1. Introduction
Monitoring young plant growth is of particular interest for the selection of more robust or resistant varieties. Tests are regularly carried out on the first days of the plant’s life to compare these varieties. During these experiments, a large number of plants are studied in parallel, often in restricted spaces. Moreover, a large number of dispersed replicates are required in the experimental designs [
1], increasing the need for space at the same time. One is able to analyze the growth of these plants in order to conduct variety analyses if they are physically separated seen from the sensor [
2]. The space restriction required for high-throughput experiments in industrial settings implies a very high density of seedlings under the cameras. Such constraints produce large number of occlusions during the growth. At first rare, these occlusions become more and more frequent until the plants form a unique canopy. Most current tools and methods for monitoring plant growth are limited by these occlusions [
3,
4,
5,
6]. The combination of occlusions and plant self-similarity makes it challenging to use standard tracking methods [
7,
8,
9]. Some rare tools are proposed to extend the monitoring despite the overlap, as in [
10] via watershed or extended feature space (color, texture, depth) as in [
11,
12] or via advanced machine learning object detection tools [
13]. Here, we investigate a distinct approach to disentangle overlapped plants.
A possibility to separate occluded objects in computer vision is to acquire multiple views from different angles of the scene. This is a common approach in plant imaging [
14,
15,
16]. A disadvantage of this approach is that it may reduce the throughput. Indeed, it may not be compatible with the synchronized imaging of a set of plants if the camera has to move around each plant. To preserve the throughput, we would need the plant to move. This can be achieved by conveying the plant, but it is a costly solution which can also cause some stress or risk contamination in case of plant–pathogens interactions. In this article, we propose to access synchronized multiple views of touching plants via the sole use of natural movements of plants to disentangle occluded objects.
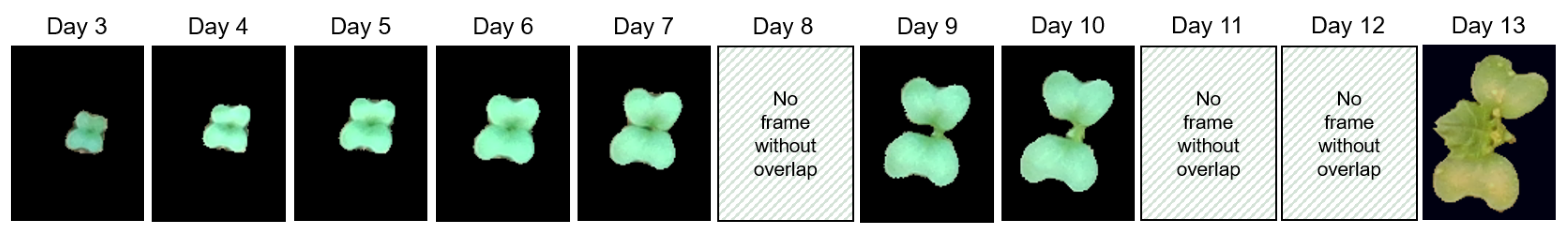
Plants have an internal timekeeper known as a circadian clock that anticipates environmental cues such as light, temperature and regulates photoperiodic rhythmicity for the proper growth and fitness of the plants [
17,
18,
19]. This circadian clock causes oscillations of the leaves synchronized with the photoperiod. Therefore, in controlled conditions, when plant are imaged at a sample rate much higher than the photoperiod, it is possible to acquire them from different perspectives. Circadian movements have already been analyzed via time lapse videos in the literature, since their amplitude can be related to stress [
20,
21,
22]. Here, alternatively, we propose to use the presence of these circadian movements with simple sampling approaches to use such sequences of images in order to extend the duration of the individual monitoring of young plants as illustrated in
Figure 1.
The sampling methods we present are based on simple sampling criterion merely without any image processing. There are existing image processing methods which reach higher growth stages than the one we propose [
3,
4,
5,
6,
10,
11,
14,
15]. These related works are, however, operating at much lower throughput. In most of these related works, the plant under study is isolated and analyzed individually with the additional help of depth sensors [
3,
11,
14,
15]. In other related works, several plants are monitored, but the model is not compatible with possible overlaps between plants [
6] or directly isolated in bounding boxes [
4]. In [
5], the tips of the leaves must be visible from the sensor which is not always compatible with occlusions. As most related work, the method presented in [
10] addresses the overlap problem with image processing techniques. This approach differs from ours, which is based on the selection of non-overlapping moments.
