Feature Importance Ranking of Random Forest-Based End-to-End Learning Algorithm
Abstract
:1. Introduction
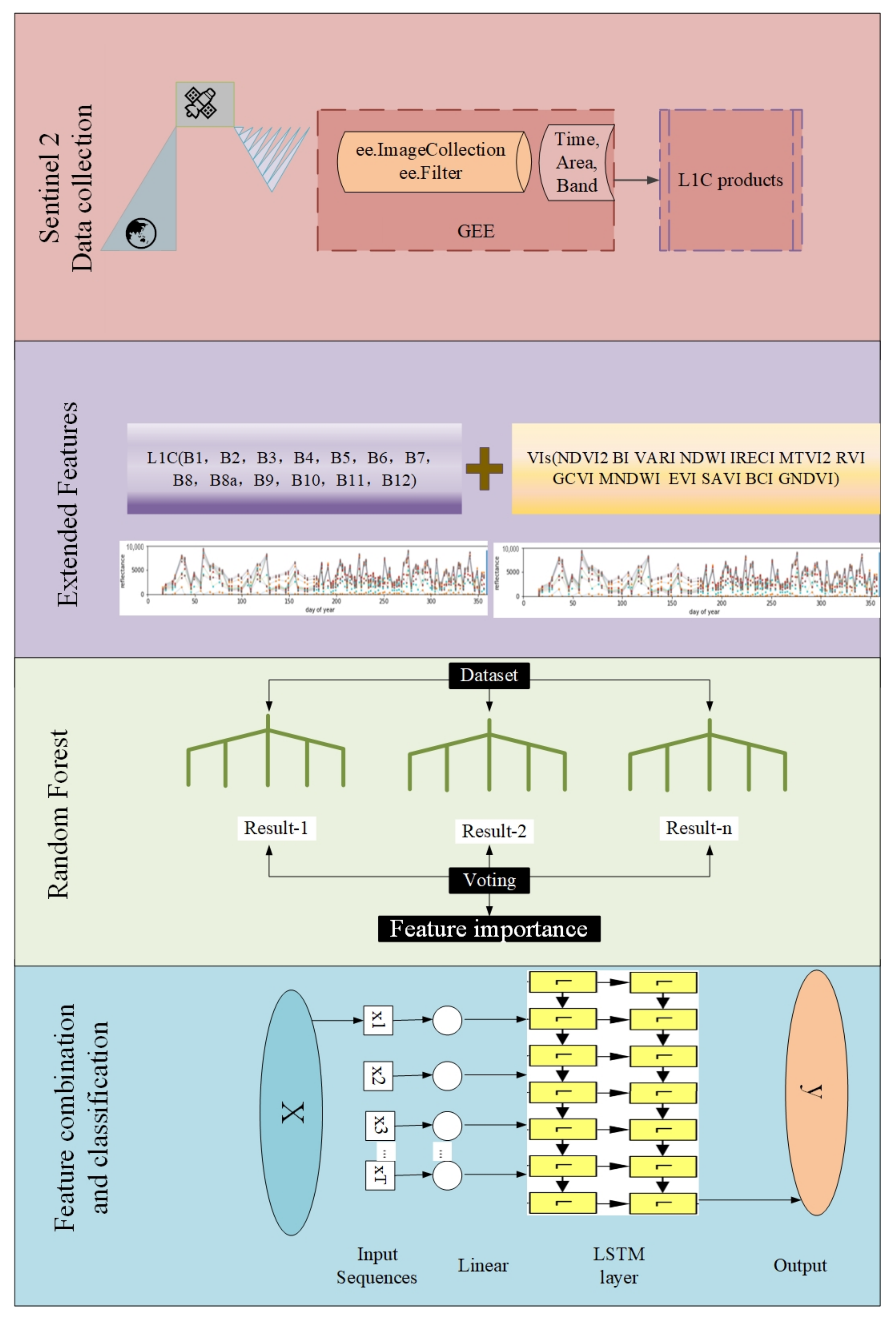
- Feature optimization: vegetation index features such as the normalized vegetation water index (NDWI), ratio vegetation index (RVI), enhanced vegetation index (EVI), and brightness index (BI) have been added to expand the features of each L1C band in the original time series. Then, the RF have been used to score the importance of features, and the important features of each time series have been selected to form a new dataset.
- Classification: Use an end-to-end deep learning model, a two-layer unidirectional LSTM, for training.
