Study of Genetic Variation in Bermuda Grass along Longitudinal and Latitudinal Gradients Using Spectral Reflectance
Abstract
1. Introduction
2. Materials and Methods
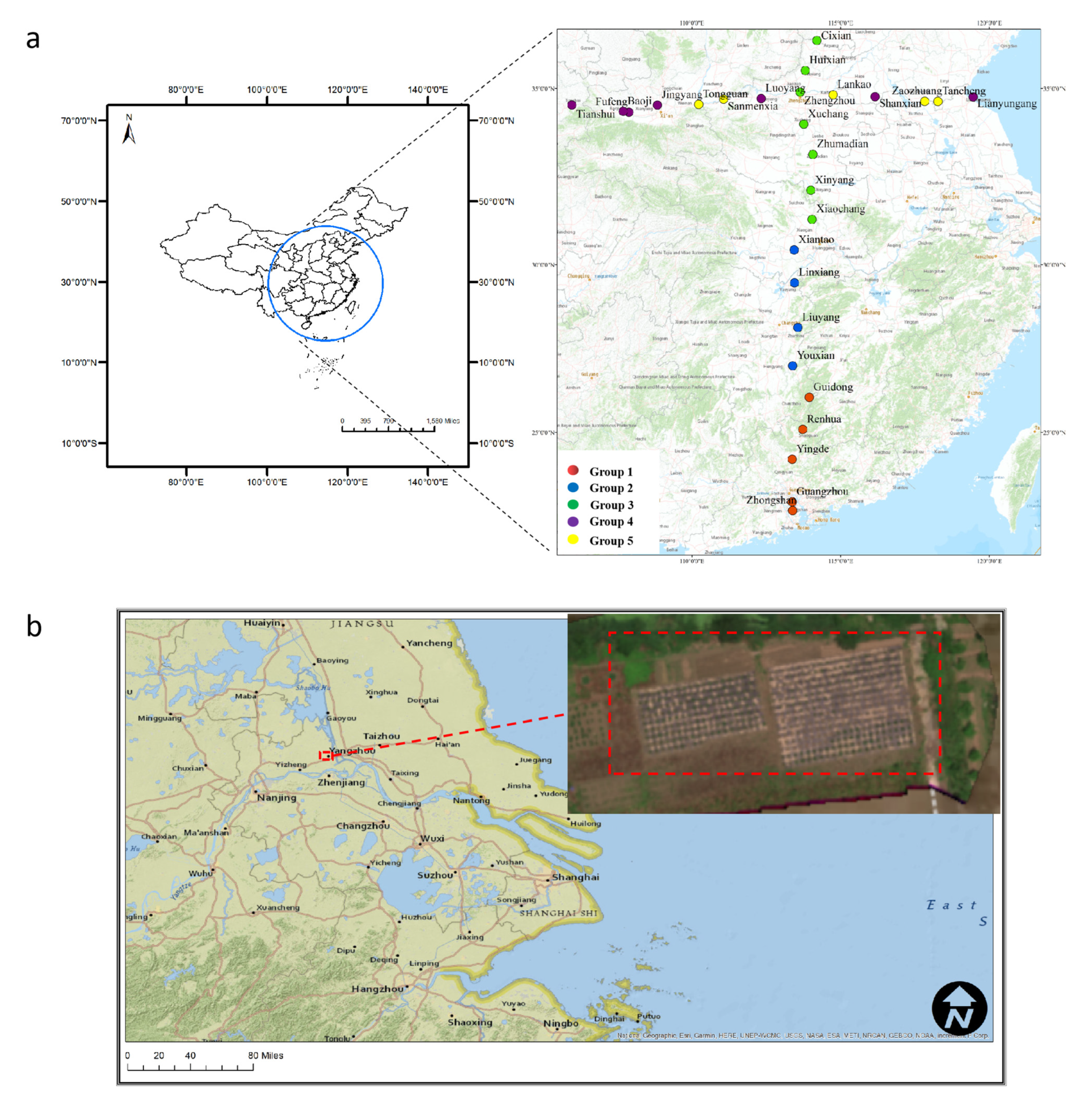
2.1. Study Area and Sample Collection
2.2. Genotypic Analysis
2.3. Multispectral Image Acquisition and Processing
2.4. Hyperspectral Data Acquisition
2.5. Data Analysis and Model Development
3. Results
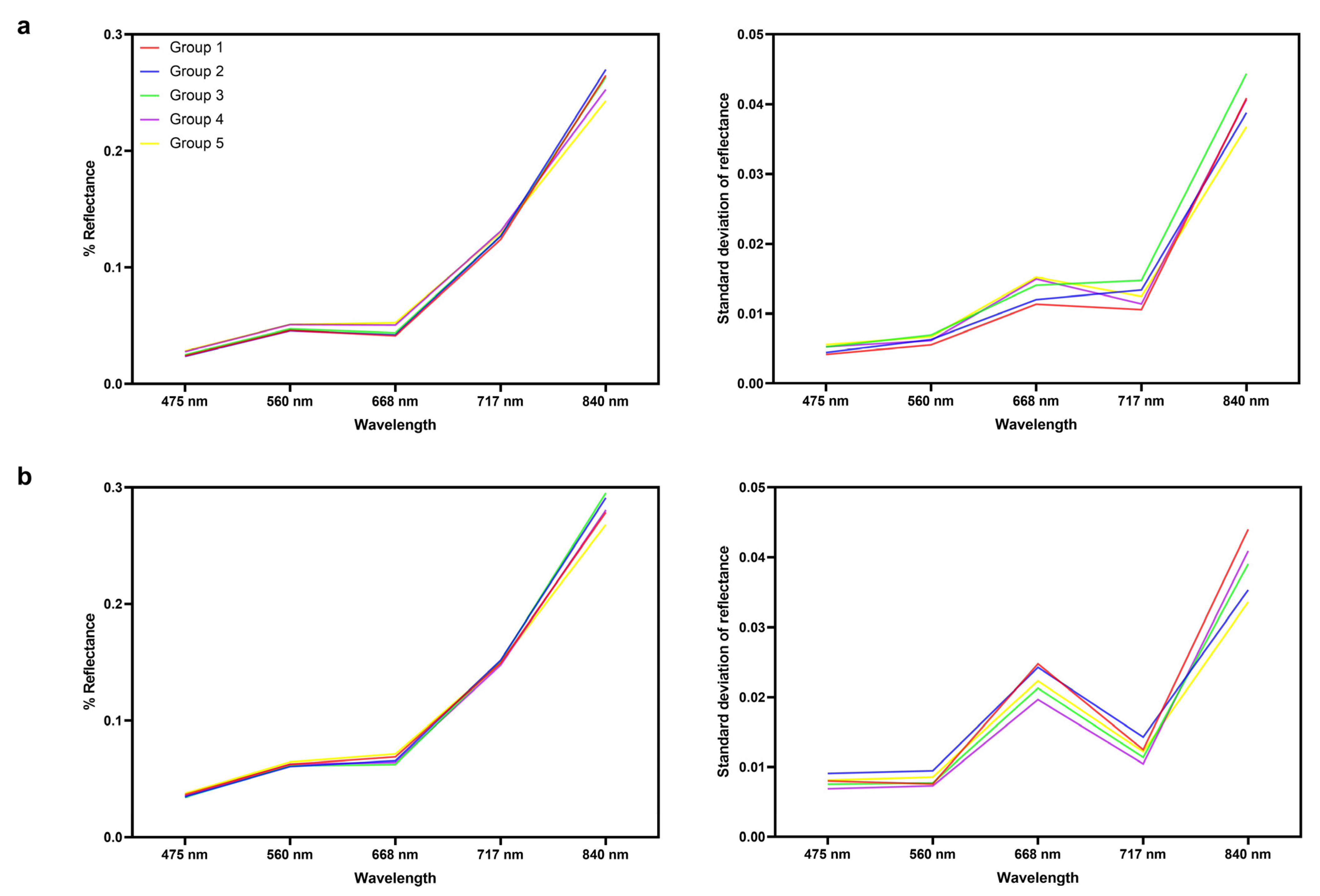
3.1. Spectral Variability among Populations at the Phylogeographic Level
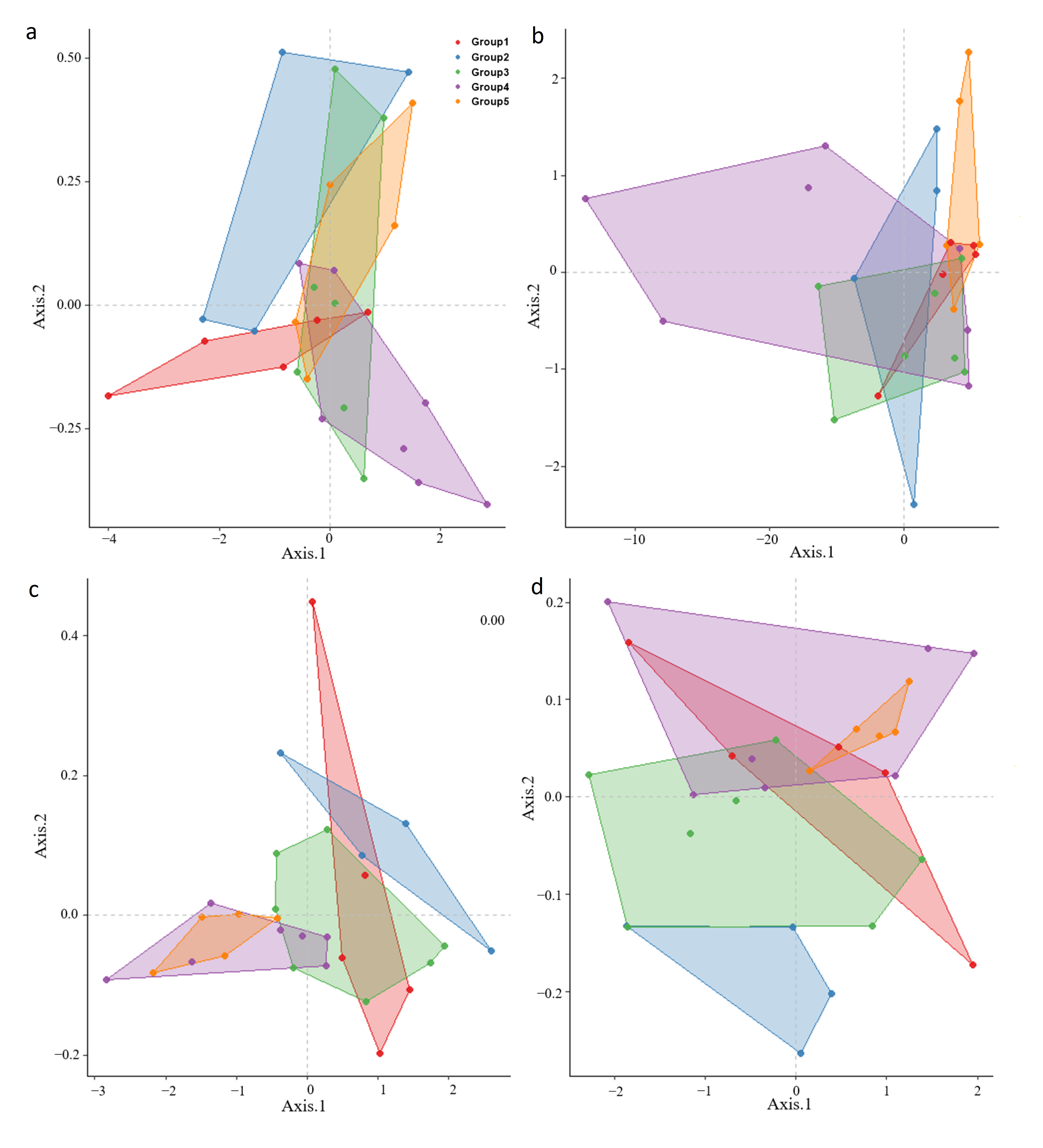
3.2. Classification of Major Genetic Groups Using Spectral Reflectance
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Couture, J.J.; Serbin, S.P.; Townsend, P.A. Spectroscopic sensitivity of real-time, rapidly induced phytochemical change in response to damage. New Phytol. 2013, 198, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.A.; Serbin, S.P.; Kruger, E.L.; Gamon, J.A. Disentangling the contribution of biological and physical properties of leaves and canopies in imaging spectroscopy data. Proc. Natl. Acad. Sci. USA 2013, 110, E1074. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Carranza-Jiménez, L.; Sinca, F.; Tupayachi, R.; Anderson, C.B.; Martinez, P. Functional and biological diversity of foliar spectra in tree canopies throughout the Andes to Amazon region. New Phytol. 2014, 204, 127–139. [Google Scholar] [CrossRef]
- Serbin, S.P.; Singh, A.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Spectroscopic determination of leaf morphological and biochemical traits for northern temperate and boreal tree species. Ecol. Appl. 2014, 24, 1651–1669. [Google Scholar] [CrossRef]
- Yoder, B.J.; Pettigrew-Crosby, R.E. Predicting nitrogen and chlorophyll content and concentrations from reflectance spectra (400–2500 nm) at leaf and canopy scales. Remote Sens. Environ. 1995, 53, 199–211. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef]
- Singh, A.; Serbin, S.P.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecol. Appl. 2015, 25, 2180–2197. [Google Scholar] [CrossRef]
- Gutierrez, M.; Reynolds, M.P.; Klatt, A.R. Association of water spectral indices with plant and soil water relations in contrasting wheat genotypes. J. Exp. Bot. 2010, 61, 3291–3303. [Google Scholar] [CrossRef]