3. Methods
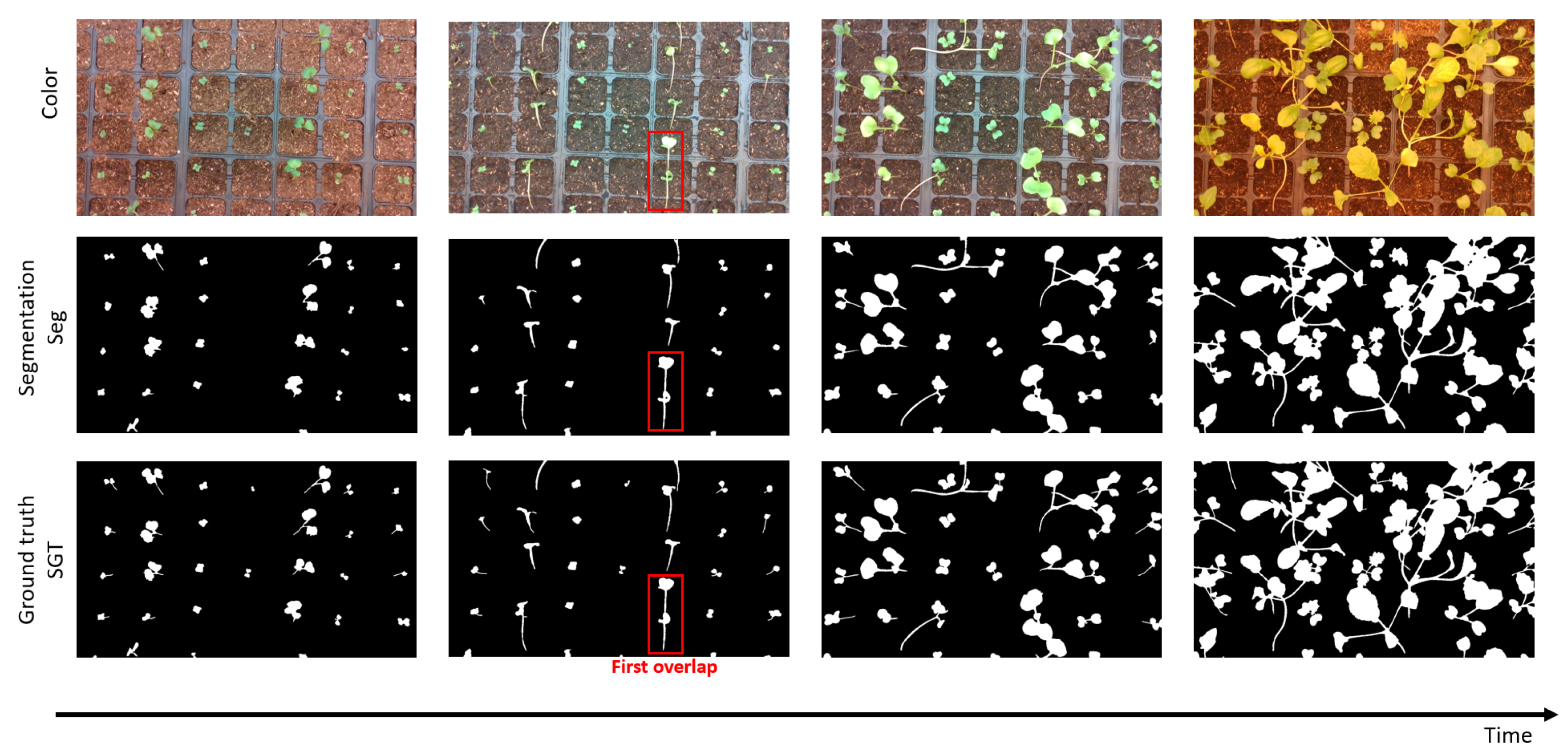
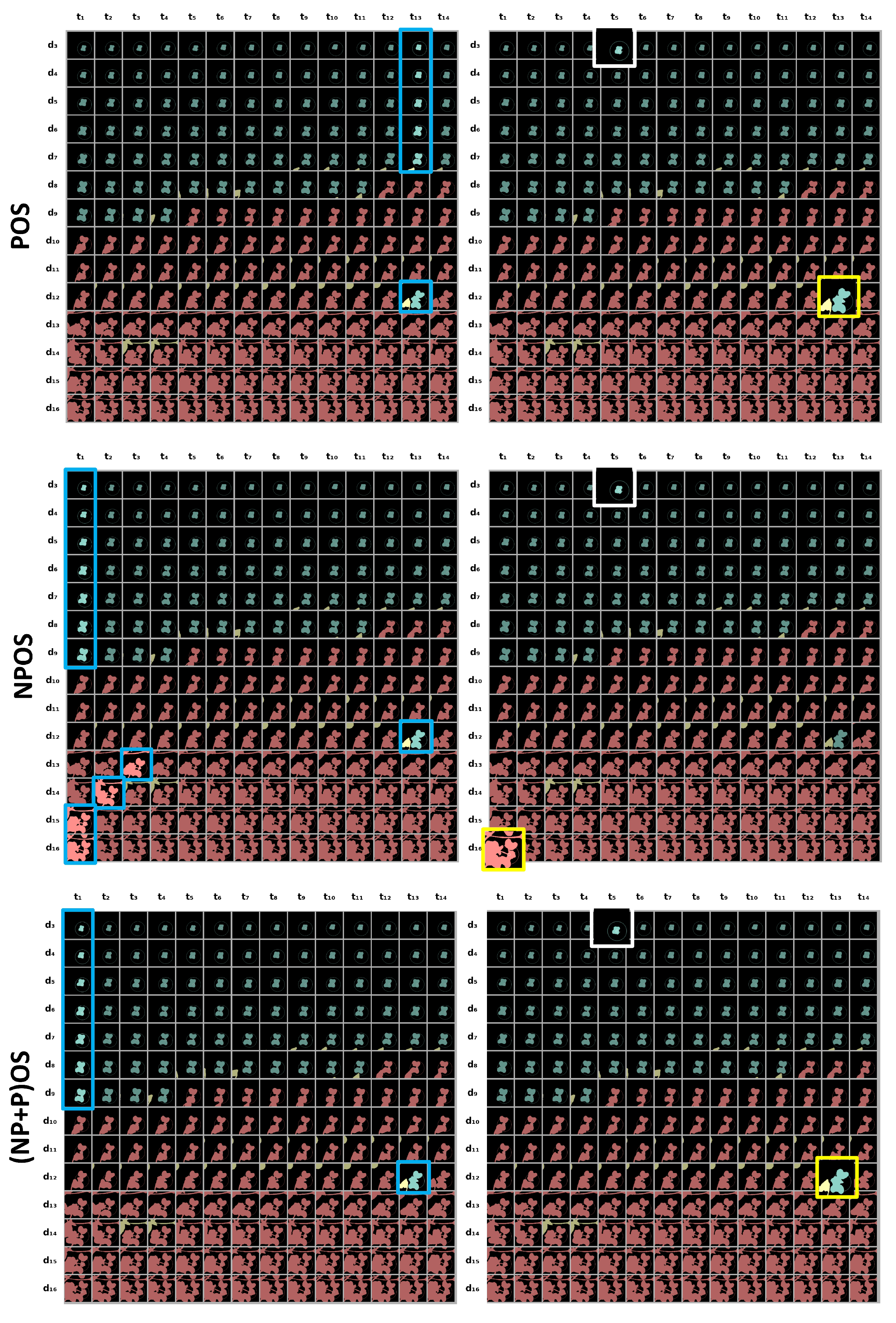
In this section, we present the set of sampling algorithms developed to monitor the individual growth of young plants in time sequences of binary images thanks to circadian cycles movements. We define a frame as a binary image from the temporal sequence of binary images and an object as a connected component among a frame. We propose two types of methods: frame sampling and object sampling. The frame sampling methods process each frame as a whole to select the best frames in the temporal sequences. The object sampling methods process each object in the frame separately to produce a reconstructed temporal sequence for each plant in the end.
3.1. Frame Sampling
Let be the number of detected objects, i.e., connected component, in the frame at day and time . We hypothesize here that a decrease in the number of detected objects indicates the presence of overlaps between plants.
