A Systematic Literature Review on Crop Yield Prediction with Deep Learning and Remote Sensing
Abstract
1. Introduction
2. Research Methods
2.1. Review Methodology
2.2. Research Questions
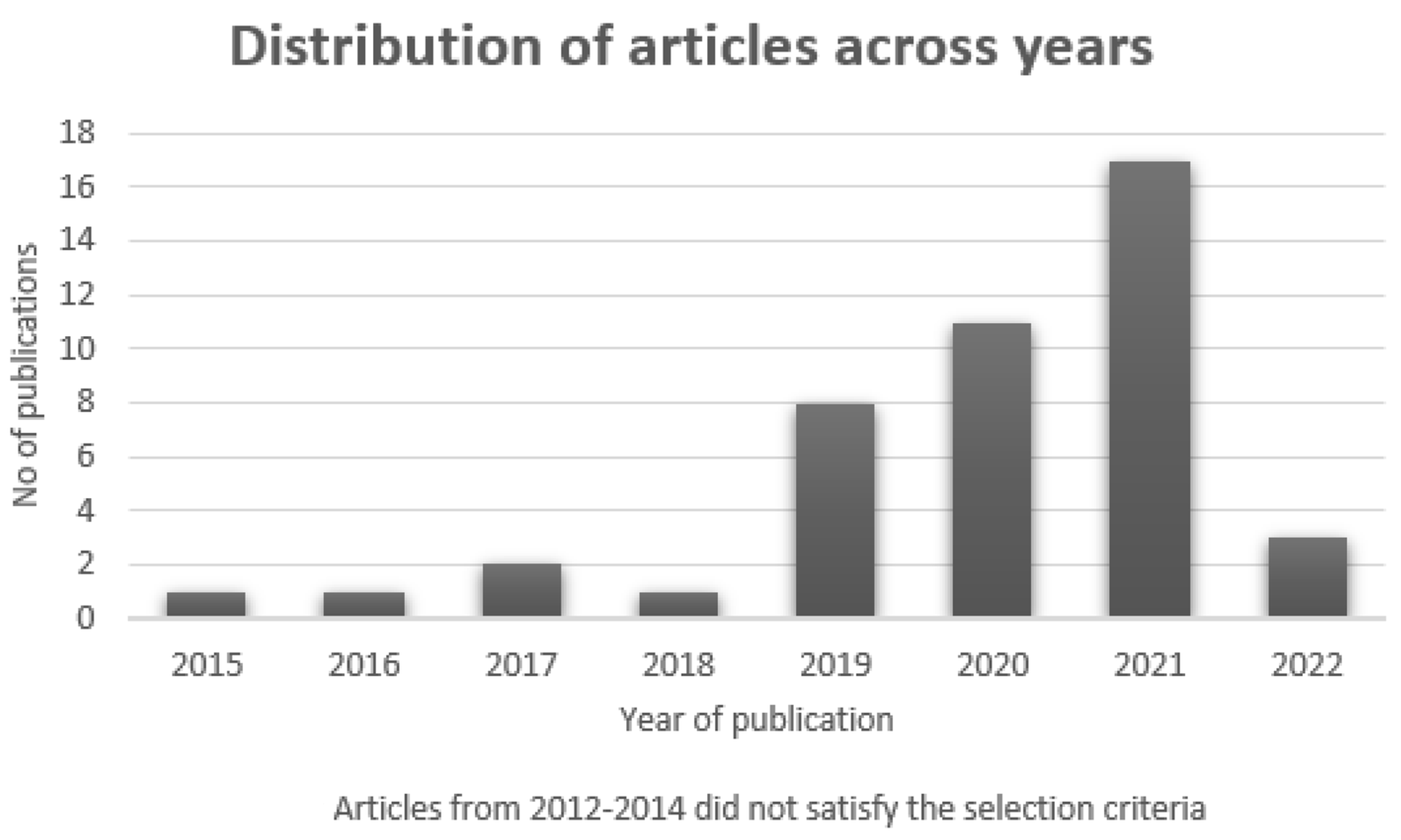
2.3. Procedure for Article Search
2.4. Article Selection Criteria
- Articles that belong to the agricultural sector but that do not fall under crop yield prediction;
- Publications that include machine learning approaches for crop yield prediction;
- Publications that have no open access;
- Literature search for articles that are published before 2012;
- Articles in different languages other than English.