2. Study Areas and Data
2.1. Study Areas
2.2. Data
3. Methods
3.1. Sample Composition
3.2. Extended Features
3.3. Feature Importance Optimization
3.4. Classifier
3.5. Experimental Design
3.6. Metrics for Model’s Performance
4. Experimental Results
4.1. Applicability of Extended Features
4.2. Accuracy of Extended Features across Sequence Lengths
4.3. Feature Ranked by RF at Optimal Sequence Lengths
4.4. Availability of Early Prediction
5. Discussion
Model’s Performance Comparison and Analysis
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| B1 | Band 1 |
| B2 | Band 2 |
| B3 | Band 3 |
| B4 | Band 4 |
| B5 | Band 5 |
| B6 | Band 6 |
| B7 | Band 7 |
| B8 | Band 8 |
| B8a | Band 8A |
| B9 | Band 9 |
| B10 | Band 10 |
| B11 | Band 11 |
| B12 | Band 12 |
| BI | Brightness Index |
| BCI | Brightness Composite Index |
| CNN | Convolutional Neural Network |
| DL | Deep Learning |
| EVI | Enhanced Vegetation Index |
| GEE | Google Earth Engine |
| GNDVI | Green Normalized Difference Vegetation Index |
| GCVI | Green Chlorophyll Vegetation Index |
| IRECI | Inverted Red-Edge Chlorophyll Index |
| L1C | Level-1C |
| L2A | Level-2A |
| LSTM | Long Short-Term-Memory |
| MNDWI | Modified Normalized Difference Water Index |
| MODIS | Moderate-Resolution Imaging Spectroradiometer |
| MTVI2 | Modified Triangular Vegetation Index 2 |
| NDVI | Normalized Difference Vegetation Index |
| NDWI | Normalized Vegetation Water Index |
| RF | Random Forest |
| RS | Remote Sensing |
| RVI | Ratio Vegetation Index |
| RNN | Recurrent Neural Network |
| S2 | Sentinel 2 |
| SAVI | Soil Adjusted Vegetation Index |
| TOA | Top-Of-Atmosphere |
| TempCNNs | Temporal Convolutional Neural Networks |
| VIs | Vegetation Indexs |
| VARI | Visible Atmosphere Resistance Index |
| 3-D CNN | Three-Dimensional Convolutional Neural Network |
References
- Wulder, M.A.; Roy, D.P.; Radeloff, V.C.; Loveland, T.R.; Anderson, M.C.; Johnson, D.M.; Healey, S.P.; Zhu, Z.; Scambos, T.A.; Pahlevan, N.; et al. Fifty years of Landsat science and impacts. Remote Sens. Environ. 2022, 228, 113195. [Google Scholar] [CrossRef]
- Emery, W.; Camps, A. Chapter 10—Land Applications. In Introduction to Satellite Remote Sensing; Emery, W., Camps, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 701–766. [Google Scholar] [CrossRef]
- Tong, X.; Zhao, W.; Xing, J.; Fu, W. Status and development of China High-Resolution Earth Observation System and application. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 3738–3741. [Google Scholar] [CrossRef]
- Liang, S. (Ed.) 8.01—Volume 8 Overview: Progress in Ocean Remote Sensing. In Comprehensive Remote Sensing; Elsevier: Oxford, UK, 2018; pp. 1–42. [Google Scholar] [CrossRef]
- Schumann, G.J.P. (Ed.) Chapter 2—An Automatic System for Near-Real Time Flood Extent and Duration Mapping Based on Multi-Sensor Satellite Data. In Earth Observation for Flood Applications; Earth Observation; Elsevier: Amsterdam, The Netherlands, 2021; pp. 7–37. [Google Scholar] [CrossRef]
- Martimort, P.; Arino, O.; Berger, M.; Biasutti, R.; Carnicero, B.; Del Bello, U.; Fernandez, V.; Gascon, F.; Greco, B.; Silvestrin, P.; et al. Sentinel-2 optical high resolution mission for GMES operational services. In Proceedings of the 2007 IEEE International Geoscience and Remote Sensing Symposium, Barcelona, Spain, 23–27 July 2007; pp. 2677–2680. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, J.F.; Qin, Z.; Qin, F. Winter wheat identification by integrating spectral and temporal information derived from multi-resolution remote sensing data. J. Integr. Agric. 2019, 18, 2628–2643. [Google Scholar] [CrossRef]
