Abstract
Accurate detection and delineation of individual trees and their crowns in dense forest environments are essential for forest management and ecological applications. This study explores the potential of combining leaf-off and leaf-on structure from motion (SfM) data products from unoccupied aerial vehicles (UAVs) equipped with RGB cameras. The main objective was to develop a reliable method for precise tree stem detection and crown delineation in dense deciduous forests, demonstrated at a structurally diverse old-growth forest in the Hainich National Park, Germany. Stem positions were extracted from the leaf-off point cloud by a clustering algorithm. The accuracy of the derived stem co-ordinates and the overall UAV-SfM point cloud were assessed separately, considering different tree types. Extracted tree stems were used as markers for individual tree crown delineation (ITCD) through a region growing algorithm on the leaf-on data. Stem positioning showed high precision values (0.867). Including leaf-off stem positions enhanced the crown delineation, but crown delineations in dense forest canopies remain challenging. Both the number of stems and crowns were underestimated, suggesting that the number of overstory trees in dense forests tends to be higher than commonly estimated in remote sensing approaches. In general, UAV-SfM point clouds prove to be a cost-effective and accurate alternative to LiDAR data for tree stem detection. The combined datasets provide valuable insights into forest structure, enabling a more comprehensive understanding of the canopy, stems, and forest floor, thus facilitating more reliable forest parameter extraction.
1. Introduction
Precise forest parameters play a pivotal role in facilitating efficient forest management and conservation [1]. Understanding the structure, health, and composition of a forest stand not only improves its management but also enables effective forecasting of future trends. This becomes especially crucial due to the increasing stressors on forest ecosystems induced by the effects of climate change and human-driven land use conversions, which lead to a decline in forest health and coverage in several parts of the world [2,3]. In particular, managing at the individual tree level is gaining increasing importance for conducting targeted interventions, guiding forest development, and ensuring sustainable forest management under the intensifying impacts of climate change and usage pressure [4].
With the growing availability of earth observation data and techniques, forest monitoring from local to global scales is becoming feasible. Satellite remote sensing data, with their respective spectral and temporal resolution, can be utilized for a wide range of forest applications [5]. However, their coarser pixel size limits their use for forest monitoring analysis on an individual tree level. Light Detection and Ranging (LiDAR) techniques, such as mobile laser scanning (MLS), have been shown to provide high-resolution data with detailed information on the structure of individual trees (please refer to the Abbreviations) [6,7]. Nonetheless, the use of MLS is impractical for large areas, and its cost-intensive equipment makes it inaccessible to many users. Although LiDAR tools mounted on unoccupied aerial vehicles (UAV) can overcome some of the spatial limitations of MLS and generate high-density forest point clouds [8,9], their accessibility within the forestry sector is still limited due to cost constraints.
For local forest monitoring, UAVs equipped with continuously improving optical camera systems have demonstrated their potential as a cost-effective and user-friendly tool for deriving various forest parameters [10,11]. Operating at low flight altitudes, they provide images with very high geometrical resolution of a few centimeters, enabling analysis on individual tree level and even capturing within-variability of single tree crowns [12]. UAV data have, therefore, been used to derive a range of forest parameters quantifying the vertical and horizontal forest structure—such as tree height, breast height diameter, crown shapes, and canopy gaps—categorizing tree species, estimating tree health and above-ground biomass, or detecting diseases [13,14,15].
1.1. UAV Imagery Processing Using Structure from Motion
One of the great advantages of UAV data is their ability to generate 3D information, such as 3D point clouds, from 2D drone imagery applying photogrammetric processing steps, commonly known as structure from motion (SfM) [16]. During data acquisition, highly overlapping images are captured, providing different perspectives on the same ground spots. Prominent feature points are extracted from each image, and features corresponding to the same 3D point are matched in the overlapping regions of different images. Aerial triangulation, such as bundle adjustment, is then applied to define camera positions and orientations, as well as to obtain 3D geometry, creating tracks from a set of matched features. Based on these estimated camera and image positions, densification algorithms (dense stereo matching) can be used to generate dense 3D point clouds. From the point cloud, a digital surface model (DSM) can be derived and, by projecting the single images using the DSM, an orthomosaic can be generated [16,17]. These data products can be further used to extract specific forest parameters. The UAV-SfM approach has the potential to provide both geometric and spectral datasets, serving as input data for various forest parameter extraction algorithms. While methods integrating geometrical point and spectral image data are increasingly used in the field of forestry, most studies rely on LiDAR data rather than on UAV-SfM point clouds [18,19].
1.2. UAV-Data-Based Products for Tree Crown Delineation
In order to analyze tree parameters on an individual tree level within a forest stand, it is necessary to first segment single trees. This involves delineating the projected tree crown, which can be identified in the orthomosaic or height models, as separate objects. This individual tree crown delineation (ITCD) serves as the foundation for various subsequent analysis steps, including tree species classification, environmental and forest monitoring at the tree level, and the extraction of individual tree parameters directly from remote sensing data [20,21,22,23]. Over the past few decades, numerous ITCD methods have developed, based on generalized characteristics of trees or forests. Most of these methods can be applied to either 2D raster such as orthomosaic, 2.5D raster such as canopy height models (CHMs), or 3D data products such as point clouds. However, there are certain methods that specifically rely on 3D data and cannot be used when only 2D data are available. These methods often originate from the field of laser scanning and are increasingly being tested for point clouds derived from SfM as well [24].
Most methods typically assume a similar—hemispherical to conical—tree shape, with one tree top located at the center of the crown. Due to its exposed position, the top of the tree receives the highest solar radiation, resulting in the highest intensity and brightness values [20,22]. Algorithms are expected to yield better results for forests with a sparse canopy, lower species diversity, and similar age structure. These characteristics are more commonly found in managed forests and in coniferous, savannas, or tundra forest systems [22].
The analysis is often divided into two parts: tree detection, which involves identifying the position of a tree trunk, and delineating the (entire) tree crown [20]. In the following, some of the more commonly used methods for tree detection and delineation will be presented.
Local maxima and region growing: This method builds upon the previously mentioned canopy characteristics. Initially, individual trees are detected by identifying local maxima, which ideally represent tree midpoints. These maxima can be based on both CHM values and brightness values [20,22]. Difficulties may arise when defining the search radius for local maxima, which is derived from pixel size and average crown diameter and due to the fact that, in reality, tree crowns are not symmetrically aligned around a central point. Smoothing filters can reduce unwanted noise within the maxima [23,25,26,27]. Wulder et al. [28] propose using varying local maxima search radii, each based on the semivariance of the pixel.
Starting from these initial seed points, neighboring pixels or objects that exhibit similarity are added to the crown objects until a termination criterion is met, indicating that the crown has been delineated. This process is known as region growing [20].
Valley-following approach: The valley-following approach, initially introduced by Gougeon [29], consists of two parts. The first part involves classifying the areas between the individual tree canopies, while the second part utilizes a rule-based method to refine these classified areas [29]. These intermediate areas are referred to as valleys. They are characterized by higher shading, resulting in lower intensity and brightness values compared to the surrounding areas or represent local minima values [25,30]. According to Ke and Quackenbush [20] and Workie [23], relying solely on this shade-following approach often leads to incomplete separation of individual trees because, depending on tree density, not all crowns are adequately separated from each other by “valleys”.
Watershed segmentation: The watershed segmentation, first described by Beucher and Lantuéjoul [31], is a form of image processing segmentation. It also draws upon the topological analogy of the canopy described earlier. In this method, the values of a gray-scale layer are inverted and “flooded”, starting from local minima (tree tops). The resulting individual watersheds are separated from each other by dams, creating distinct segments [21,22,32]. Markers can be used in this process, representing the local minima from which the “flooding” originates, ideally representing tree tops. The resulting segments are supposed to delimit the individual tree tops [23]. According to Derivaux et al. [32], a common challenge in this method is over-segmentation, which can be mitigated through targeted marker placement, application of smoothing filters, or through the combined use with region-merging techniques and other methods.
Template matching: Template matching is a method that can be employed to detect individual tree crowns when the tree crowns exhibit similar shapes and comparable spectral values. Templates are created based on a gray value layer, representing patterns of typical tree shapes and values, mostly averaged. Both radiometric and geometric properties of the crown are utilized, and different viewing angles can be considered. The template is compared to all possible tree points, with high correlation values above a defined threshold representing individual trees [21,22,25,33].
Deep learning methods: Deep learning methods, such as instance segmentation, are increasingly being utilized for the detection and delineation of single tree crowns [34]. Commonly used model architectures are Mask R-CNN [35], artificial neural networks [15], or U-nets [36]. These methods offer advantages such as the ability to use multi-band images as input, instead of relying solely on a single band, and a focus on textural features, which is beneficial as adjacent trees might have very similar spectral properties. However, a disadvantage is the need for training data, which are typically obtained through time-consuming manual crown delineation and/or field work.