- Fritsche-Neto, R.; Borém, A.; Cobb, J.N. Phenomics; Springer International Publishing: New York, NY, USA, 2015. [Google Scholar]
- Larigauderie, A.; Prieur-Richard, A.-H.; Mace, G.M.; Lonsdale, M.; Mooney, H.A.; Brussaard, L.; Cooper, D.; Cramer, W.; Daszak, P.; Díaz, S.; et al. Biodiversity and ecosystem services science for a sustainable planet: The DIVERSITAS vision for 2012–20. Curr. Opin. Environ. Sustain. 2012, 4, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Alberto, F.J.; Aitken, S.N.; Alía, R.; González-Martínez, S.C.; Hänninen, H.; Kremer, A.; Lefèvre, F.; Lenormand, T.; Yeaman, S.; Whetten, R.; et al. Potential for evolutionary responses to climate change—Evidence from tree populations. Glob. Chang. Biol. 2013, 19, 1645–1661. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.C.; Bocedi, G.; Hendry, A.P.; Mihoub, J.-B.; Peer, G.; Singer, A.; Bridle, J.R.; Crozier, L.G.; Meester, L.D.; Godsoe, W.; et al. Improving the forecast for biodiversity under climate change. Science 2016, 353, aad8466. [Google Scholar] [CrossRef]
- Messer, P.W.; Ellner, S.P.; Hairston, N.G. Can population genetics adapt to rapid evolution? Trends Genet. 2016, 32, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Rosas, U.; Banta, J.; Bhambhra, N.; Purugganan, M.D. Genome-wide patterns of Arabidopsis gene expression in nature. PloS Genet. 2012, 8, e1002662. [Google Scholar] [CrossRef]
- Morinaga, S.; Iwasaki, T.; Suyama, Y. Integrative Observations and Assessments; Springer: Tokyo, Japan, 2014. [Google Scholar]
- Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Hobbie, S.E.; Madritch, M.D.; Wang, R.; Tilman, D.; Gamon, J.A. Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nat. Ecol. Evol. 2018, 2, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Gamon, J.A. Remote sensing of terrestrial plant biodiversity. Remote Sens. Environ. 2019, 231, 111218. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Ustin, S.L.; Roberts, D.A.; Gamon, J.A.; Asner, G.P.; Green, R.O. Using imaging spectroscopy to study ecosystem processes and properties. AIBS Bull. 2004, 54, 523–534. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Vaughnand, N.R.; Llactayo, W. Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation. Science 2017, 355, 385–389. [Google Scholar] [CrossRef]