3. Overview of the Existing Approaches
3.1. Deep Learning
3.1.1. Artificial Neural Networks (ANN)
3.1.2. Deep Neural Networks (DNN)
3.1.3. Bayesian Neural Networks (BNN)
3.1.4. Convolution Neural Network (CNN)
3.1.5. 2D-CNN and 3D-CNN
3.1.6. Faster R-CNN
3.1.7. Long Short-Term Memory (LSTM)
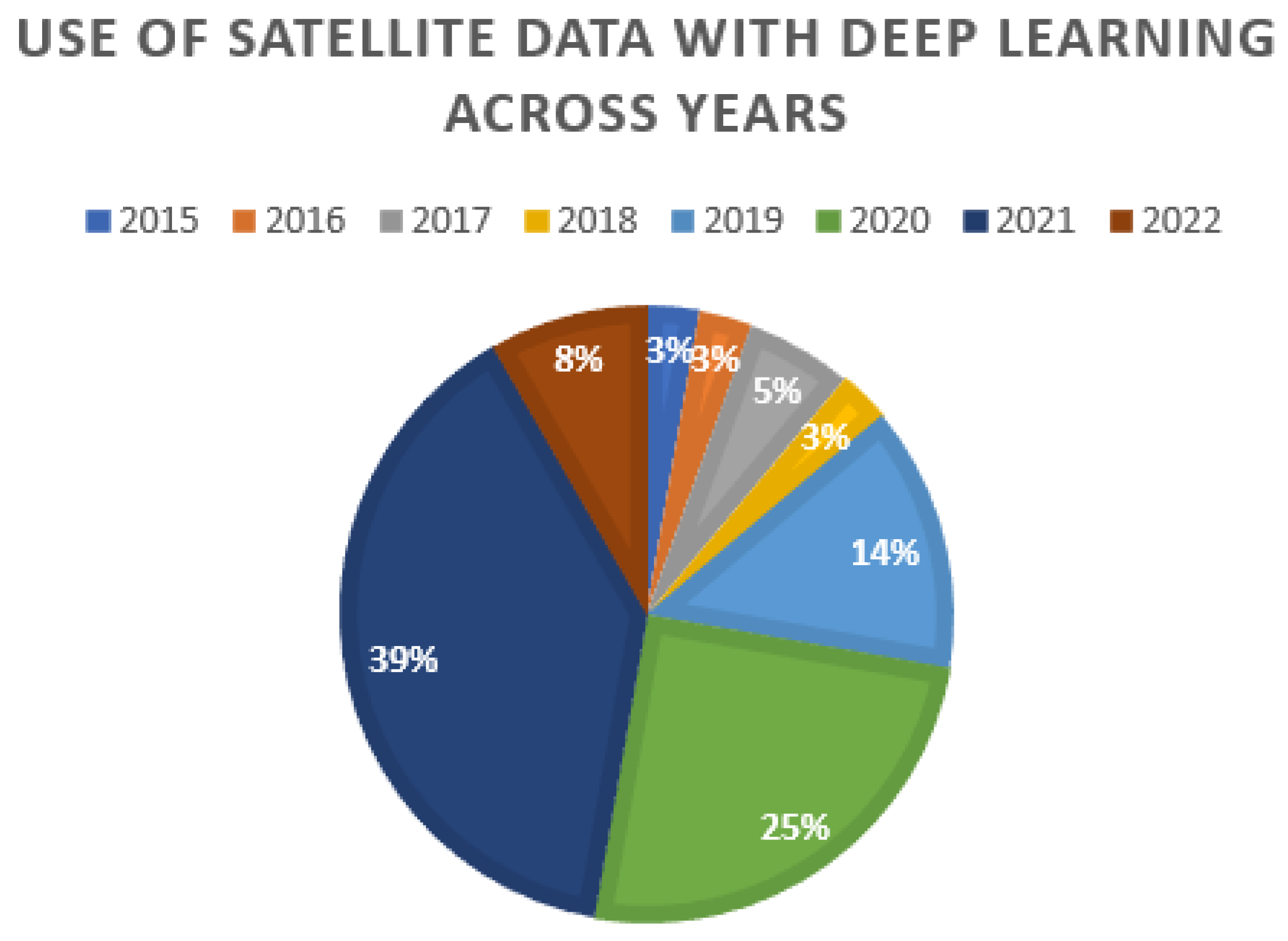
3.2. Remote Sensing for Data Acquisition
3.3. Impact of Vegetation Indices and Environmental Factors
4. Results and Discussion
- RQ1
- —Approaches used in literature discussion:
- TIMESAT;
- Neurosolution version 7.1.1.1;
- Use of Orthoimages;
- The labeling program developed by the Computer Science and Artificial Intelligence Laboratory (MIT, Massachusetts, USA);
- FarmWorks;
- Google Earth Engine-based tensor generation;
- Orthomosaic map generation (RGB images)—Agisoft PhotoScan Professional 1.2.5;
- Orthomosaic reflectance map (multispectral images)—Pix4Dmapper 4.0;
- Georeferencing—Esri ArcGIS 10.3;
- The clipping process—“ExtractByMask” function from Arcpy Python module;
- Layer stacking of images;
- Mosaic and orthorectify, lens distortion, and vignetting issue correction (UAV RGB images)—Pix4Dmapper software;
- Drawing of shapefile—plotshpcreate of R library;
- Conversion of Geotiff plots to Numpy arrays—Python script;
- Dimension transform technique—irregular shaped images;
- MODIS products;
- Image pre-processing—Spectronon software (version 2.134; Resonon, Inc., Bozeman, Montana);
- Spectral data denoising—wavelet transform technique.