Point cloud-based methods: Terrestrial/airborne laser scanning and UAV-SfM provide 3D point cloud data that can be directly used for the single crown delineation. Most studies focusing on 3D methods utilize LiDAR data, as UAV-SfM does not penetrate dense crown structures well, especially during leaf-on season. As a result, UAV-SfM leaf-on point clouds tend to be less dense for lower canopy and forest floor areas. Various point-cloud-based approaches have been applied for ITCD, including K-means clustering [37], template matching [38], voxel space approaches [39], and mean-shift segmentation [40]. Deep learning models are trained using reference point clouds and have been used to segment single trees [39], employing frameworks such as PointNet [41].
Several recent studies applied one or multiple of the above-mentioned methods to detect tree positions and perform crown delineation using UAV-based imagery data products. A selection of relevant studies is presented in Table 1.

Table 1.
Selection of recent studies on tree detection and crown delineation in forest ecosystems using UAV optical data.
As emphasized by several of these studies, detection and delineation of tree crowns in complex forest structures with dense canopy closure and overlapping tree crowns remains challenging [35,51]. Some studies focus on ITCD in forest with low to moderate canopy closure or open forest stands [42,48,53], as well as on forest plantations characterized by a more regular tree spacing and structure [44,46]. These types of forest tend to facilitate the detection of crown boundaries and the number of trees, as most algorithms perform better in homogeneous forest stands with lower canopy closure [19].
The objective of this study was to develop a method that focuses on the utilization and combination of leaf-on and leaf-off UAV-SfM data to derive stand parameters such as stem positions and crown delineation of individual trees within dense, structurally diverse deciduous forest environments. An approach was devised to extract stem positions using the leaf-off point cloud. These stem co-ordinates were subsequently combined with leaf-on datasets to derive crown delineations. Furthermore, a systematic accuracy assessment has been implemented to distinguish error sources in stem detection. This assessment involves comparing the extracted stem positions, a manually digitized UAV stem position dataset, and an independent MLS-based reference dataset.
2. Materials and Methods
Figure 1 illustrates the overall workflow that was developed and applied in this study. UAV RGB imagery of the forest was acquired twice: once on 4 August 2021 during leaf-on conditions (May–October) and once on 29 March 2022 during leaf-off conditions (November–April). Using an SfM approach, point clouds, orthomosaic, and a DSM were computed. A ground classification from the leaf-off data was used to normalize the point cloud and generate a digital terrain model (DTM). From the DTM and DSM, a CHM was extracted. A slice of the normalized leaf-off point cloud is used as input data for the tree stem detection method. Different reference datasets have been acquired for accuracy assessment. The ability of the UAV data and the proposed algorithm to detect stems has also been analyzed based on the respective tree type, such as overstory, understory, or tilted trees. In a second step, the extracted stem co-ordinates, leaf-on orthomosaic, and CHM are used as markers to derive individual crown delineation through a region growing algorithm.
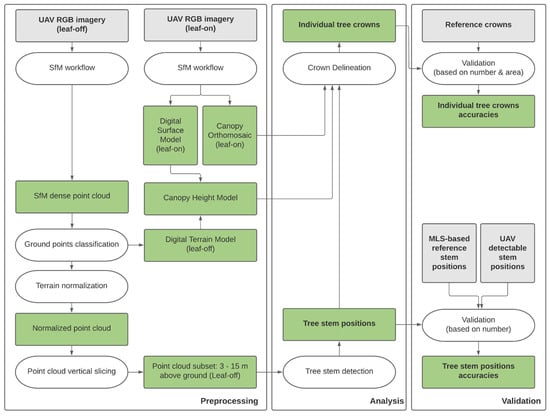
Figure 1.
General workflow of the methodology combining leaf-off and leaf-on UAV-SfM datasets to generate tree stem positions and individual tree crowns. Color coding: grey = input data, green = intermediate and final data products, white = processing steps.
In the following paragraphs, the workflow will be described in detail, outlining the steps taken to acquire and process the UAV imagery, collect reference data, derive stem positions and ITCD, and validate the obtained results.
2.1. Study Site Hainich National Park
The present analysis was conducted within the Hainich National Park in Thuringia, central Germany (Figure 2). The study area, known as “Huss site”, is located within the forest stand of the national park and has an area of 28.2 ha with an average tree cover density of 91% [54]. The center of the area is located at 10°26′5″E and 51°4′47″N. Within the test side Huss, a wide range of ecological investigations are conducted and several instruments for long-term forest monitoring have been established [55]. Surrounding the national park is the Hainich forest hill chain, which spans an area of approximately 20,000 ha. To the south, the mountain range borders the Thuringian Forest. To the east, it slopes towards the Thuringian Basin, while, to the west, it slopes relatively steeply to the valley of the Werra River. The elevations in the area range between 225 and 493 m a.s.l. [55,56,57].
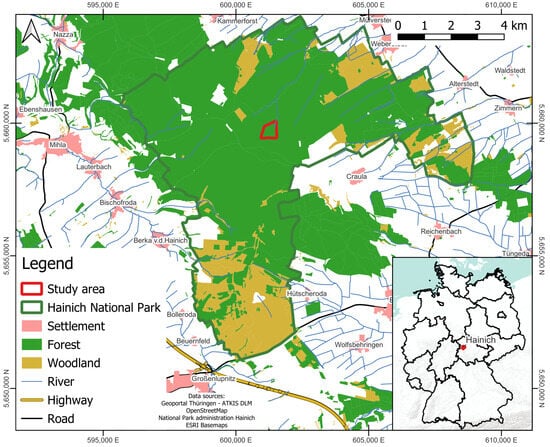
Figure 2.
Overview of the study site “Huss” within the Hainich National Park located in the state of Thuringia in central Germany. The co-ordinate system used for all figures is ETRS89/UTM 32N (EPSG: 25832).
The Hainich National Park covers an area of 7520 ha and includes the southern part of the natural area. It was established in December 1997 [58] and has been a UNESCO World Heritage Site of ancient and primeval beech forests of the Carpathians and other regions of Europe since June 2011. The national park area has been used for military purposes since 1935, leading to large-scale clearing in some areas. Due to military use, there were almost no other human factors of influence. Following the withdrawal of the Soviet army in 1991, the area became a succession area where reforestation is taking place without human regulation [59]. The current objective of the national park is to allow the natural dynamics to shape these forest areas and to study them. As a result, 90% of the national park is free from anthropogenic use and subject to natural processes [56,59]. The majority of the Huss site is covered by deciduous forests; based on an inventory by the Hainich National Park administration from 2013, it is primarily composed of beech trees (Fagus sylvatica) accounting for 78% of the trees. Other tree species interspersed in the forest include sycamore (Acer pseudoplatanus) with 12% share, ash (Fraxinus excelsior) with 5% share, hornbeam (Carpinus betulus) with 2% share, Norway maple (Acer platanoides) with 2% share, wych elm (Ulmus glabra), and oak (Quercus spec.). As per the inventory, the mean diameter at breast height (DBH) of the predominant and dominant stock is 56.77 cm, while the mean diameter of all trees (with a DBH greater than 7 cm) is 32.20 cm. Since 2018, the area has experienced severe drought events, particularly during the vegetation periods, that have caused water stress and widespread tree damage events on localized scales [60]. The impact extends not only to the forest structure but also to the diverse fauna, which includes approximately 10,000 species dependent on the forest habitat. Notable examples include the European wildcat, roe deer, wild boar, and various bat species [56].
2.2. Acquisition and Preprocessing of UAV Imagery
UAV images were acquired for the study area at two time points: 4 August 2021 (leaf-on) and 29 March 2022 (leaf-off). Both datasets were captured using a DJI (Da-Jiang Innovations Science and Technology Co., Ltd., Shenzhen, China) Phantom 4 Pro equipped with a real-time kinematic (RTK) receiver and an RGB camera. The camera features a 1-inch CMOS sensor with 20 MP. It utilizes a wide-angle lens with a focal length of 24 mm (equivalent to 35 mm) and a field of view spanning 84°. The mechanical shutter speed ranges from 8 to 1/2000 s, and the supported data format is JPEG [61]. Throughout the entire flight period, a correction signal from a reference station was received via mobile internet connection using the German satellite positioning service (SAPOS). This enabled highly accurate real-time positioning with errors in the range of a few centimeters, eliminating the need for ground control points [11].
The acquisition and camera parameters for the two UAV missions are summarized in Table 2. The leaf-on data consist of a single overflight of the area, capturing images from a nadir perspective. The leaf-off dataset comprises three overflights of the area: one from a nadir perspective and two using an oblique camera perspective (70° or 20° off-nadir), each taken from an opposite flight direction. Oblique flights were included as they provide additional viewing angles on tree stems and forest floor, and ensure a more complete point cloud. For all flights, a standard parallel-flight line pattern was chosen. These flight parameters were selected with the aim of obtaining comprehensive information about the forest stand and its components (canopy, stems, and forest floor) during the respective acquisition date. The flux tower within the Hainich National Park [62] was selected as take-off and landing spot to ensure a direct line of sight throughout the flights.

Table 2.
UAV mission and acquisition parameters (in bold). Wind speed was measured at the weather station Weberstedt/Hainich located 5 km to the NE of the test site. The mission footprint (i.e., the entire area covered by the UAV campaign) is noticeably larger than the Huss site because the flight planning accounted for a buffer surrounding the site. Abbreviation: ISO = International Organization for Standardization.