- Madritch, M.D.; Kingdon, C.C.; Singh, A.; Mock, K.E.; Lindroth, R.L.; Townsend, P.A. Imaging spectroscopy links aspen genotype with below-ground processes at landscape scales. Philosophical Transactions of the Royal Society of London. Series B Biol. Sci. 2014, 369, 20130194. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Meireles, J.E.; Couture, J.J.; Kaproth, M.A.; Kingdon, C.C.; Singh, A.; Serbin, S.P.; Center, A.; Zuniga, E.; Pilz, G.; et al. Associations of leaf spectra with genetic and phylogenetic variation in oaks: Prospects for remote detection of biodiversity. Remote Sens. 2016, 8, 221. [Google Scholar] [CrossRef]
- Meireles, J.E.; Cavender-Bares, J.; Townsend, P.A.; Ustin, S.; Gamon, J.A.; Schweiger, A.K.; Schaepman, M.E.; Asner, G.P.; Martin, R.E.; Singh, A.; et al. Leaf reflectance spectra capture the evolutionary history of seed plant. New Phytol. 2020, 228, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Maeda, M.; Chang, A.; Bhandari, M.; Ashapure, A.; Landivar-Bowles, J. The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr. Opin. Biotechnol. 2021, 70, 15–22. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Tang, H.; Brolly, M.; Zhao, F.; Strahler, A.H.; Schaaf, C.L.; Ganguly, S.; Zhang, G.; Dubayah, R. Deriving and validating Leaf Area Index (LAI) at multiple spatial scales through lidar remote sensing: A case study in Sierra National Forest, CA. Remote Sens. Environ. 2014, 143, 131–141. [Google Scholar] [CrossRef]
- Jay, S.; Maupas, F.; Bendoula, R.; Gorretta, N. Retrieving LAI, chlorophyll and nitrogen contents in sugar beet crops from multi-angular optical remote sensing: Comparison of vegetation indices and PROSAIL inversion for field phenotyping. Field Crops Res. 2017, 210, 33–46. [Google Scholar] [CrossRef]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential biodiversity variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gamon, J.A. Remote sensing of plant functional types. New Phytol. 2010, 186, 795–816. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, A.E.; Warner, T.A.; Fang, F. Implementation of machine-learning classification in remote sensing: An applied review. Int. J. Remote Sens. 2018, 39, 2784–2817. [Google Scholar] [CrossRef]
- Burton, G.W. Breeding Bermuda grass for the Southeastern United States. Agron. J. 1947, 39, 8. [Google Scholar] [CrossRef]