3.1.1. Baseline
We iterate over successive frames until an overlap is detected. In this case, the last frame obtained is considered as the last one available without plant overlapping, as represented in
Figure 3. This basic approach corresponds to a trivial baseline that we propose to improve via more advanced criteria of selections for the frame or for the objects.
3.1.2. Periodic Frame Sampling
When monitoring dynamic processes, a standard approach is to have a periodic sampling rate. This type of sampling is designated notch filter in electrical engineering. Ambient conditions change significantly within a day. By synchronizing the sampling rate to the photoperiod, we obtain time series with the closest conditions between the time points. As the circadian rhythm acts on the inclination of the leaves, we expect that there exists an optimal time of the day to separate each plant on the longest possible duration.
We therefore tested all possible sample times of the day and select the one which enables maximum duration while keeping as long as possible the same number of objects in frames. As illustrated in
Figure 3, this method corresponds to a down-sampling of fixed periodicity equal to the photoperiod and synchronized to the optimal time of the day
.
3.1.3. Non-Periodic Frame Sampling
The first overlap of objects might not be the last opportunity to have no overlap in the rest of the time sequence. In addition, the frame with no overlap might not be positioned at periodic time points. Therefore, as illustrated in
Figure 3, an alternative to the two previous sampling methods consists of selecting all the frames in which no overlap is detected.
3.2. Object Sampling
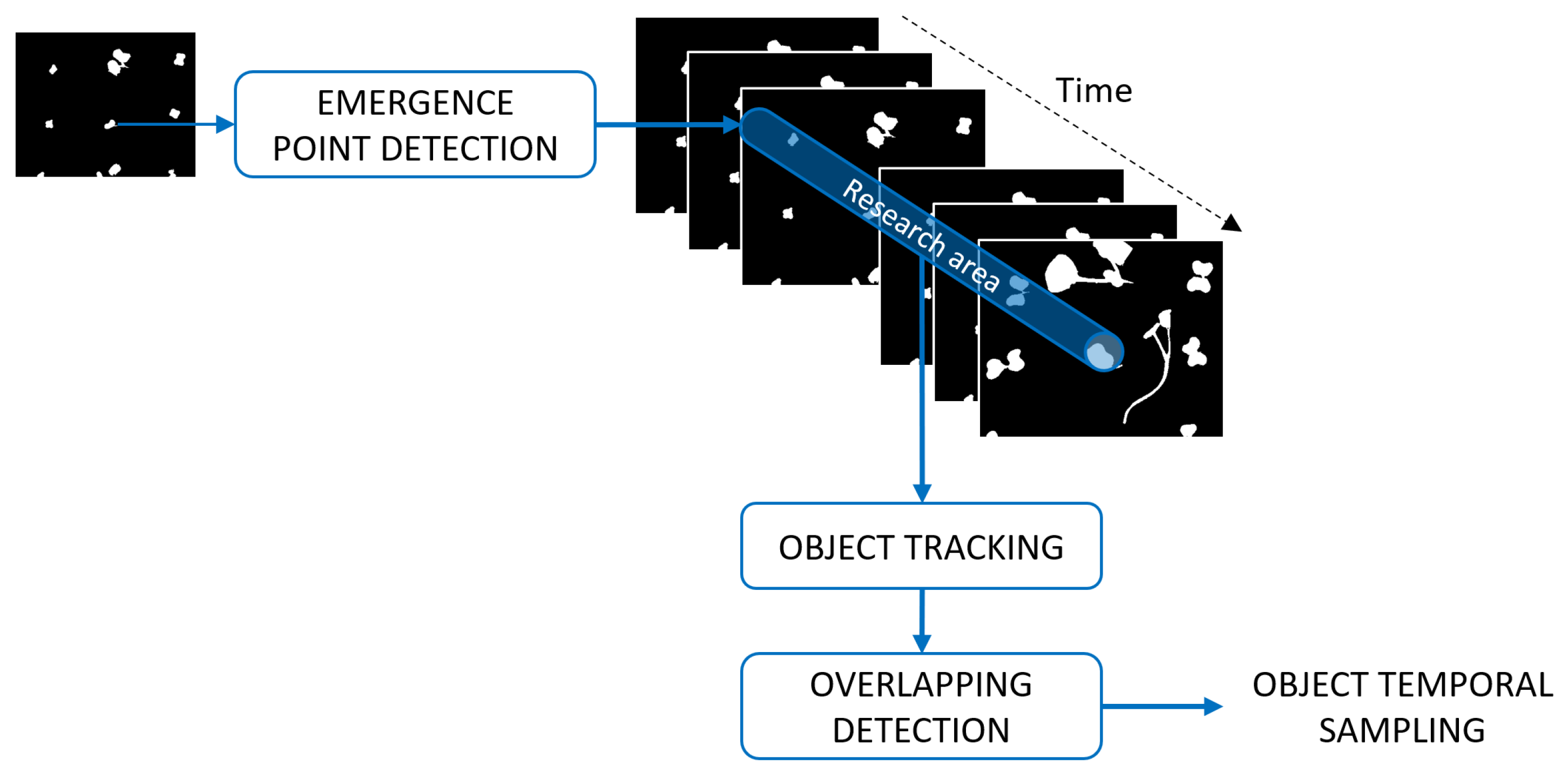