The UAV images were preprocessed using an SfM workflow in the software Agisoft Metashape (version 1.8.0). An overview of the applied parameters can be found in Table 3. The RGB images from both flight campaigns were processed without prior modification, resulting in 3D point clouds and orthomosaics (raster) for both the leaf-off and the leaf-on season. All images of the leaf-off flights (nadir and oblique) were processed together within a single SfM workflow. The orthomosaics were resampled to a geometric resolution of 0.05 m. The ETRS89/UTM 32N co-ordinate system (EPSG: 25832) was employed for the entire study and all associated data. The geometric accuracy of the processing data products was assessed through visual inspection to ensure they do not contain any unwanted artifacts. Additionally, calculated errors, such as the average error of camera positions and the reprojection error, were examined. The results indicate good geometric accuracy and a very high alignment of the bitemporal data products (Table 3).

Table 3.
Parameters of the Structure from Motion (SfM) processing steps (Agisoft Metashape) (in bold) using the UAV RGB imagery as input.
The leaf-off point cloud was used to perform a ground classification, detecting points belonging to the forest floor. Out of the 174 million points of the leaf-off point cloud, 28 million were classified as ground points. Using these ground points, the leaf-off point cloud was terrain normalized. For accuracy assessment, the ground classification was compared to the elevation of the airborne lidar dataset from the Thuringian State Office of Land Management and Geological Information, which was acquired in 2017 and has a resolution of 1 m. The ground points from both datasets were converted to raster layers with a resolution of 5 m. Pixel-wise differences were calculated, resulting in an overall RMSE of 0.450 m.
The elevation of the classified leaf-off ground points (DTM) and the leaf-on canopy points (DSM) were used to generate a CHM with a geometric resolution of 0.1 m. All the processing steps were carried out using the software LASTools (version 211112). Figure 3 illustrates a profile cross-section of the two point clouds and their combination. UAV leaf-off data alone from the same study site two years earlier (2019) have already been used by Thiel et al. [63] to delineate dead wood debris and have been proven to be suitable for deriving information from the forest floor.

Figure 3.
Profile cross-section of the normalized point clouds of the study area in the Hainich National Park generated from UAV RGB images applying an SfM approach. The upper section shows the leaf-off point cloud from 29 March 2022, the middle section the leaf-on point cloud from 4 August 2021, and the lower section the combination of both datasets.
2.3. Reference Data
- (a)
- Inventory by the Hainich National Park administration
The Huss site was manually inventoried by the Hainich National Park Administration in 2013 during a field campaign. The data include the number of all trees for the entire study site, along with various tree parameters. A subset of the inventory was used, containing a total of 3251 living trees with pronounced and well-developed crowns, which form the predominant and dominant stock of the forest stand. This corresponds to classes 1 and 2 according to tree classes proposed by Kraft [64]. The data were used to compare the total numbers of overstory tree stems.
- (b)
- Stem co-ordinates
Two datasets were generated to validate the detected stem positions and analyze error sources. For both, a subset of the study site located in the north-eastern part was used, referred to as the validation area (Figure 4). The first one is based on an MLS point cloud acquired in August 2022 and is used as an independent (MLS-based) reference dataset. The second one was obtained from the UAV leaf-off point cloud and represents all detectable tree stems within the used UAV data; hereafter, it is referred to as “UAV detectable” (Figure 4).
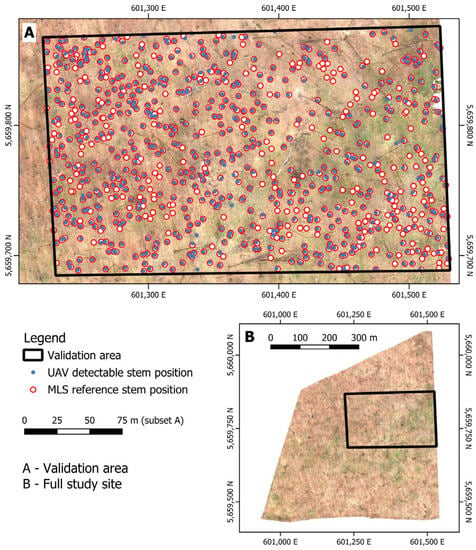
Figure 4.
Overview of the two reference datasets obtained from the UAV leaf-off point cloud and an MLS scan. Overstory tree trunks being part of the canopy were manually digitized within the respective area.
For both point clouds, trees with visible trunks and bases were carefully digitized manually. To do so, the whole point cloud as well as point cloud slices from different height levels were inspected visually to detect stems. Each selected stem point had to correspond to an overstory tree, in order to exclude small understory trees in the reference dataset. A marker was placed at the base of the stem and the stem co-ordinates were derived from it. These stem base co-ordinates were then used as stem co-ordinates, even if the stem did not grow vertically. This means that the trunk co-ordinate is not the midpoint between the base of the trunk and the crown. The accuracy of the individual digitized stem base co-ordinates from the MLS datasets was further verified in the field by manually georeferencing and conducting an inventory of a sample of 145 overstory trees within a subset of the validation area. Within the validation area, a total of 802 overstory trees were selected from the MLS dataset, while a total of 578 overstory trees were selected from the UAV dataset.
In order to use the MLS dataset, certain preprocessing steps were performed. After data acquisition in the field by a ZEB Horizon laser scanner (GeoSLAM, Ltd., Nottingham, UK), the scan was georeferenced using three ground control points located within the MLS scan area. The exact locations of these control points, measured with a carrier phase GNSS rover, were available from previous studies. This allowed the MLS data to be georeferenced with an RMSE of 0.088 m. Further processing included the normalization of the dataset using previously classified ground points within the same MLS dataset.
Since this study focuses only on overstory trees, all selected trees in both datasets were checked to see if they form an upper canopy crown. This step is necessary as the Hainich National Park is an unmanaged forest and contains a high number of small understory trees, oftentimes just of a few meters’ height. For the UAV-detectable dataset, trees that were not selected as overstory trees were categorized additionally into one of the following groups: small trees (understory), tree stumps, and tilted stems. Point clusters that were not clearly identifiable as a tree trunk in the UAV point cloud were labeled as unclassified trees.
- (c)
- Crown shapes
Individual crown shapes were carefully digitized by hand using the RGB leaf-on orthomosaic, CHM, and leaf-on and leaf-off point clouds. This derivation process encompassed the entire Huss study site. Figure 5 shows the RGB and CHM data, along with the resulting reference tree canopy delineations.
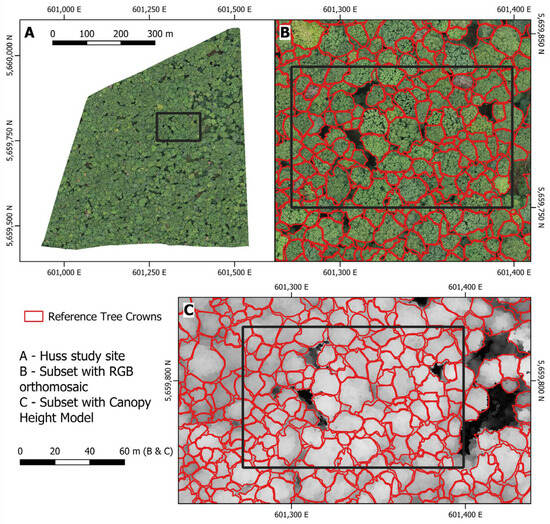
Figure 5.
Subsets of the crown reference dataset generated manually using the RGB orthomosaic and CHM data.
It is important to acknowledge that, despite expertise, challenges arose during the derivation process. At times, it was difficult to determine whether a given area contained a single tree crown with branches or multiple tree crowns.
2.4. Method for Automatic Detection of Stem Coordinates
For the derivation of trunk co-ordinates, the leaf-off point cloud is used. The approach involves grouping points that belong to a trunk into one cluster and representing the center of each cluster as a single point with co-ordinates. From these clusters, various metrics are computed to subsequently identify tree trunks whose crown also appear in the leaf-on point cloud. To focus on relevant points, the normalized leaf-off point cloud is trimmed to include only points within the height range of 3 to 15 m (LASTools command las2las) (Figure 6). The lower threshold of 3 m was chosen to prevent ground points and downed dead wood debris from being included, while the upper threshold of 15 m was selected to avoid including lower branches of the canopy and to facilitate the removal of smaller trees measuring just a few meters in heights. These thresholds may need to be adjusted based on the specific characteristics of the study site and were customized for the forest of the Huss site.
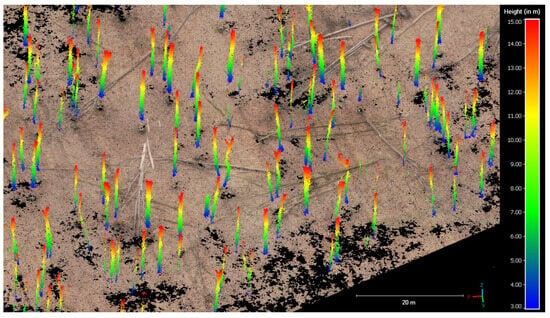
Figure 6.
Normalized UAV-SfM point cloud (leaf-off season) at a height of 3 to 15 m used for tree trunk detection. Blue points represent low points and red points represent higher points.