- Casler, M.D.; Duncan, R.R.; Casler, M.D.; Duncan, R.R. Turfgrass biology, genetics and breeding. Q. Rev. Biol. 1976, 51, 286–287. [Google Scholar]
- Wu, Y.Q.; Taliaferro, C.M.; Bai, G.H.; Anderson, M.P. AFLP analysis of Cynodon dactylon (L.) Pers. Var. dactylon genetic variation. Genome 2004, 47, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Q.; Liu, G.D.; Bai, C.J.; Wang, W.Q.; Zhou, S.Y.; Yu, D. Estimation of genetic variation in Cynodon dactylon accessions using the ISSR technique. Biochem. Syst. Ecol. 2010, 38, 993–999. [Google Scholar] [CrossRef]
- Jewell, M.; Frère, C.H.; Harris-Shultz, K.; Anderson, W.F.; Godwin, I.D.; Lambrides, C.J. Phylogenetic analysis reveals multiple introductions of Cynodon species in Australia. Mol. Phylogenetics Evol. 2012, 65, 390–396. [Google Scholar] [CrossRef]
- Zhang, J.X.; Wang, M.L.; Guo, Z.P.; Guan, Y.Z.; Guo, Y.X.; Yan, X.B. Variations in morphological traits of Bermuda grass and relationship with soil and climate along latitudinal gradient. Hereditas 2018, 155, 31. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.L.; Zhang, J.X.; Guo, Z.P.; Guan, Y.Z.; Qu, G.; Liu, J.Y.; Guo, Y.X.; Yan, X.B. Morphological variation in Cynodon dactylon (L.) Pers. And its relationship with the environment along a longitudinal gradient. Hereditas 2020, 157, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Wang, M.L.; Guo, Z.P.; Guan, Y.Z.; Liu, J.Y.; Guo, Y.X.; Yan, X.B. Genetic diversity and population structure of Bermuda grass [Cynodon dactylon (L.) Pers.] along latitudinal gradient and the relationship with polyploidy level. Diversity 2019, 11, 135. [Google Scholar] [CrossRef]
- Zhang, J.X.; Wang, M.L.; Fan, J.; Guo, Z.P.; Guan, Y.Z.; Qu, G.; Zhang, C.J.; Guo, Y.X.; Yan, X.B. Non-linear genetic diversity and notable population differentiation caused by low gene flow of Bermuda grass [Cynodon dactylon (L.) Pers.] along longitude gradient. Peer J. 2021, 9, e11953. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Stamatakis, A. RaxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Smith, G.M.; Milton, E.J. The use of the empirical line method to calibrate remotely sensed data to reflectance. Int. J. Remote Sens. 1999, 20, 2653–2662. [Google Scholar] [CrossRef]