- Arvor, D.; Jonathan, M.; Meirelles, M.S.P.; Dubreuil, V.; Durieux, L. Classification of MODIS EVI time series for crop mapping in the state of Mato Grosso, Brazil. Int. J. Remote Sens. 2011, 32, 7847–7871. [Google Scholar] [CrossRef]
- Salvatore Praticò, S.; Solano, F.; Di Fazio, S.; Modica, G. Machine Learning Classification of Mediterranean Forest Habitats in Google Earth Engine Based on Seasonal Sentinel-2 Time-Series and Input Image Composition Optimisation. Remote Sens. 2021, 13, 586. [Google Scholar] [CrossRef]
- Peñá-Barragán, J.M.; Ngugi, M.K.; Plant, R.E.; Six, J. Object-based crop identification using multiple vegetation indices, textural features and crop phenology. Remote Sens. Environ. 2011, 115, 1301–1316. [Google Scholar] [CrossRef]
- Brown, J.C.; Kastens, J.H.; Coutinho, A.C.; de Castro Victoria, D.; Bishop, C.R. Classifying multiyear agricultural land use data from Mato Grosso using time-series MODIS vegetation index data. Remote Sens. Environ. 2013, 130, 39–50. [Google Scholar] [CrossRef]
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Conrad, C.; Fritsch, S.; Zeidler, J.; Rücker, G.; Dech, S. Per-Field Irrigated Crop Classification in Arid Central Asia Using SPOT and ASTER Data. Remote Sens. 2010, 2, 1035–1056. [Google Scholar] [CrossRef]
- Zhong, L.; Gong, P.; Biging, G.S. Efficient corn and soybean mapping with temporal extendability: A multi-year experiment using Landsat imagery. Remote Sens. Environ. 2014, 140, 1–13. [Google Scholar] [CrossRef]
- Pelletier, C.; Webb, G.I.; Petitjean, F. Temporal Convolutional Neural Network for the Classification of Satellite Image Time Series. Remote Sens. 2019, 11, 523. [Google Scholar] [CrossRef]
- Fawaz, H.I.; Lucas, B.; Forestier, G.; Pelletier, C.; Schmidt, D.F.; Weber, J.; Webb, G.I.; Idoumghar, L.; Muller, P.; Petitjean, F. InceptionTime: Finding AlexNet for Time Series Classification. arXiv 2019, arXiv:1909.04939. [Google Scholar]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Amato, F.; Guignard, F.; Robert, S.; Kanevski, M.F. A novel framework for spatio-temporal prediction of environmental data using deep learning. Sci. Rep. 2020, 10, 22243. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Meng, S.; Hu, X.; Wang, X.; Zhong, Y. Cropnet: Deep Spatial-Temporal-Spectral Feature Learning Network for Crop Classification from Time-Series Multi-Spectral Images. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 4187–4190. [Google Scholar] [CrossRef]
- Zhou, Y.; Luo, J.; Feng, L.; Yang, Y.; Chen, Y.; Wu, W. Long-short-term-memory-based crop classification using high-resolution optical images and multi-temporal SAR data. GISci. Remote Sens. 2019, 56, 1170–1191. [Google Scholar] [CrossRef]
- Rußwurm, M.; Courty, N.; Emonet, R.; Lefèvre, S.; Tuia, D.; Tavenard, R. End-to-end learned early classification of time series for in-season crop type mapping. ISPRS J. Photogramm. Remote Sens. 2023, 196, 445–456. [Google Scholar] [CrossRef]
- Rußwurm, M.; Lefèvre, S.; Körner, M. BreizhCrops: A Satellite Time Series Dataset for Crop Type Identification. arXiv 2019, arXiv:1905.11893. [Google Scholar]
- Rußwurm, M.; Körner, M. Self-attention for raw optical Satellite Time Series Classification. ISPRS J. Photogramm. Remote Sens. 2020, 169, 421–435. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. 1974. Available online: https://ntrs.nasa.gov/citations/19740022614 (accessed on 1 January 2023).
- EOS Data Analytics. 2022. Available online: https://eos.com/blog/vegetation-indices (accessed on 1 January 2023).