To reduce noise in the leaf-off point cloud, the Statistical Outlier Filter (SOR) operation was applied using CloudCompare (version 2.12.0). The filtered point cloud exhibits a 1.67% reduction in points, equivalent to approximately 400,000 points. The remaining points were then grouped into clusters using the Label Connected Components (LCC) function (Figure 7). The parameter settings for the SOR algorithms are as follows: number of points = 250, standard deviation = 3. For LLC function, the settings are number of points = 100, octree level = 12. This process yields a total of 10,110 potential tree trunks for the entire study area.
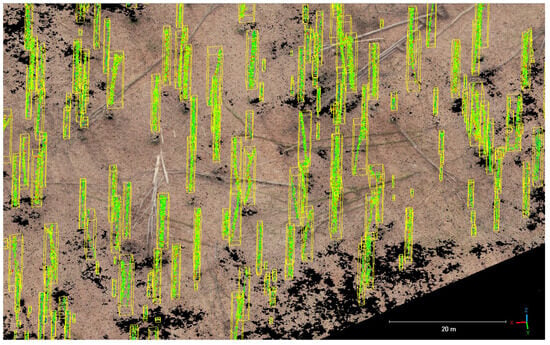
Figure 7.
Clustered points of tree stem candidates after applying a Label Connected Components (LCC) function (CloudCompare) as a pre-step to obtain tree stem positions.
A Python code is employed to calculate the following parameters that describe each cluster: minimum, maximum, and mean X, Y, and Z co-ordinates, center co-ordinates (co-ordinate half way between the minimum and maximum X and Y co-ordinate), number of points, difference between the minimum and maximum Z height, and orientation angle (orientation of the points within the cluster using the 3-dimensional distance between the lowest point and the highest point).
With the calculation of individual co-ordinates for each cluster, the clusters can now be described by their center co-ordinate representing potential stem co-ordinates and can be further filtered for actual tree trunks according to their parameter values. Only points associated with a cluster containing more than 750 points, a difference between the maximum z-height and the minimum z-height exceeding three meters, and an orientation angle less than 15 degrees are considered. The angle threshold was chosen not only to exclude clusters that do not represent stems due to their orientation but also to exclude standing deadwood, which often is tilted. To avoid overlapping stem candidates belonging to the same tree, each stem candidate co-ordinate was buffered by the distance of 0.375 m. In cases where two or more co-ordinates overlap, the centroid of the buffered area is calculated and used as the co-ordinate of the tree trunk. This process results in 3739 trunk co-ordinates for the entire study area, which are subsequently used to derive the tree crowns.
2.5. Method for Automatic Delineation of Tree Crown Shapes
The objective of this step is the delimitation of the individual tree crowns within the study area by segmenting the input data, aiming to represent each tree crown as a separate object. Initially, a tree mask was generated to exclude areas such as understory and non-forest areas (e.g., open clearings). This was accomplished by applying a segmentation to the study area and removing objects with a mean height lower than 20 m. Various algorithms were tested using the extracted tree stem positions as input data for crown delineation within the generated tree mask. Ultimately, a region growing method implemented in the software Trimble eCognition (version 10.2) was chosen (Figure 8).
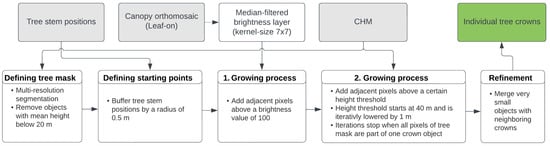
Figure 8.
Workflow of the ITCD using the derived tree stem positions, the CHM, the canopy orthomosaic (leaf-on), and a region growing approach that comprises two distinct growing processes (Software eCognition).
The selected method utilizes the previously derived tree positions as starting points, which ideally also correspond to the highest and brightest point within each crown. Employing specific growing criteria, each starting point is iteratively expanded by including surrounding pixels until it forms an object that represents a complete crown. To prevent starting points from falling within a small local minimum inside the crown, they were expanded first to form a circle with radius of 50 cm. Two criteria were employed for the growing process. Initially, a median-filtered brightness layer was used, calculated by averaging the intensities of all three RGB layers. Adjacent pixels above a certain brightness threshold were added to the growing crown. Secondly, the CHM was utilized, enlarging the crowns by including pixels above a certain tree height threshold. The same thresholds were applied to all objects. The height threshold started with the highest CHM values of the study site (40 m) and then iteratively lowered by 1 m until every pixel was added to one crown object. To refine the results, very small objects with an area below 7.5 m2 were merged with their neighboring crowns, resulting in a total number of objects that is less than the number of detected tree stem positions (Figure 8).
The results of the crown delineation were compared to ITCD methods that do not integrate markers. Among these methods were a CHM-based watershed segmentation without markers (using Trimble eCognition software, version 10.2) and a pretrained convolutional neural network on the leaf-on RGB orthomosaic (using the Python package DeepForest, version 1.2.1).
2.6. Accuracy Assessment
- (a)
- Stem detection
The extracted tree stem positions were validated against the UAV-detectable stem points and the MLS reference stem points within the validation area (Figure 4). For each extracted tree position, the nearest tree position from the respective validation dataset was detected, and each reference point was linked to the closest extracted tree position. A distance threshold of 1.5 m was used between the nearest extracted and reference stem point to determine correctly extracted stem positions. This threshold was chosen considering the average stem diameter at breast height of overstory trees within the study site, which is 49.2 cm. This value was obtained from a subset of 159 overstory trees where the diameter was manually measured. Due to the manner in which the reference points were generated through manual digitization, they are positioned on one site of the trunk rather than in the middle. As a result, minor shifts between stem positions of different datasets can occur. Various additional threshold values between 1.0 and 2.0 m were tested, but they only resulted in slight changes in the accuracy values. Therefore, 1.5 m was chosen as a stable threshold that captures correctly extracted stem points without mismatching not corresponding stem positions. Thus, any extracted stem point with a reference point within a radius of 1.5 m is considered as true positive (TP). False positives (FP) are extracted stem points outside the respective radius, and false negatives (FN) are reference stem points with no corresponding extracted stem point within the distance thresholds. Recall, Precision, and F1-score were calculated for all combinations of datasets, whereby Recall, Precision, and F1-score are defined as following [65]:
- (b)
- Crown delineation
To assess the accuracy of the extracted tree crown delineation compared to the reference crowns, the intersection over union (IoU) metric was used. The IoU is calculated by comparing the overlapping area between matching extracted crown polygons and reference crown polygons (intersection) with the total area of both polygons combined (union). For each extracted polygon, the reference polygon with the highest overlap was considered the matching polygon. An IoU score of 1 indicates an identical reference and extracted crown; 0 means there is no overlapping area between them. The mean value of all IoU polygon scores is calculated, as well as the percentage of crowns with an IoU ≥ 0.5. This threshold is commonly used in various studies to define true positive tree crowns [36,51,66,67].
In addition, precision, recall, and F1-score (Equations (1)–(3)) were calculated for each crown and as mean values. These values were computed by comparing the correctly delineated crown area to the area of extracted and reference crown, respectively. The correctly delineated crown area (TP) is defined as the overlapping area (intersection) of reference and extracted areas. The area of the extracted crown equals the sum of TP and FP, while the area of the reference crown equals the sum of TP and FN.
3. Results
3.1. Stem Detection
A total of 3739 tree trunks were detected by the algorithm for the whole study site (Figure 9), which aligns well with the inventory conducted by the National Park Administration, counting 3251 trees for the Huss site. However, it is important to consider the time difference of 9 years. The number of extracted trees also closely matches the number of reference crowns, with 3700 crowns delineated within the study site.
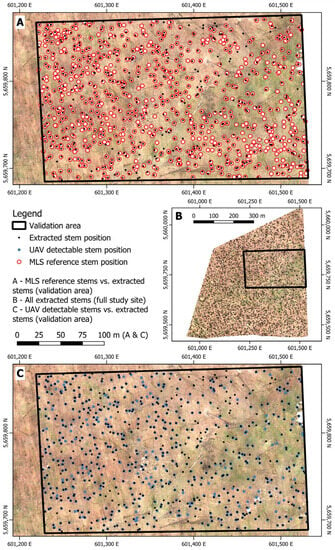
Figure 9.
Comparison of the detected stem positions applying the proposed methodology (black) with the MLS reference dataset (red) and the UAV-detectable dataset (blue) within the validation area. Accuracy metrices were calculated using these three datasets.
As mentioned in Section 2.3, precision, recall, and F1-score have been calculated for the extracted stem positions compared to both the MLS reference dataset and the UAV-detectable dataset, as well as for the two validation datasets compared to each other. The comparison between extracted trees and UAV-detectable trees, both based on the same UAV-SfM dataset, evaluates the algorithm’s performance in detecting tree stems present in the UAV data (Table 4, row (a)). Validating the extracted positions against the MLS reference data demonstrates how many correct trees can be detected out of all present trees (Table 4, row (c)). The comparison between the UAV-detectable and MLS reference datasets reflects the ability of drone data in generating a correct and complete digital copy of tree positions (Table 4, row (b)). In total, 626 of the extracted tree positions lie within the validation area, compared to a total of 578 UAV-detectable trees and a total of 802 MLS reference trees within the same subset. The F1-scores for the extracted tree stems are 0.773 when validating with the MLS reference dataset and 0.801 when validating with the UAV-detectable stems.