- Liebisch, F.; Kirchgessner, N.; Schneider, D.; Walter, A.; Hund, A. Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods 2015, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Duan, T.; Chapman, S.C.; Guo, Y.; Zheng, B. Dynamic monitoring of NDVI in wheat agronomy and breeding trials using an unmanned aerial vehicle. Field Crops Res. 2017, 210, 71–80. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Blackburn, G.A. Quantifying chlorophylls and 2arotenoids at leaf and canopy scales: An evaluation of some hyperspectral approaches. Remote Sens. Environ. 1998, 66, 273–285. [Google Scholar] [CrossRef]
- Hollberg, J.; Schellberg, J. Distinguishing Intensity Levels of Grassland Fertilization Using Vegetation Indices. Remote Sens. 2017, 9, 81. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Jiang, Z.Y.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Gauch, H.G. Multivariate Analysis in Community Ecology; Cambridge University Press: Cambridge, UK, 1982. [Google Scholar]
- Sammut, C.; Webb, G.I. Encyclopedia of Machine Learning; Springer: Boston, MA, USA, 2010. [Google Scholar]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- F’eret, J.B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]
- Cozzolino, D. Use of Infrared Spectroscopy for In-Field Measurement and Phenotyping of Plant Properties: Instrumentation, Data Analysis, and Examples. Appl. Spectrosc. Rev. 2014, 49, 564–584. [Google Scholar] [CrossRef]
- AOAC. Official Methods and Analysis, 15th ed.; Method 990-03; AOAC: Arlington, VA, USA, 1990. [Google Scholar]
- Zhang, J.X.; Chen, M.H.; Gan, L.; Zhang, C.J.; Shen, Y.; Qian, J.; Han, M.L.; Guo, Y.X.; Yan, X.B. Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China. Plants 2020, 9, 1778. [Google Scholar] [CrossRef]
- Jorde, P.E.; SoVik, G.; Westgaard, J.I.; Albretsen, J.; André, C.; Hvingel, C.; Johansen, T.; Sandvik, A.D.; Kingsley, M.; Jørstad, K.E. Genetically distinct populations of northern shrimp, Pandalus borealis, in the North Atlantic: Adaptation to different temperatures as an isolation factor. Mol. Ecol. 2015, 24, 1742–1757. [Google Scholar] [CrossRef]