For frame sampling methods, a single overlap is enough to not select a frame even if the other objects are not overlapped. To overcome the constraint of studying all objects simultaneously at the level of the frame, we now propose individualizing the monitoring of each plants following the two steps in the pipeline of
Figure 4. First, the connected components in each frame are tracked. Overlaps are then detected among the sequence of individual tracked objects.
3.2.1. Plant Tracking
Objects are initialized at the emergence of the plants from the soil. The location of the emergence points is then used to define a research area. We selected this area as a disk centered on the emergence point and with the radius empirically chosen as the radius of the pots, as illustrated in
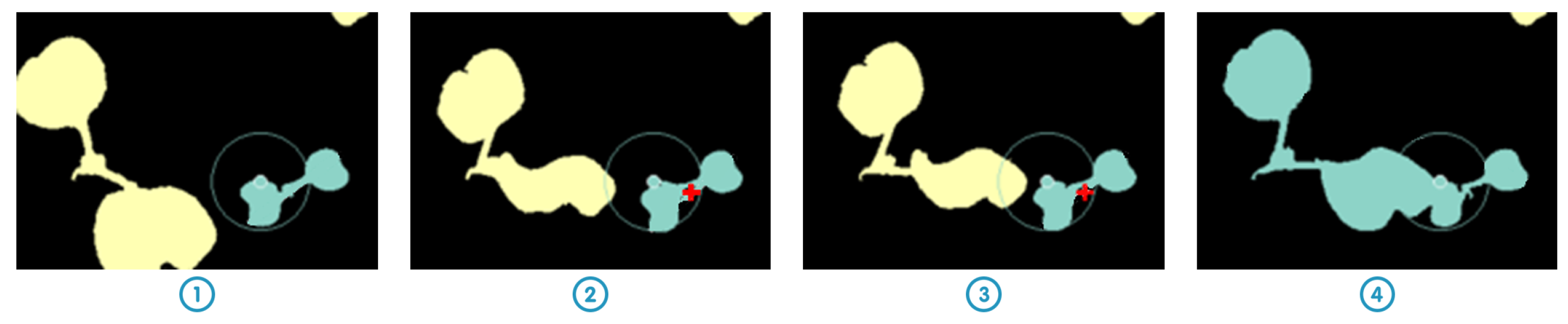
Figure 4. Objects are then tracked within this research area as described in
Figure 5. If several objects are found in the research area, the one with the center of mass the closest to the previous center of mass of the tracked object is selected.
3.2.2. Overlapping Detection
The individual objects might include some overlapping of plants at some times. As for frame sampling methods, we define temporal strategies to select non-overlapped plants in the sequence of each object.
We assume that when plants overlap, the evolution of the objects should be abrupt. The evolution of an object
is extracted as a sequence of binary masks, noted
. For illustration in this article, we have characterized the size and shape of objects by their surface
and their diameter
, where
measures the cardinality,
measures the maximum, and
measures the Euclidian norm. To analyze object evolution, we then defined
the temporal features of interest as
with
representing the time interval used. Here again as a disclaimer, we advocate that many different feature vectors
could be chosen and that the specific choice of parameters could be further optimized without loss of general interest of the proposed sampling methods.