Table 4.
Results of the accuracy assessment for stem detection, comparing the three available datasets with each other (detected stem positions, UAV-detectable stem positions, and MLS reference stem positions). TP = true positive, FN = false negative, FP = false positive. The color coding (dark green–green–yellow–orange–red) indicates the value range within which each respective number lies between the highest (green) and lowest (red) accuracy values.
In addition to the 578 labeled UAV overstory trees, tree stems present in the UAV datasets classified into small trees (understory), tree stumps, tilted stems, and point clusters that are not clearly recognizable as a tree trunk (unclassified trees) were gradually added to the dataset and used for the accuracy assessment (Table 5).

Table 5.
Results of the accuracy assessment for the stem detection comparing the detected stem positions using the proposed method with the UAV-detectable dataset, considering different types of trees labeled within the UAV point cloud (overstory trees, small/understory trees, tree stumps, tilted trees, and unclassified trees). TP = true positive, FN = false negative, FP = false positive. The color coding (dark green–green–yellow–orange–red) indicates the value range within which each respective number lies between the highest (green) and lowest (red) accuracy values.
3.2. Crown Delineation
Individual tree crowns were generated for the entire study site. Two subsets comparing the extracted crowns to the reference crowns are shown in Figure 10. A total of 3414 crowns were delineated for the entire Huss study site, compared to 3700 crowns in the crown delineation reference dataset. The deviation to the detected stem positions (3739 tree positions were detected) can be explained by the fact that 325 tree positions did not have a corresponding delineated crown. The IoU score was calculated for all extracted crowns, resulting in a mean score of 0.442. A proportion of 42.9% of all crowns had a score ≥ 0.5.
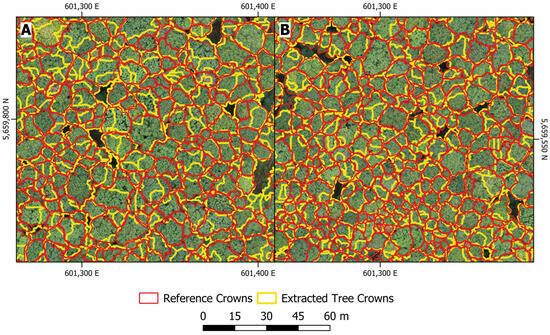
Figure 10.
Results of the crown delineation using the proposed method (yellow) compared to the reference crowns (red) for two selected subsets (A,B) within the study area.
The mean precision value for all crowns was 0.680, and the mean recall value was 0.614, resulting in an F1-score of 0.645. As a high number of false positives (area) indicates over-segmentation, while a high number of false negatives (area) indicates under-segmentation, low precision values indicate a tendency of over-segmented crowns and low recall values indicate a tendency of under-segmented crowns. Therefore, no strong trend of either under- or over-segmentation can be observed in the applied method, as precision and recall have similar value ranges. However, slightly lower recall values may indicate partial under-segmentation of crowns, which aligns well with the slightly lower number of extracted crowns compared to the reference crowns.
The results of the approaches, which did not include tree stem markers, resulted in a mean IoU score of 0.393 for the watershed segmentation and 0.238 for the DeepForest segmentation.
Moreover, tree height was calculated for each of the extracted crowns utilizing the CHM. This analysis exemplifies the use of individual tree crowns for deriving further forest parameters. It resulted in a mean tree crown height per object of 30.45 m (using the mean height value per crown) and a mean tree height of 33.72 m (using the maximum height value per crown).
4. Discussion
4.1. Stem Detection
The total number of trees detected by the algorithm aligns well with the count of trees recorded by the National Park Administration. The minor discrepancies in the numbers could stem from differing definitions: the inventory data classify trees in classes based on dominance [64], while our categorization distinguishes between overstory and understory. For instance, if co-dominant trees are also included in the inventory data, the count increases to 4876. Comparing the extracted trees to the MLS reference dataset, the results showed a slight underestimation of the total number of trees, approximately by 22%. The precision value of 0.867 indicates a high reliability of the extracted trees. However, the recall value of 0.697 underscores that some of the trees were missed.
Interestingly, when comparing the extracted tree stems with the UAV-detectable data, the situation is reversed: recall is slightly higher than precision. The high recall value of 0.834 suggest that the developed algorithm performs well and is able to detect most of the trees present in the used UAV dataset. A considerable percentage of trees not detected by the algorithm can, therefore, be attributed to an incomplete UAV dataset, where the drone may not be able to detect all relevant tree stems. The slightly lower precision values are mainly due to the algorithm detecting point clusters as trees that are outside the defined scope (i.e., trees that are not overstory trees). The Hainich National Park represents an unmanaged and heterogeneous forest environment, where various tree types of different ages and growth forms, which together form a complex structure, are present. Apart from dominant overstory trees, the area is characterized by an understory layer of smaller trees, remaining tree stumps from broken or fallen trees, and standing dead wood. When using a remote-sensing-based approach in structurally diverse forest environments, it is important to consider these distinct tree types. Defining the tree type to be detected is crucial, as the number of trees can vary significantly depending on factors such as the inclusion of understory trees. As this study employs stem positions for crown delineations, the goal of the tree detection was to detect overstory trees. Since, so far, it is challenging to derive reliable stem parameters such as DBH directly from the UAV-SfM point cloud, detected stem positions cannot be filtered by DBH class. However, by analyzing the 3D extend of the point clusters, we were able to identify tilted stems based on their growing angle and excluded them. Although tilted stems are not of interest in this case, the same approach could be further explored to detect snags, standing dead trees, which are often tilted. However, as mentioned before, identifying only overstory trees was not met in every case. The results in Table 5 illustrate in more detail that some of the extracted trees are likely not part of the canopy. The precision values increase gradually when understory trees, tree stumps, tilted stems, and unclassified stems are included. When all types of tree stems are considered, nearly all extracted tree stems are correct (precision of 0.976). This indicates that the extracted tree positions are very reliable but do not in every case represent overstory trees. When matched with their closest UAV-detectable tree (max. distance 1.5 m) among the 626 extracted trees within the validation area, 129 of them do not correspond to a classified overstory tree but to one of the other categorized tree types, most of them to understory trees. To improve the detection of only overstory trees, it is possible to use a point cloud slice from a higher forest layer, which could exclude smaller understory trees and tree stumps. However, this approach may increase the number of missed overstory trees as the point cloud becomes sparser with elevated tree height. To be able to filter trees not only by the above tree categories but also by tree size, it would be of great interest to derive parameters such as DBH directly from the point cloud. To improve the UAV-SfM point cloud, different flight parameters were tested during this study, but further investigation regarding the acquisition parameters could be conducted, e.g., by adding more oblique flights or changing the camera settings [68].
While most studies using optical UAV data focus on raster-based tree detection methods (Table 1), such as methods based on local maxima filtering from CHMs [27,69], it has been demonstrated that point cloud approaches are also suitable for producing accurate stem positions using UAV-SfM point clouds. These methods often stem from the laser scanning sector; their potential for UAV-SfM point clouds, especially under leaf-off conditions, could be further investigated.
To evaluate the suitability of UAV-SfM point clouds in general for detecting single tree positions, independent of the chosen stem detection approach, we compared the UAV-detectable stem positions with the MLS reference stem positions. This comparison was performed visually and by validating the UAV-detectable dataset with the MLS reference dataset (Table 4, row (b)). The accuracy assessment between the two datasets demonstrated very high precision values, indicating the high reliability of trees visible in the SfM point cloud. However, recall values are considerably lower, highlighting the already mentioned fact that tree stems are missing in the SfM dataset. Nevertheless, it could be demonstrated that point clouds based on UAV-SfM approaches are a cost-effective and accurate substitute to LiDAR in tree stem detection. This goes along with several studies that compared UAV-SfM and airborne LiDAR point clouds of forests and stated that UAV-SfM can be an adequate substitute to airborne LiDAR point clouds for forest applications [70,71,72].
4.2. Crown Delineation
By applying the proposed method using detected stems for crown delineation, a moderate F1-score could be achieved. The resulting IoU score remains relatively low. As mentioned earlier, the model does tend to slightly under-segment the crowns.
The moderate accuracy values reflect the challenging nature of defining crown boundaries in a dense, structurally diverse, and complex forest such as the Hainich National Park. Even with careful observation, it is often difficult to distinguish visually individual crowns within the orthomosaic or point cloud. For example, neighboring trees of the same tree species can appear as either two individual trees or two branches belonging to the same tree. These ambiguities are hard to resolve using only leaf-on data, whether through visual interpretation or ITCD algorithms based on canopy information. To address these limitations, we analyzed the combination of leaf-off and leaf-on data for crown delineation, reasoning that every individual tree should have a detectable tree stem, while crown parts without a detected tree stem would correspond to branches. We compared the crown delineation with several commonly used ITCD methods that do not employ markers. The results of the marker-less approaches yielded lower accuracy values compared to the proposed method, which involves including tree stem positions from leaf-off data as markers. This indicates that the integration of stem positions as markers improved the results. Regarding the comparison of both tree stems and crowns with the reference data within the corresponding subset, the numbers of reference crowns (606), extracted crowns (617), and extracted tree stems (626) match very well. However, the number of MLS reference tree stems is higher (802). Considering this, it can be assumed that the visual delineation of crowns in the UAV data, as well as extracted tree stems and crowns, underestimate the number of overstory trees/crowns by approximately 22–24% in dense forest stands. As most ITCD studies validate their crowns by comparing them to crowns generated by visual interpretation of the data [19,20], it is likely that many remote-sensing-based studies underestimate the number of overstory trees/crowns in dense forest environments.