- Yamasaki, E.; Altermatt, F.; Cavender-Bares, J.; Schuman, M.C.; Zuppinger-Dingley, D.; Garonna, I.; Schneider, F.D.; Escribà, C.G.; Moorsel, S.J.; Hahl, T.; et al. Genomics meets remote sensing in global change studies: Monitoring and predicting phenology, evolution and biodiversity. Curr. Opin. Environ. Sustain. 2017, 29, 177–186. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Moran, C.J. The AmericaView Classification Methods Accuracy Project: A Rigorous Approach for Model Selection. Remote Sens. Environ. 2015, 170, 115–120. [Google Scholar] [CrossRef]
- Han, L.; Yang, G.; Yang, H.; Xu, B.; Li, Z.; Yang, X. Clustering field-based maize phenotyping of plant-height growth and canopy spectral dynamics using a UAV remote-sensing approach. Front. Plant Sci. 2018, 9, 1638. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.D.; Morsdorf, F.; Schmid, B.; Petchey, O.L.; Hueni, A.; Schimel, D.S.; Schaepman, M.E. Mapping functional diversity from remotely sensed morphological and physiological forest traits. Nat. Commun. 2017, 8, 1441. [Google Scholar] [CrossRef]
- Kursar, T.A.; Dexter, K.G.; Lokvam, J.; Pennington, R.T.; Richardson, J.E.; Weber, M.G.; Murakamia, E.T.; Draked, C.; McGregord, R.; Coleya, P.D. The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proc. Natl. Acad. Sci. USA 2009, 106, 18073–18078. [Google Scholar] [CrossRef]
- Bush, A.; Sollmann, R.; Wilting, A.; Bohmann, K.; Cole, B.; Balzter, H.; Martius, C.; Zlinszky, A.; Calvignac-Spencer, S.; Cobbold, C.A.; et al. Connecting Earth observation to high-throughput biodiversity data. Nat. Ecol. Evol. 2017, 1, 176. [Google Scholar] [CrossRef]
- Shang, J.; Liu, J.; Ma, B.; Zhao, T.; Jiao, X.; Geng, X.; Huffman, T.; Kovacsc, J.M.; Walters, D. Mapping spatial variability of crop growth conditions using RapidEye data in Northern Ontario, Canada. Remote Sens. Environ. 2015, 168, 113–125. [Google Scholar] [CrossRef]
- Dong, T.; Liu, J.; Shang, J.; Qian, B.; Ma, B.; Kovacs, J.M.; Walters, D.; Jiao, X.; Geng, X.; Shi, Y. Assessment of red-edge vegetation indices for crop leaf area index estimation. Remote Sens. Environ. 2019, 222, 133–143. [Google Scholar] [CrossRef]
- Kokaly, F.; Clark, N.; Swayze, A.; Livo, E.; Hoefen, M.; Pearson, C.; Wise, A.; Benzel, M.; Lowers, A.; Driscoll, L.; et al. Usgs Spectral Library Version 7 Data: Us Geological Survey Data Release; United States Geological Survey (USGS): Reston, VA, USA, 2017.