To detect if an object is composed of a single plant or an overlapping of several plants, we set a threshold on the absolute and relative growth of the selected features defined in Equation (
1). Absolute thresholding is first used to detect if physical quantities are sufficiently high for an abrupt evolution. Then, relative thresholding is used to analyze this evolution in relation to the size of the object.
As for frame sampling strategies defined in
Figure 3, we detect abrupt evolution with or without a periodic constraint. A first approach is to use a non-periodic object sampling by detecting abrupt growth on successive objects. Another approach is to use a periodic object sampling by detecting abrupt growth on objects separated by one photoperiod, i.e.,
. Let NPOS be the method of non-periodic object sampling and POS be the method of periodic object sampling.
As shown in
Figure 3, the sequences returned by the NPOS and POS sampling methods are distinct, since they operate at different time scales for the exclusion of abrupt changes. Thus, it is possible to further apply an optimal periodic sampling on the non-periodic object sampling. We tested this combination of non-periodic sampling followed by a periodic sampling at the level of each object that we called (NP+P)OS.
3.3. Metrics
To compare the frame sampling and object sampling methods, one can analyze the maximum duration of plant monitoring. An obvious metric would be to use the gain of time by comparison with the baseline method. However, due to possible errors in overlapping detection, some methods can produce final output exceeding the temporal ground truth (TGT). To take this issue into account, we consider the signed Mean Absolute Error (sMAE) as the metric
where
is the temporal ground truth for maximum duration of monitoring and
is the maximum duration of monitoring for the concerned method. We look for the minimal absolute value of the sMAE and have a negative sign to avoid the monitoring of overlapped plants.
One can also analyze the source of errors committed by the different sampling methods. This is given by the rate of wrong object selection for the tracking part and the rate of wrong frame classification for the overlapping detection part. An overall analysis of the daily output sequence quality can be given by its rate of wrong frames. It is also meaningful to look at the number of empty frames in the output sequence. Counting empty frames is relevant to quantify the number of days without frames containing the object detected as non-overlapped.
3.4. Parameter Tuning
Some parameters are used for the overlapping detection part of the object sampling methods. They correspond to the thresholds of the absolute and relative features defined in Equation (
1). In order to maximize the duration of monitoring and optimize the metric defined in Equation (
2), we need to tune these parameters. For a first proof of interest of the sampling methods proposed here, the thresholds were set empirically on a subset of three objects from the Cabbage
dataset. All remaining objects of Cabbage
, Cabbage
and Cabbage
are used to validate the methods.
The growth rate of plants directly depends on the species studied. This has an impact on the optimal values of the thresholds. To ensure the transferability of the methods to the change of species, we also use two objects from the Pepper dataset to fine-tune the corresponding parameters.
4. Results
We are now ready to compare the different sampling methods proposed to monitor individual plant growth. The comparison on the criterion of maximum duration of the output sequence is provided in
Table 1 for the Cabbage datasets.
An overall result expressed by the maximum monitoring duration is the superiority of the object sampling methods compared to the frame sampling one. The best results are obtained by (NP+P)OS with a gain of 7.23 days on the segmentation ground truth compared to the baseline method. This represents a considerable advance in the monitoring of plants. We go from 3 days of monitoring to a monitoring of about 10 days. This leads to a possible analysis time of the sequence 2.41 times longer than for the basic method. This method optimizes the
, defined in Equation (
2), with an average of 0.23 days lower than the temporal ground truth on segmentation ground truth. On these examples, for none of the objects do we exceed the temporal ground truth, which gives an insight into the ability of the method not to extend the final sequence to wrongly selected frames. This observation is not the same for the NPOS and POS methods, which in addition to presenting larger deviations from the temporal ground truth tend to exceed the maximum possible real duration, which is reflected by the positive sign of the
.
Let us focus on the errors produced by object sampling methods in
Table 2. For each dataset, the (NP+P)OS method brings less errors in the output sequence. The combination of nearest temporality and growth seasonality outperforms the use of each one separately. Applied on segmentation ground truth, no output sequence has a misdetected frame. Even with segmentation errors, the (NP+P)OS method ensures a low amount of wrong sampled frames.