To improve the crown delineation results, the use of other algorithms could be further explored. Several deep learning approaches have shown promising results [34,36,51,66,73] and new approaches are constantly developed. Additionally, it would be worth investigating how prior-derived stem positions from leaf-off data could be used as additional input for deep learning crown delineation.
4.3. Combination of Leaf-off and Leaf-on UAV Data to Model Forest Structure
Building on our findings, we recommend combining leaf-off and leaf-on SfM point clouds, especially for dense deciduous forest stands. Our study has demonstrated that these datasets contain highly complementary information, as shown in Figure 3. Leaf-on data provide limited under-canopy information due to the density of the canopy, while relying solely on leaf-off data makes it challenging to accurately estimate crown parameters and tree heights [74]. However, by leveraging both datasets, a more comprehensive picture of the forest canopy, tree stems, and forest floor can be obtained, enabling more accurate derivation of forest parameters. This option, of course, is only possible for areas with deciduous forests experiencing a leaf-off season. This finding aligns with the results of other studies that have combined leaf-off and leaf-on SfM data primarily to generate accurate height models [74,75,76]. For instance, Nasiri et al. [75] generated a CHM by utilizing both leaf-off and leaf-on data from UAV imagery to derive tree height and crown diameter. Moudrý et al. [76] reported comparable accuracy between DTMs derived from leaf-off SfM data and those from airborne LiDAR. In our study, we not only used leaf-off and leaf-on data to generate height models but also used the tree detection results from the leaf-off point cloud to enhance the accuracy of other forest-related parameters that rely on accurate tree detection and identification, such as crown delineation in our case.
Several ITCD methods, such as region growing approaches, require markers as starting points for crown delineation. Ideally, these markers should represent the highest points of the crowns. Although watershed segmentation can be applied without markers, it has been reported to work better when markers are included [32]. In dense deciduous forest environments, obtaining these markers solely from CHMs is challenging. However, our method, which combines leaf-off and leaf-on data, provides accurate tree stem positions that can serve as marker points. Although, it must be acknowledged that, even with highly accurate stem positions, crown delineation is limited if the leaf-on data do not allow the distinction between neighboring crowns. Nevertheless, to accurately determine the number and location of trees in a dense forest site, it is essential to look beneath the canopy. Some studies are exploring the possibility of under-canopy UAV flights for the data retrieval of stem and forest floor [77,78]. However, currently, automated flight missions under dense forest canopies cannot be realized.
5. Conclusions
This study emphasizes the significance of integrating leaf-off and leaf-on SfM point clouds for single-tree-based monitoring and management using UAVs equipped with optical RGB camera systems. Our focus was on deriving overstory tree stem positions and crown delineations using UAV-SfM data products.
Tree stem detection was carried out by applying a clustering and rule-based approach on the leaf-off point cloud. Accuracy assessment involved comparing three datasets: the extracted stems, manually detected UAV stems, and MLS reference stems. We found that the proposed method delivers highly reliable stem positions (precision of 0.867). Recall values were slightly lower, indicating that approximately 22% of the present trees were missed. Categorizing different tree types allowed us to analyze the type of tree stem being detected by the algorithm. This analysis revealed that the distinction between overstory and understory tree stems is not always met. When working in dense structurally diverse forest environments, the high number of small understory trees has to be considered. Further investigations could explore the potential of UAV-SfM leaf-off data for detecting understory trees. Validating the UAV detectable stems against the MLS reference stems demonstrated high accuracy values (F1-score of 0.811), confirming the potential of UAV-SfM point clouds in general as a cost-effective alternative to LiDAR data. Using the extracted stem positions as markers for ITCD improved the results compared to methods without markers. However, the moderate accuracy values (F1-score of 0.645, mean IoU of 0.442) indicate the challenges of delineating crowns in dense forests. Promising improvements are expected with the development of new deep learning approaches. The results highlighted that both the number of overstory tree stems and crowns tends to be underestimated, probably even within the crown reference data, indicating potential underestimations in many studies focusing on dense deciduous forests. This underscores the challenges in collecting reliable reference data, as neither the view of the canopy from above via remote sensing data nor from below via field surveys seem to provide the complete picture.
Overall, we advocate for the use of a combined leaf-off and leaf-on SfM approach in future forest monitoring and management tasks. While our focus was on tree stem detection and ITCD, the potential of a combined leaf-off–leaf-on point cloud could be further explored for the derivation of other forest parameters and methods.
Author Contributions
Conceptualization, S.D., C.T. and M.M.M.; methodology, S.D., C.T. and M.M.M.; software, S.D., M.M.M. and F.B.; validation, S.D., M.M.M., F.B., M.N., J.Z., F.M. and M.G.H.; formal analysis, S.D., M.M.M. and F.B.; investigation, S.D., M.M.M., F.B., M.N., J.Z. and F.M.; resources, S.D. and C.T.; data curation, S.D., F.B., J.Z., F.M., M.G.H. and F.K.; writing—original draft preparation, S.D. and F.B.; writing—review and editing, S.D., M.M.M., C.T., C.D., F.K. and S.H.; visualization, S.D., F.B. and M.N.; supervision, C.T., C.D. and S.H.; project administration, C.T., S.D., C.D. and S.H.; funding acquisition, C.T. and S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy reasons.
Acknowledgments
The authors would like to thank the Hainich National Park Administration for their co-operation and provision of the data.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AMS | Adaptive-kernel bandwidth Mean-Shift |
| CHM | Canopy Height Model |
| CNN | Convolutional Neural Network |
| DBH | Diameter at Breast Height |
| DSM | Digital Surface Model |
| DTM | Digital Terrain Model |
| FMS | Fixed-kernel bandwidth Mean-Shift |
| FN | False Negative |
| FP | False Positive |
| IoU | Intersection over Union |
| ISO | International Organization for Standardization |
| ITCD | Individual Tree Crown Delineation |
| LCC | Label Connected Components |
| LiDAR | Light Detection and Ranging |
| MLS | Mobile Laser Scanning |
| RGB | Red Green Blue |
| RMSE | Root Mean Square Error |
| RTK | Real-Time Kinematic |
| SfM | Structure from Motion |
| SOR | Statistical Outlier Filter |
| SSD | Single Shot Detector |
| TP | True Positive |
| UAV | Unoccupied Aerial Vehicle |
| VWF | Variable Window Filter |
References
- Fassnacht, F.E.; White, J.C.; Wulder, M.A.; Næsset, E. Remote sensing in forestry: Current challenges, considerations and directions. For. Int. J. For. Res. 2023, cpad024. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef]
- Thonfeld, F.; Gessner, U.; Holzwarth, S.; Kriese, J.; da Ponte, E.; Huth, J.; Kuenzer, C. A First Assessment of Canopy Cover Loss in Germany’s Forests after the 2018–2020 Drought Years. Remote Sens. 2022, 14, 562. [Google Scholar] [CrossRef]
- Zuidema, P.A.; van der Sleen, P. Seeing the forest through the trees: How tree-level measurements can help understand forest dynamics. New Phytol. 2022, 234, 1544–1546. [Google Scholar] [CrossRef]
- Almeida, C.; Maurano, L.; Valeriano, D.; Câmara, G.; Vinhas, L.; Gomes, A.; Monteiro, A.; Souza, A.; Rennó, C.; e Silva, D.; et al. Methodology for Forest Monitoring Used in PRODES and DETER Projects; Instituto Nacional de Pesquisas Espaciais (INPE): São José dos Campos, Brazil, 2021. [Google Scholar]
- De Conto, T.; Olofsson, K.; Görgens, E.B.; Rodriguez, L.C.E.; Almeida, G. Performance of stem denoising and stem modelling algorithms on single tree point clouds from terrestrial laser scanning. Comput. Electron. Agric. 2017, 143, 165–176. [Google Scholar] [CrossRef]
- Yépez-Rincón, F.D.; Luna-Mendoza, L.; Ramírez-Serrato, N.L.; Hinojosa-Corona, A.; Ferriño-Fierro, A.L. Assessing vertical structure of an endemic forest in succession using terrestrial laser scanning (TLS). Case study: Guadalupe Island. Remote Sens. Environ. 2021, 263, 112563. [Google Scholar] [CrossRef]
- Kuželka, K.; Slavík, M.; Surový, P. Very High Density Point Clouds from UAV Laser Scanning for Automatic Tree Stem Detection and Direct Diameter Measurement. Remote Sens. 2020, 12, 1236. [Google Scholar] [CrossRef]
- Chen, Q.; Gao, T.; Zhu, J.; Wu, F.; Li, X.; Lu, D.; Yu, F. Individual Tree Segmentation and Tree Height Estimation Using Leaf-Off and Leaf-On UAV-LiDAR Data in Dense Deciduous Forests. Remote Sens. 2022, 14, 2787. [Google Scholar] [CrossRef]
- Yao, H.; Qin, R.; Chen, X. Unmanned Aerial Vehicle for Remote Sensing Applications—A Review. Remote Sens. 2019, 11, 1443. [Google Scholar] [CrossRef]