- Hueni, A.; Nieke, J.; Schopfer, J.; Kneubuhler, M.; Itten, K. The spectral database SPECCHIO for improved long-term usability and data sharing. Comput. Geosci. 2009, 35, 557–565. [Google Scholar] [CrossRef]
- Awad, M.M.; Alawar, B.; Jbeily, R. A New Crop Spectral Signatures Database Interactive Tool (CSSIT). Data 2019, 4, 77. [Google Scholar] [CrossRef]


| Spectral Data | Number of Genetic Samples | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Wavelength Range | Spatial Resolution | |
| Hyperspectral leaf data | 446 | 58 | 66 | 118 | 115 | 89 | 400 to 2500 nm | |
| Hyperspectral canopy data | 310 | 39 | 47 | 78 | 83 | 63 | 410 to 1300 nm | Field of view (FOV) of 25° at approximately 30–40 cm height |
| Multispectral day 1 | 445 | 57 | 66 | 118 | 115 | 89 | 475 nm, 560 nm, 668 nm, 717 nm and 840 nm | 8 cm (3.1in) per pixel |
| Multispectral day 2 | 438 | 57 | 65 | 115 | 114 | 87 | 476 nm, 560 nm, 668 nm, 717 nm and 840 nm | 8 cm (3.1in) per pixel |
| Vegetation Index | Reference |
|---|---|
| Indices calculated from multispectral data DSI = NIR − Red | [47] |
| RSI = NIR/Red | [48] |
| NDVI = (NIR − Red)/(NIR + Red) | [49] |
| CI red edge = (NIR/Rededge) − 1 | [50] |
| MTCI = (NIR − Rededge)/(Rededge − Red) | [51] |
| EVI = 2.5 × (NIR − Red)/(NIR+2.4 × Red + 1) | [52] |
| OSAVI = 1.16 × (NIR − Red)/(NIR + Red + 0.16) | [53] |
| Indices calculated from hyperspectral data DSI = R796 − R679 | [47] |
| RSI = R796 / R679 | [48] |
| NDVI = (R796 − R679)/(R796 + R679) | [49] |
| CI red edge = R796/R719 − 1 | [50] |
| MTCI = (R796 − R719)/(R719 + R679) | [51] |
| EVI = 2.5 × (R796 − R679)/(R796 + 2.4 × R679 + 1) | [52] |
| OSAVI = 1.16 × (R796 − R679)/(R796 + R679 + 0.16) | [53] |
| Level | Data | Df | Sum of Squares | Mean of Squares | F Value | p Value |
|---|---|---|---|---|---|---|
| Among populations | Leaf hyperspectral data | 27 | 23.056 | 0.854 | 4.111 | 0.000 |
| Canopy hyperspectral data | 27 | 431.623 | 15.986 | 2.832 | 0.000 | |
| Early multispectral data (17 May) | 27 | 7.014 | 0.260 | 3.109 | 0.000 | |
| Late multispectral data (1 June) | 27 | 5.876 | 0.218 | 2.900 | 0.000 | |
| Among groups | Leaf hyperspectral data | 4 | 7.960 | 1.990 | 8.611 | 0.000 |
| Canopy hyperspectral data | 4 | 107.027 | 26.757 | 4.258 | 0.002 | |
| Early multispectral data (17 May) | 4 | 1.598 | 0.399 | 4.366 | 0.002 | |