In
Table 2, the transition from Cabbage
to Cabbage
with the addition of segmentation errors demonstrates a strong transferability of the methods. The methods still perform well overall on datasets with segmentation errors. Transferability to another species is also shown by the stability of the results when switching to the Pepper
dataset.
It is important to note that the POS method has lower frame classification errors on segmentation predictions than the NPOS and (NP+P)OS methods. On the other hand, due to the algorithm structure, a false detection of overlap is directly propagated throughout the daily series.
In
Figure 6, we present an example of frame selection with object sampling methods and the gain of the methods compared to the baseline method. This example is relevant to the comparison of the methods because there is a long period with overlapping on the studied plant. The gain can therefore be significant. The two methods POS and (NP+P)OS return correct sequences and bring a gain of 9 days. The periodic sampling of (NP+P)OS fixed the errors of the NPOS method. This example shows the improvement brought by the combination of the periodic and non-periodic sampling.
5. Discussion
The methods presented provide a significant extension of the duration of the plant monitoring. However, there are some remaining errors. We propose to analyze them for the best method, i.e., the (NP+P)OS method, as provided in
Table 3. For the (NP+P)OS method applied on segmentation ground truth, the main source of errors corresponds to the simultaneous occurrence of overlaps. When several overlaps appear in the time interval between two frames, the abrupt change is well detected, and the object is well identified as overlapped. However, we do not know how many plants are actually overlapped. When the loss of an overlap on the object is detected, we can conclude that the object is non-overlapped when it still is, as shown in
Figure 7. To avoid this kind of error, we could increase the acquisition frequency. In addition, one could estimate the surface of leaves at each time frame over the population and detect such simultaneous overlaps.
The second source of error corresponds to a wrong overlapping detection due to the fact that objects can reach the limits of the frames. Then, objects are cut, and it results in an artificial abrupt growth. For each plant monitored, we should then restrict the application of the sampling methods to a spatio-temporal domain in which we are not certain of reaching the limits of the frames.
The binarization of the predicted images was produced by a standard Random Forest algorithm available under open source software [
28]. However, any alternative solution providing correct segmentation is likely to be used. Depending on the segmentation accuracy, different gains can be obtained. This impact of the quality of the segmentation on the performance of our sampling algorithms is an interesting perspective for further work.
The closest work carried out in [
10] proposes a different approach. The acquisition frequency is much lower and the few overlaps are processed by image processing methods, in particular watershed [
29]. The claimed gain for the method in [
10] is 28%, which is equivalent to 2 days on the dataset considered. The (NP+P)OS method seems to be better for each of our studied datasets with a gain presented in
Table 2 in the range of 10 days. Interestingly, our method does not perform spatial image processing but rather focuses on the temporal selection of the frame or the object in the images. These two approaches are complementary and compatible with each others. Their coupling could be studied in order to further extend the duration of monitoring of overlapped plants. In addition, additional sensors such as the depth sensor used in [
3,
11,
14,
15,
23] could be used to enhance the contrast between overlapping plants.
Our sampling methods require an oversampling of the plant compared to the time scale of the circadian movement of the plants. This oversampling comes with a cost corresponding to an increase of data flow and data storage. In our hardware setup, images were first acquired on nano-computers [
25] and then transferred to a main storing server. So far for this article, all images were transferred to the main server, and the sampling methods were simulated on data at rest. However, this hardware setup, with two processing units, offers opportunities for the deployment of our sampling methods during the acquisition process. The segmentation of the images could be first deployed on the nano-computers together with the sampling methods proposed in this article. Frames or objects detected as overlapped could be erased directly. Then, only the sampled frames or objects could be transferred to the main server. The overlapping detection of our sampling methods requires a buffer which varies from one method to another. Only the last frame is needed for non-periodic sampling when a full sliding day needs to be buffered for periodic sampling. Embedding our sampling method in nano-computers together with advanced watershed approaches and enhanced contrast with depth cameras constitutes a practical perspective currently under development in our group.
The proposed sampling methods are presented in controlled conditions. Segmentation is facilitated by an overall even and stable illumination. This also ensures that the movements correspond to natural movements of the plants. However, the proposed sampling methods could be extended to other growth conditions. For instance, they could be applied in outdoor conditions benefiting from other sources of movements (due to wind). In such conditions, more elaborated segmentation algorithms should be used, but the sampling methods would remain valuable.