- Thiel, C.; Müller, M.M.; Berger, C.; Cremer, F.; Dubois, C.; Hese, S.; Baade, J.; Klan, F.; Pathe, C. Monitoring Selective Logging in a Pine-Dominated Forest in Central Germany with Repeated Drone Flights Utilizing a Low Cost RTK Quadcopter. Drones 2020, 4, 11. [Google Scholar] [CrossRef]
- Johansen, K.; Raharjo, T.; McCabe, M. Using Multi-Spectral UAV Imagery to Extract Tree Crop Structural Properties and Assess Pruning Effects. Remote Sens. 2018, 10, 854. [Google Scholar] [CrossRef]
- Dainelli, R.; Toscano, P.; Di Gennaro, S.F.; Matese, A. Recent Advances in Unmanned Aerial Vehicle Forest Remote Sensing—A Systematic Review. Part I: A General Framework. Forests 2021, 12, 327. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sens. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Ahmadi, P.; Mansor, S.; Farjad, B.; Ghaderpour, E. Unmanned Aerial Vehicle (UAV)-Based Remote Sensing for Early-Stage Detection of Ganoderma. Remote Sens. 2022, 14, 1239. [Google Scholar] [CrossRef]
- Jiang, S.; Jiang, C.; Jiang, W. Efficient structure from motion for large-scale UAV images: A review and a comparison of SfM tools. ISPRS J. Photogramm. Remote Sens. 2020, 167, 230–251. [Google Scholar] [CrossRef]
- Schonberger, J.L.; Frahm, J.-M. Structure-from-Motion Revisited. In Proceedings of the 29th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2016, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 4104–4113, ISBN 978-1-4673-8851-1. [Google Scholar]
- Deluzet, M.; Erudel, T.; Briottet, X.; Sheeren, D.; Fabre, S. Individual Tree Crown Delineation Method Based on Multi-Criteria Graph Using Geometric and Spectral Information: Application to Several Temperate Forest Sites. Remote Sens. 2022, 14, 1083. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation—Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Wang, L.; Gong, P.; Biging, G.S. Individual Tree-Crown Delineation and Treetop Detection in High-Spatial-Resolution Aerial Imagery. Photogramm. Eng. Remote Sens. 2004, 70, 351–357. [Google Scholar] [CrossRef]
- Tochon, G.; Féret, J.B.; Valero, S.; Martin, R.E.; Knapp, D.E.; Salembier, P.; Chanussot, J.; Asner, G.P. On the use of binary partition trees for the tree crown segmentation of tropical rainforest hyperspectral images. Remote Sens. Environ. 2015, 159, 318–331. [Google Scholar] [CrossRef]
- Workie, T.G. Estimating forest above-ground carbon using object-based analysis of very high spatial resolution satellite images. Afr. J. Environ. Sci. Technol. 2017, 11, 587–600. [Google Scholar] [CrossRef]
- Lindberg, E.; Holmgren, J. Individual Tree Crown Methods for 3D Data from Remote Sensing. Curr. Forestry Rep. 2017, 3, 19–31. [Google Scholar] [CrossRef]
- Erikson, M.; Olofsson, K. Comparison of three individual tree crown detection methods. Mach. Vis. Appl. 2005, 16, 258–265. [Google Scholar] [CrossRef]
- Hirschmugl, M.; Ofner, M.; Raggam, J.; Schardt, M. Single tree detection in very high resolution remote sensing data. Remote Sens. Environ. 2007, 110, 533–544. [Google Scholar] [CrossRef]
- Ottoy, S.; Tziolas, N.; van Meerbeek, K.; Aravidis, I.; Tilkin, S.; Sismanis, M.; Stavrakoudis, D.; Gitas, I.Z.; Zalidis, G.; de Vocht, A. Effects of Flight and Smoothing Parameters on the Detection of Taxus and Olive Trees with UAV-Borne Imagery. Drones 2022, 6, 197. [Google Scholar] [CrossRef]
- Wulder, M.; Niemann, K.; Goodenough, D.G. Local Maximum Filtering for the Extraction of Tree Locations and Basal Area from High Spatial Resolution Imagery. Remote Sens. Environ. 2000, 73, 103–114. [Google Scholar] [CrossRef]
- Gougeon, F.A. A Crown-Following Approach to the Automatic Delineation of Individual Tree Crowns in High Spatial Resolution Aerial Images. Can. J. Remote Sens. 1995, 21, 274–284. [Google Scholar] [CrossRef]
- Gougeon, F.A.; Leckie, D.G. The Individual Tree Crown Approach Applied to Ikonos Images of a Coniferous Plantation Area. Photogramm. Eng. Remote Sens. 2006, 72, 1287–1297. [Google Scholar] [CrossRef]
- Beucher, S.; Lantuéjoul, C. Use of Watersheds in Contour Detection. In Proceedings of the International Workshop on Image Processing: Real-time Edge and Motion Detection/Estimation, Rennes, France, 17–21 September 1979. [Google Scholar]
- Derivaux, S.; Forestier, G.; Wemmert, C.; Lefèvre, S. Supervised image segmentation using watershed transform, fuzzy classification and evolutionary computation. Pattern Recognit. Lett. 2010, 31, 2364–2374. [Google Scholar] [CrossRef]
- Olofsson, K.; Wallerman, J.; Holmgren, J.; Olsson, H. Tree species discrimination using Z/I DMC imagery and template matching of single trees. Scand. J. For. Res. 2006, 21, 106–110. [Google Scholar] [CrossRef]
- Zhao, H.; Morgenroth, J.; Pearse, G.; Schindler, J. A Systematic Review of Individual Tree Crown Detection and Delineation with Convolutional Neural Networks (CNN). Curr. Forestry Rep. 2023, 9, 149–170. [Google Scholar] [CrossRef]
- Yu, K.; Hao, Z.; Post, C.J.; Mikhailova, E.A.; Lin, L.; Zhao, G.; Tian, S.; Liu, J. Comparison of Classical Methods and Mask R-CNN for Automatic Tree Detection and Mapping Using UAV Imagery. Remote Sens. 2022, 14, 295. [Google Scholar] [CrossRef]
- Freudenberg, M.; Magdon, P.; Nölke, N. Individual tree crown delineation in high-resolution remote sensing images based on U-Net. Neural Comput. Applic. 2022, 34, 22197–22207. [Google Scholar] [CrossRef]
- Dalponte, M.; Frizzera, L.; Gianelle, D. Individual tree crown delineation and tree species classification with hyperspectral and LiDAR data. PeerJ 2019, 6, e6227. [Google Scholar] [CrossRef] [PubMed]
- Weiser, H.; Schäfer, J.; Winiwarter, L.; Krašovec, N.; Fassnacht, F.E.; Höfle, B. Individual tree point clouds and tree measurements from multi-platform laser scanning in German forests. Earth Syst. Sci. Data 2022, 14, 2989–3012. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, K.; Zhu, Y.; Wang, X.; Yun, T. Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning. Forests 2021, 12, 131. [Google Scholar] [CrossRef]
- Xiao, W.; Zaforemska, A.; Smigaj, M.; Wang, Y.; Gaulton, R. Mean Shift Segmentation Assessment for Individual Forest Tree Delineation from Airborne Lidar Data. Remote Sens. 2019, 11, 1263. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. PointNet: Deep Learning on Point Sets for 3D Classification and Segmentation. arXiv 2016, arXiv:1612.00593. [Google Scholar]
- Swayze, N.C.; Tinkham, W.T.; Vogeler, J.C.; Hudak, A.T. Influence of flight parameters on UAS-based monitoring of tree height, diameter, and density. Remote Sens. Environ. 2021, 263, 112540. [Google Scholar] [CrossRef]
- Miraki, M.; Sohrabi, H.; Fatehi, P.; Kneubuehler, M. Individual tree crown delineation from high-resolution UAV images in broadleaf forest. Ecol. Inform. 2021, 61, 101207. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; Cosenza, D.N.; Rodriguez, L.C.E.; Silva, M.; Tomé, M.; Díaz-Varela, R.A.; González-Ferreiro, E. Comparison of ALS- and UAV(SfM)-derived high-density point clouds for individual tree detection in Eucalyptus plantations. Int. J. Remote Sens. 2018, 39, 5211–5235. [Google Scholar] [CrossRef]
- Young, D.J.N.; Koontz, M.J.; Weeks, J. Optimizing aerial imagery collection and processing parameters for drone-based individual tree mapping in structurally complex conifer forests. Methods Ecol. Evol. 2022, 13, 1447–1463. [Google Scholar] [CrossRef]
- Mohan, M.; Leite, R.V.; Broadbent, E.N.; Wan Mohd Jaafar, W.S.; Srinivasan, S.; Bajaj, S.; Dalla Corte, A.P.; do Amaral, C.H.; Gopan, G.; Saad, S.N.M.; et al. Individual tree detection using UAV-lidar and UAV-SfM data: A tutorial for beginners. Open Geosci. 2021, 13, 1028–1039. [Google Scholar] [CrossRef]
- Komárek, J.; Klápště, P.; Hrach, K.; Klouček, T. The Potential of Widespread UAV Cameras in the Identification of Conifers and the Delineation of Their Crowns. Forests 2022, 13, 710. [Google Scholar] [CrossRef]
- Mohan, M.; Silva, C.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, A.; Dia, M. Individual Tree Detection from Unmanned Aerial Vehicle (UAV) Derived Canopy Height Model in an Open Canopy Mixed Conifer Forest. Forests 2017, 8, 340. [Google Scholar] [CrossRef]
- Lei, L.; Yin, T.; Chai, G.; Li, Y.; Wang, Y.; Jia, X.; Zhang, X. A novel algorithm of individual tree crowns segmentation considering three-dimensional canopy attributes using UAV oblique photos. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102893. [Google Scholar] [CrossRef]
- Pleșoianu, A.-I.; Stupariu, M.-S.; Șandric, I.; Pătru-Stupariu, I.; Drăguț, L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Chadwick, A.J.; Goodbody, T.R.H.; Coops, N.C.; Hervieux, A.; Bater, C.W.; Martens, L.A.; White, B.; Röeser, D. Automatic Delineation and Height Measurement of Regenerating Conifer Crowns under Leaf-Off Conditions Using UAV Imagery. Remote Sens. 2020, 12, 4104. [Google Scholar] [CrossRef]
- Qin, H.; Zhou, W.; Yao, Y.; Wang, W. Individual tree segmentation and tree species classification in subtropical broadleaf forests using UAV-based LiDAR, hyperspectral, and ultrahigh-resolution RGB data. Remote Sens. Environ. 2022, 280, 113143. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Zhang, L.; Fan, X.; Ye, Q.; Fu, L. Individual tree segmentation and tree-counting using supervised clustering. Comput. Electron. Agric. 2023, 205, 107629. [Google Scholar] [CrossRef]
- Copernicus Land Monitoring Service. Tree Cover Density. 2018. Available online: https://land.copernicus.eu/pan-european/high-resolution-layers/forests/tree-cover-density (accessed on 12 December 2022).