- RQ2
- —Remote sensing used with deep learning:
- RQ3
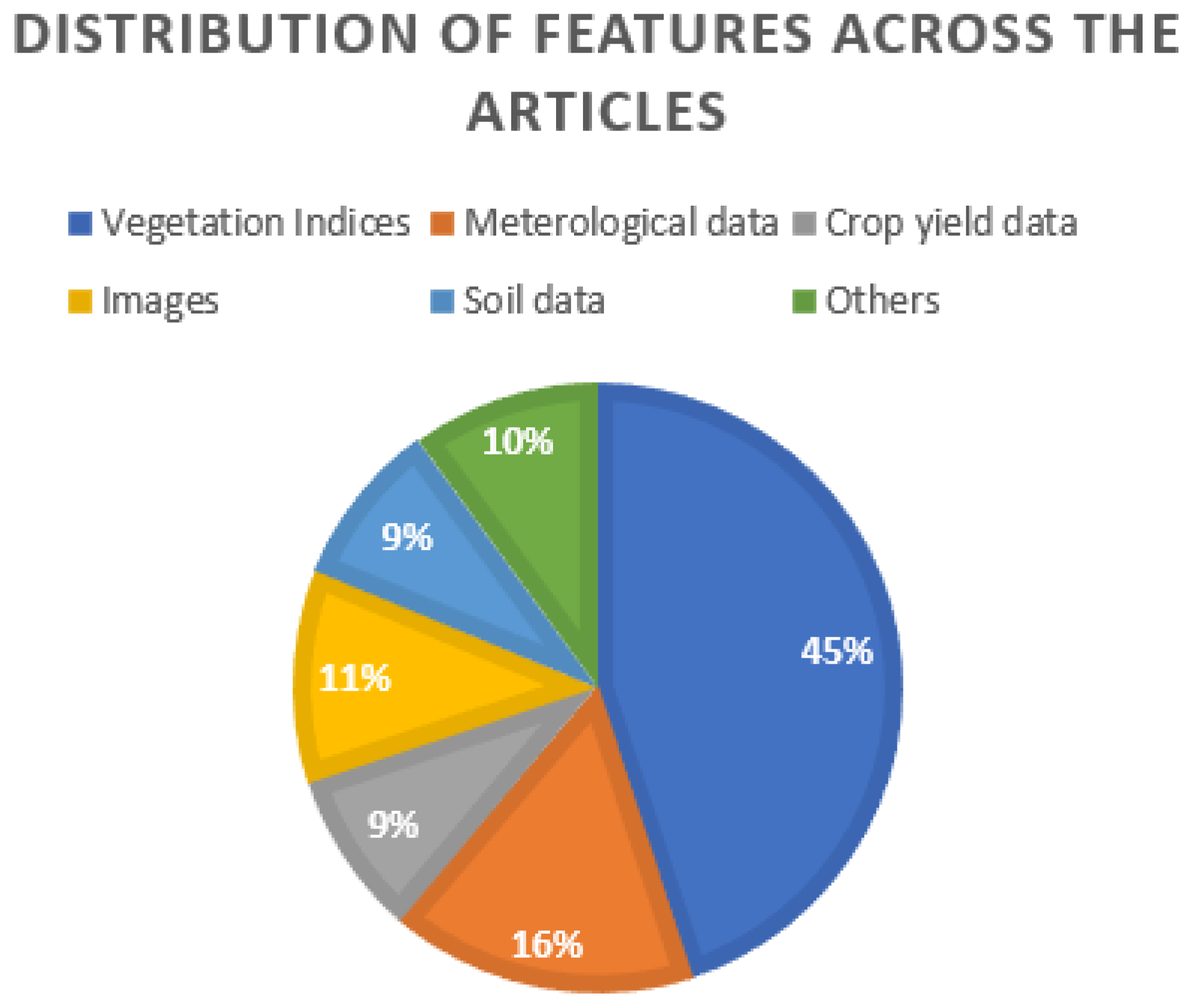
- —Features used in crop yield prediction
- RQ4
- —Challenges in using deep learning approaches and remote sensing for crop yield prediction
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- WHO. World Hunger Is Still Not Going Down after Three Years and Obesity Is Still Growing—UN Report. Available online: https://www.who.int/news/item/15-07-2019-world-hunger-is-still-not-going-down-after-three-years-and-obesity-is-still-growing-un-report (accessed on 15 December 2021).