| Late multispectral data (1 June) | 4 | 2.490 | 0.622 | 7.640 | 0.000 |
| Classification Error Rates: Mean (SD) | F1 Scores: Mean (SD) | Cohen’s Kappa Scores: Mean (SD) | ||
|---|---|---|---|---|
| Leaf hyperspectral dataset | Among 5 groups | 0.45 (0.02) | 0.52 (0.03) | 0.45 (0.04) |
| Between longitude and latitude | 0.19 (0.01) | 0.81 (0.04) | 0.62 (0.08) | |
| Among 2 groups at longitude | 0.18 (0.01) | 0.80 (0.03) | 0.58 (0.08) | |
| Among 3 groups at latitude | 0.32 (0.03) | 0.61 (0.07) | 0.47 (0.09) | |
| Canopy hyperspectral dataset | Among 5 groups | 0.69 (0.02) | 0.31 (0.06) | 0.16 (0.07) |
| Between longitude and latitude | 0.27 (0.02) | 0.72 (0.05) | 0.43 (0.10) | |
| Among 2groups at longitude | 0.26 (0.03) | 0.74 (0.04) | 0.48 (0.08) | |
| Among 3groups at latitude | 0.57 (0.05) | 0.42 (0.04) | 0.14 (0.05) | |
| Early multispectral dataset (May 17) | ||||
| Among 5 groups | 0.04 (0.02) | 0.96 (0.02) | 0.95 (0.03) | |
| Late multispectral dataset (June 1) | ||||
| Among 5 groups | 0.03 (0.02) | 0.97 (0.02) | 0.96 (0.03) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Han, M.; Wang, L.; Chen, M.; Chen, C.; Shen, S.; Liu, J.; Zhang, C.; Shang, J.; Yan, X. Study of Genetic Variation in Bermuda Grass along Longitudinal and Latitudinal Gradients Using Spectral Reflectance. Remote Sens. 2023, 15, 896. https://doi.org/10.3390/rs15040896
Zhang J, Han M, Wang L, Chen M, Chen C, Shen S, Liu J, Zhang C, Shang J, Yan X. Study of Genetic Variation in Bermuda Grass along Longitudinal and Latitudinal Gradients Using Spectral Reflectance. Remote Sensing. 2023; 15(4):896. https://doi.org/10.3390/rs15040896
Chicago/Turabian StyleZhang, Jingxue, Mengli Han, Liwen Wang, Minghui Chen, Chen Chen, Sicong Shen, Jiangui Liu, Chao Zhang, Jiali Shang, and Xuebing Yan. 2023. "Study of Genetic Variation in Bermuda Grass along Longitudinal and Latitudinal Gradients Using Spectral Reflectance" Remote Sensing 15, no. 4: 896. https://doi.org/10.3390/rs15040896
APA StyleZhang, J., Han, M., Wang, L., Chen, M., Chen, C., Shen, S., Liu, J., Zhang, C., Shang, J., & Yan, X. (2023). Study of Genetic Variation in Bermuda Grass along Longitudinal and Latitudinal Gradients Using Spectral Reflectance. Remote Sensing, 15(4), 896. https://doi.org/10.3390/rs15040896