- Huss, J.; Butler-Manning, D. Entwicklungsdynamik eines buchendominierten „Naturwald“-Dauerbeobachtungsbestands auf Kalk im Nationalpark Hainich/Thüringen. Wald. Online 2006, 3, 67–81. [Google Scholar]
- Fritzlar, D.; Henkel, A.; Profft, I. Einführung in den Naturraum. In Exkursionsführer. Wissenschaft im Hainich; Hainich-Tagung, Bad Langensalza, 27–29 April; Nationalparkverwaltung Hainich: Gotha, Germany, 2016; pp. 5–11. [Google Scholar]
- Schramm, H. Naturräumliche Gliederung der Exkursionsgebiete. In Exkursionsführer zur Tagung der AG Forstliche Standorts- und Vegetationskunde vom 18. bis 21. Mai 2005 in Thüringen; Thüringer Landesanstalt für Wald, Jagd und Fischerei: Gotha, Germany, 2005; pp. 8–19. [Google Scholar]
- Nationale Naturlandschaften. Thüringer Gesetz über den Nationalpark Hainich: ThürNPHG; Nationale Naturlandschaften: Berlin, Germany, 1997. [Google Scholar]
- Biehl, R. Der Nationalpark Hainich "Urwald mitten in Deutschland". In Exkursionsführer zur Tagung der AG Forstliche Standorts- und Vegetationskunde vom 18. bis 21. Mai 2005 in Thüringen; Thüringer Landesanstalt für Wald, Jagd und Fischerei: Gotha, Germany, 2005; pp. 44–47. [Google Scholar]
- Henkel, A.; Hese, S.; Thiel, C. Erhöhte Buchenmortalität im Nationalpark Hainich? AFZ—Der Wald. 2021, 3, 26–29. [Google Scholar]
- DJI. DJI Phantom 4 RTK: User Manual v1.4; DJI: Shenzen, China, 2018. [Google Scholar]
- Knohl, A.; Schulze, E.-D.; Kolle, O.; Buchmann, N. Large carbon uptake by an unmanaged 250-year-old deciduous forest in Central Germany. Agric. For. Meteorol. 2003, 118, 151–167. [Google Scholar] [CrossRef]
- Thiel, C.; Mueller, M.M.; Epple, L.; Thau, C.; Hese, S.; Voltersen, M.; Henkel, A. UAS Imagery-Based Mapping of Coarse Wood Debris in a Natural Deciduous Forest in Central Germany (Hainich National Park). Remote Sens. 2020, 12, 3293. [Google Scholar] [CrossRef]
- Kraft, G. Beiträge zur Lehre von den Durchforstungen, Schlagstellungen und Lichtungshieben; Klindworth: Hannover, Germany, 1884. [Google Scholar]
- Ye, Z.; Wei, J.; Lin, Y.; Guo, Q.; Zhang, J.; Zhang, H.; Deng, H.; Yang, K. Extraction of Olive Crown Based on UAV Visible Images and the U2-Net Deep Learning Model. Remote Sens. 2022, 14, 1523. [Google Scholar] [CrossRef]
- Braga, G.J.R.; Peripato, V.; Dalagnol, R.; Ferreira, P.M.; Tarabalka, Y.; Aragão, O.C.L.E.; de Campos Velho, F.H.; Shiguemori, E.H.; Wagner, F.H. Tree Crown Delineation Algorithm Based on a Convolutional Neural Network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual Tree-Crown Detection in RGB Imagery Using Semi-Supervised Deep Learning Neural Networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Hese, S. Oblique and Cross-Grid UAV Imaging Flight Plans—A Sneak Preview of the Analysis of Resulting 3D Point Cloud Properties for Deciduous Forest Surfaces—Low Cost 3D Mapping with the Phantom 4R (RTK). 2022. Available online: https://jenacopterlabs.de/?p=1711 (accessed on 10 July 2023).
- Harikumar, A.; D’Odorico, P.; Ensminger, I. Combining Spectral, Spatial-Contextual, and Structural Information in Multispectral UAV Data for Spruce Crown Delineation. Remote Sens. 2022, 14, 2044. [Google Scholar] [CrossRef]
- Thiel, C.; Schmullius, C. Comparison of UAV photograph-based and airborne lidar-based point clouds over forest from a forestry application perspective. Int. J. Remote Sens. 2017, 38, 2411–2426. [Google Scholar] [CrossRef]
- Ghanbari Parmehr, E.; Amati, M. Individual Tree Canopy Parameters Estimation Using UAV-Based Photogrammetric and LiDAR Point Clouds in an Urban Park. Remote Sens. 2021, 13, 2062. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Malenovský, Z.; Turner, D.; Vopěnka, P. Assessment of Forest Structure Using Two UAV Techniques: A Comparison of Airborne Laser Scanning and Structure from Motion (SfM) Point Clouds. Forests 2016, 7, 62. [Google Scholar] [CrossRef]
- Lassalle, G.; Ferreira, M.P.; La Rosa, L.E.C.; de Souza Filho, C.R. Deep learning-based individual tree crown delineation in mangrove forests using very-high-resolution satellite imagery. ISPRS J. Photogramm. Remote Sens. 2022, 189, 220–235. [Google Scholar] [CrossRef]
- Ni, W.; Dong, J.; Sun, G.; Zhang, Z.; Pang, Y.; Tian, X.; Li, Z.; Chen, E. Synthesis of Leaf-on and Leaf-off Unmanned Aerial Vehicle (UAV) Stereo Imagery for the Inventory of Aboveground Biomass of Deciduous Forests. Remote Sens. 2019, 11, 889. [Google Scholar] [CrossRef]
- Nasiri, V.; Darvishsefat, A.A.; Arefi, H.; Pierrot-Deseilligny, M.; Namiranian, M.; Le Bris, A. Unmanned aerial vehicles (UAV)-based canopy height modeling under leaf-on and leaf-off conditions for determining tree height and crown diameter (case study: Hyrcanian mixed forest). Can. J. For. Res. 2021, 51, 962–971. [Google Scholar] [CrossRef]
- Moudrý, V.; Gdulová, K.; Fogl, M.; Klápště, P.; Urban, R.; Komárek, J.; Moudrá, L.; Štroner, M.; Barták, V.; Solský, M. Comparison of leaf-off and leaf-on combined UAV imagery and airborne LiDAR for assessment of a post-mining site terrain and vegetation structure: Prospects for monitoring hazards and restoration success. Appl. Geogr. 2019, 104, 32–41. [Google Scholar] [CrossRef]
- Kuželka, K.; Surový, P. Mapping Forest Structure Using UAS inside Flight Capabilities. Sensors 2018, 18, 2245. [Google Scholar] [CrossRef] [PubMed]
- Krisanski, S.; Taskhiri, M.; Turner, P. Enhancing Methods for Under-Canopy Unmanned Aircraft System Based Photogrammetry in Complex Forests for Tree Diameter Measurement. Remote Sens. 2020, 12, 1652. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).