- Gao, B.C. NDWI-A Normalized Difference Water Index for Remote Sensing of Vegetation Liquid Water from Space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.; Pattey, E.; Zarco-Tejada, P.; Strachan, I. Hyperspectral vegetation indices and Novel Algorithms for Predicting Green LAI of crop canopies: Modeling and Validation in the Context of Precision Agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.; Keydan, G.P.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, Y.; Ling, F.; Wang, Q.; Li, W.; Li, X. Water Bodies Mapping from Sentinel-2 Imagery with Modified Normalized Difference Water Index at 10-m Spatial Resolution Produced by Sharpening the SWIR Band. Remote Sens. 2016, 8, 354. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.; Gao, X.; Ferreira, L. Overview of the Radiometric and Biophysical Performance of the MODIS Vegetation Indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Nedkov, R. Orthogonal transformation of segmented images from the satellite sentinel-2. Comptes Rendus L’Académie Bulg. Sci. Sci. Math. Nat. 2017, 70, 687–692. [Google Scholar]
- Gitelson, A.; Kaufman, Y.; Merzlyak, M. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Pasquarella, V.J.; Holden, C.E.; Woodcock, C.E. Improved mapping of forest type using spectral-temporal Landsat features. Remote Sens. Environ. 2018, 210, 193–207. [Google Scholar] [CrossRef]
- Teluguntla, P.G.; Thenkabail, P.S.; Oliphant, A.J.; Xiong, J.; Gumma, M.K.; Congalton, R.G.; Yadav, K.; Huete, A.R. A 30-m landsat-derived cropland extent product of Australia and China using random forest machine learning algorithm on Google Earth Engine cloud computing platform. ISPRS J. Photogramm. Remote Sens. 2018, 144, 325–340. [Google Scholar] [CrossRef]
- Pelletier, C.; Valero, S.; Inglada, J.; Champion, N.; Dedieu, G. Assessing the robustness of Random Forests to map land cover with high resolution satellite image time series over large areas. Remote Sens. Environ. 2016, 187, 156–168. [Google Scholar] [CrossRef]
- Józefowicz, R.; Zaremba, W.; Sutskever, I. An Empirical Exploration of Recurrent Network Architectures. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Sak, H.; Senior, A.W.; Beaufays, F. Long Short-Term Memory Based Recurrent Neural Network Architectures for Large Vocabulary Speech Recognition. arXiv 2014, arXiv:1402.1128. [Google Scholar]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer Normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]






| Authors | Method | Highlights |
|---|---|---|
| Conrad et al. (2010) [13] | The tassels cap indices greenness (representing the density of green vegetation cover) and brightness (soil moisture) | Using very high-resolution satellite data to define field boundaries; Multi-temporal medium-resolution satellite data were classified to distinguish between crops and crop rotations within each field object. |
| Brown et al. (2013) [11] | NDVI | The 5-year classification accuracy is over 80% under optimal conditions. Year-to-year changes in crop phenology highlight the need for multi-year studies. |
| Arvor et al. (2011) [11] | EVI | These classes represent agricultural practices involving three commercial crops (soybean, maize and cotton) planted in single or double cropping systems. |
| Zhong et al. (2014) [14] | EVI | Using phenology; Phenological indices improve the scalability of the random forest classifier. |
| Peñá-Barragán et al. (2011) [10] | 12 VIs | Texture features improve the discrimination of heterogeneous permanent crops; Information from NIR and SWIR bands is required for detailed crop identification. |
| Salvatore et al. (2021) [9] | 3 VIs | Exploiting GEE. |
| Kussul et al. (2017) [17] | CNNs | Using Landsat-8 and Sentinel-1A time series; High accuracy. |
| Luo et al. (2020) [19] | LSTM | High accuracy. |
| Zhou et al. (2019) [20] | 3-D CNN named CropNet | High accuracy. |
| Train | Validate | Test | Crops | |
|---|---|---|---|---|
| BavarianCrops | 16,600 | 3057 | 7813 | 7: meadow, summer barley, corn, winter, wheat, winter barley, clover, and winter triticale. |
| BreizhCrops | 21,000 | 4000 | 3000 | 9: barley, wheat, rapeseed, corn, sunflowers, orchards, nuts, permanent grass, and temporary grass |
| Feature Variables | Calculation Formula | Resolution (m) |
|---|---|---|
| NDVI2 [24] | 10 | |
| BI [23] | 10 | |
| VARI [25] | 10 | |
| NDWI [26] | 20 | |
| IRECI [23] | 20 | |
| MTVI2 [27] | 20 | |
| RVI [25] | 10 | |
| GCVI [28] | 10 | |
| MNDWI [29] | 20 | |
| EVI [30] | 10 | |
| SAVI [31] | 10 | |
| BCI [32] | 10 | |
| GNDVI [33] | 10 |
| BavarianCrops | Feature Combinations | Feature Numbers | OA | |
|---|---|---|---|---|
| Sequencelength | ||||
