- Wang, H.; Chen, X.Z.; Zhang, T.X.; Xu, Z.Y.; Li, J.Y. CCTNet: Coupled CNN and Transformer Network for Crop Segmentation of Remote Sensing Images. Remote Sens. 2022, 14, 1956. [Google Scholar] [CrossRef]
- Jiao, X.F.; Kovacs, J.M.; Shang, J.L.; McNairn, H.; Walters, D.; Ma, B.L.; Geng, X.Y. Object-oriented crop mapping and monitoring using multi-temporal polarimetric RADARSAT-2 data. ISPRS J. Photogramm. Remote Sens. 2014, 96, 38–46. [Google Scholar] [CrossRef]
- Lambert, M.J.; Traore, P.C.S.; Blaes, X.; Baret, P.; Defourny, P. Estimating smallholder crops production at village level from Sentinel-2 time series in Mali’s cotton belt. Remote Sens. Environ. 2018, 216, 647–657. [Google Scholar] [CrossRef]
- Vuolo, F.; Neuwirth, M.; Immitzer, M.; Atzberger, C.; Ng, W. How much does multi-temporal Sentinel-2 data improve crop type classification? Int. J. Appl. Earth Obs. Geoinf. 2018, 72, 122–130. [Google Scholar] [CrossRef]
- Boschetti, M.; Busetto, L.; Manfron, G.; Laborte, A.; Asilo, S.; Pazhanivelan, S.; Nelson, A. PhenoRice: A method for automatic extraction of spatio-temporal information on rice crops using satellite data time series. Remote Sens. Environ. 2017, 194, 347–365. [Google Scholar] [CrossRef]
- Kim, M.K.; Tejeda, H.; Yu, T.E. US milled rice markets and integration across regions and types. Int. Food Agribus. Manag. Rev. 2017, 20, 623–635. [Google Scholar] [CrossRef]
- Maxwell, S.K.; Sylvester, K.M. Identification of “ever-cropped” land (1984–2010) using Landsat annual maximum NDVI image composites: Southwestern Kansas case study. Remote Sens. Environ. 2012, 121, 186–195. [Google Scholar] [CrossRef]
- Javed, M.A.; Ahmad, S.R.; Awan, W.K.; Munir, B.A. Estimation of Crop Water Deficit in Lower Bari Doab, Pakistan Using Reflection-Based Crop Coefficient. ISPRS Int. J. Geo-Inf. 2020, 9, 173. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Liu, Z.; Lin, Y.T.; Cao, Y.; Hu, H.; Wei, Y.X.; Zhang, Z.; Lin, S.; Guo, B.N. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the 18th IEEE/CVF International Conference on Computer Vision (ICCV), Electr Network, Montreal, QC, Canada, 11–17 October 2021; pp. 9992–10002. [Google Scholar]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lin, T.Y.; Dollar, P.; Girshick, R.; He, K.M.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar]
- Zhao, H.S.; Shi, J.P.; Qi, X.J.; Wang, X.G.; Jia, J.Y. Pyramid Scene Parsing Network. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar]
- Wu, S.; Wang, G.R.; Tang, P.; Chen, F.; Shi, L.P. Convolution with even-sized kernels and symmetric padding. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]





| Confusion Matrix | Prediction | ||
|---|---|---|---|
| Rice | Non-Rice | ||
| Ground Truth | Rice | TP (True Positive) | FN (False Negative) |
| Non-rice | FP (False Positive) | TN (True Negative) | |
| Model | OA | MIoU | Recall | Precision | F1 | Kappa |
|---|---|---|---|---|---|---|
| U-Net [54] | 0.8934 | 0.8127 | 0.7652 | 0.4966 | 0.5526 | 0.4883 |
| DeepLab v3 [55] | 0.9080 | 0.8338 | 0.7868 | 0.7856 | 0.7855 | 0.7219 |
| Swin Transformer [56] | 0.9547 | 0.9134 | 0.7788 | 0.8386 | 0.8076 | 0.7820 |
| Model | Training Time (Per Epoch) | Prediction Time (Per Image) | Model Parameters | |
|---|---|---|---|---|
| Amount (Million) | Size (MB) | |||
| U-Net [54] | 4 min 32 s | 3.60 s | 7.70 | 30.79 |
| DeepLab v3 [55] | 8 min 48 s | 3.90 s | 39.63 | 158.54 |
| Swin Transformer [56] | 7 min 27 s | 4.40 s | 62.31 | 249.23 |
| Soybean | Corn | Cotton | Sweet Potato | Grassland | Fallow | Forest | Woody Wetland | Open Space | Others | |
|---|---|---|---|---|---|---|---|---|---|---|
| U-Net [54] | 20.75% | 23.39% | 18.41% | 26.85% | 0.02% | 0.89% | 0.002% | 0.09% | 3.60% | 5.07% |
| DeepLab v3 [55] | 7.84% | 8.44% | 0.37% | 2.68% | 0.11% | 0.91% | 0.03% | 0.70% | 7.06% | 5.94% |
| Swin Transformer [56] | 5.08% | 11.52% | 1.41% | 5.43% | 0.03% | 0.43% | 0.002% | 0.17% | 2.93% | 5.16% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Song, J.; Zhu, Y. Evaluation and Comparison of Semantic Segmentation Networks for Rice Identification Based on Sentinel-2 Imagery. Remote Sens. 2023, 15, 1499. https://doi.org/10.3390/rs15061499
Xu H, Song J, Zhu Y. Evaluation and Comparison of Semantic Segmentation Networks for Rice Identification Based on Sentinel-2 Imagery. Remote Sensing. 2023; 15(6):1499. https://doi.org/10.3390/rs15061499
Chicago/Turabian StyleXu, Huiyao, Jia Song, and Yunqiang Zhu. 2023. "Evaluation and Comparison of Semantic Segmentation Networks for Rice Identification Based on Sentinel-2 Imagery" Remote Sensing 15, no. 6: 1499. https://doi.org/10.3390/rs15061499
APA StyleXu, H., Song, J., & Zhu, Y. (2023). Evaluation and Comparison of Semantic Segmentation Networks for Rice Identification Based on Sentinel-2 Imagery. Remote Sensing, 15(6), 1499. https://doi.org/10.3390/rs15061499