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, SAVI, BCI, GNDVI] | 26 | 0.8760 | |
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, SAVI, BCI, GNDVI] | 25 | 0.8639 | |
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, SAVI, GNDVI] | 24 | 0.8617 | |
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, GNDVI] | 23 | 0.8534 | |
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI] | 22 | 0.8541 | |
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI] | 21 | 0.8582 | |
| 65 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B9, NDVI2, BI, VARI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 20 | 0.8616 | |
| 65 | [B1, B10, B11, B12, B2, B4, B5, B6, B7, B9, NDVI2, BI, VARI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 19 | 0.8600 | |
| 65 | [B1, B10, B11, B12, B2, B4, B5, B6, B7, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 18 | 0.8513 | |
| 65 | [B1, B10, B11, B12, B2, B4, B5, B6, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 17 | 0.8662 | |
| 65 | [B1, B10, B11, B12, B2, B4, B5, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 16 | 0.8623 | |
| 65 | [B1, B10, B11, B12, B2, B4, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 15 | 0.8613 | |
| 65 | [B1, B10, B11, B12, B4, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 14 | 0.8563 | |
| 65 | [B1, B10, B11, B4, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 13 | 0.8593 | |
| 65 | [B1, B10, B11, B4, B9, NDVI2, BI, MTVI2, RVI, GCVI, MNDWI, EVI] | 12 | 0.8521 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, SAVI, BCI, GNDVI] | 26 | 0.8000 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, SAVI, BCI, GNDVI] | 25 | 0.7962 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, SAVI, GNDVI] | 24 | 0.7960 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI, GNDVI] | 23 | 0.7913 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B8A, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI] | 22 | 0.7820 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B9, NDVI2, BI, VARI, NDWI, IRECI, MTVI2, RVI, GCVI, MNDWI, EVI] | 21 | 0.7801 | |
| 50 | [B1, B10, B11, B12, B2, B3, B4, B5, B6, B7, B9, NDVI2, BI, VARI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 20 | 0.7763 | |
| 50 | [B1, B10, B11, B12, B2, B4, B5, B6, B7, B9, NDVI2, BI, VARI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 19 | 0.7712 | |
| 50 | [B1, B10, B11, B12, B2, B4, B5, B6, B7, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 18 | 0.7709 | |
| 50 | [B1, B10, B11, B12, B2, B4, B5, B6, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 17 | 0.7660 | |
| 50 | [B1, B10, B11, B12, B2, B4, B5, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 16 | 0.7615 | |
| 50 | [B1, B10, B11, B12, B2, B4, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 15 | 0.7596 | |
| 50 | [B1, B10, B11, B12, B4, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 14 | 0.7563 | |
| 50 | [B1, B10, B11, B4, B9, NDVI2, BI, NDWI, MTVI2, RVI, GCVI, MNDWI, EVI] | 13 | 0.7523 | |
| 50 | [B1, B10, B11, B4, B9, NDVI2, BI, MTVI2, RVI, GCVI, MNDWI, EVI] | 12 | 0.7500 | |
| Dataset | Minimum Sequence Length | Precision | Recall | Fscore | Kappa |
|---|---|---|---|---|---|
| BavarianCrops | 65 | 0.796 | 0.735 | 0.754 | 0.815 |
| BreizhCrops | 50 | 0.552 | 0.535 | 0.540 | 0.734 |
| Dataset | Classifier | Sequence Length | Feature Numbers | OA | Fscore |
|---|---|---|---|---|---|
| BavarianCrops | RF | 70 | 13 | 0.65 | 0.56 |
| LSTM | 70 | 13 | 0.86 | 0.77 | |
| BreizhCrops | RF | 70 | 13 | 0.62 | 0.61 |
| LSTM | 70 | 13 | 0.80 | 0.74 |
| Dataset | Classifier | Sequence Length | Feature Numbers | OA | Fscore |
|---|---|---|---|---|---|
| BavarianCrops | RNN | 65 | 26 | 0.80 | 0.71 |
| LSTM | 65 | 26 | 0.87 | 0.79 | |
| BreizhCrops | RNN | 50 | 26 | 0.76 | 0.68 |
| LSTM | 50 | 26 | 0.80 | 0.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Liu, S.; Feng, W.; Dauphin, G. Feature Importance Ranking of Random Forest-Based End-to-End Learning Algorithm. Remote Sens. 2023, 15, 5203. https://doi.org/10.3390/rs15215203
Yuan X, Liu S, Feng W, Dauphin G. Feature Importance Ranking of Random Forest-Based End-to-End Learning Algorithm. Remote Sensing. 2023; 15(21):5203. https://doi.org/10.3390/rs15215203
Chicago/Turabian StyleYuan, Xiaoguang, Shiruo Liu, Wei Feng, and Gabriel Dauphin. 2023. "Feature Importance Ranking of Random Forest-Based End-to-End Learning Algorithm" Remote Sensing 15, no. 21: 5203. https://doi.org/10.3390/rs15215203
APA StyleYuan, X., Liu, S., Feng, W., & Dauphin, G. (2023). Feature Importance Ranking of Random Forest-Based End-to-End Learning Algorithm. Remote Sensing, 15(21), 5203. https://doi.org/10.3390/rs15215203