- UN. Pathways to Zero Hunger. Available online: https://www.un.org/zerohunger/content/challenge-hunger-can-be-eliminated-our-lifetimes (accessed on 15 December 2021).
- Kheir, A.M.S.; Alkharabsheh, H.M.; Seleiman, M.F.; Al-Saif, A.M.; Ammar, K.A.; Attia, A.; Zoghdan, M.G.; Shabana, M.M.A.; Aboelsoud, H.; Schillaci, C. Calibration and validation of AQUACROP and APSIM models to optimize wheat yield and water saving in Arid regions. Land 2021, 10, 1375. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Tranfield, D.; Denyer, D.; Smart, P. Towards a methodology for developing evidence-informed management knowledge by means of systematic review. Br. J. Manag. 2003, 14, 207–222. [Google Scholar] [CrossRef]
- Kitchenham, B.A.; Charters, S. Guidelines for Performing Systematic Literature Reviews in Software Engineering (EBSE 2007-001); Keele University: Keele, UK; Durham University: Durham, UK, 2007. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef] [PubMed]
- Shen, C. A Transdisciplinary review of deep learning research and its relevance for water resources scientists. Water Resour. Res. 2018, 54, 8558–8593. [Google Scholar] [CrossRef]
- Basso, B.; Cammarano, D.; Carfagna, E. Review of crop yield forecasting methods and early warning systems. In Proceedings of the First Meeting of the Scientific Advisory Committee of the Global Strategy to Improve Agricultural and Rural Statistics, FAO, Rome, Italy, 9–10 April 2013. [Google Scholar]
- Horie, T.; Yajima, M.; Nakagawa, H. Yield forecasting. Agric. Syst. 1992, 40, 211–236. [Google Scholar] [CrossRef]
- Jeong, J.H.; Resop, J.P.; Mueller, N.D.; Fleisher, D.H.; Yun, K.; Butler, E.E.; Timlin, D.J.; Shim, K.-M.; Gerber, J.S.; Reddy, V.R. Random Forests for global and regional crop yield predictions. PLoS ONE 2016, 11, e0156571. [Google Scholar] [CrossRef]
- Islam, N.; Rashid, M.; Wibowo, S.; Xu, C.-Y.; Morshed, A.; Wasimi, S.; Moore, S.; Rahman, S. Early weed detection using image processing and machine learning techniques in an Australian chilli farm. Agriculture 2021, 11, 387. [Google Scholar] [CrossRef]
- Islam, N.; Rashid, M.M.; Wibowo, S.; Wasimi, S.; Morshed, A.; Xu, C.; Moore, S.T. Machine learning based approach for weed detection in chilli field using RGB images. In Advances in Natural Computation, Fuzzy Systems and Knowledge Discovery; Meng, H., Lei, T., Li, M., Li, K., Xiong, N., Wang, L., Eds.; Springer: Cham, Switzerland, 2021; Volume 88, pp. 1097–1105. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Khaki, S.; Wang, L.; Archontoulis, S.V. A CNN-RNN framework for crop yield prediction. Front. Plant Sci. 2020, 10, 1750. [Google Scholar] [CrossRef]
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Johnson, M.D.; Hsieh, W.W.; Cannon, A.J.; Davidson, A.; Bédard, F. Crop yield forecasting on the Canadian Prairies by remotely sensed vegetation indices and machine learning methods. Agric. For. Meteorol. 2016, 218–219, 74–84. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Nevavuori, P.; Narra, N.; Lipping, T. Crop yield prediction with deep convolutional neural networks. Comput. Electron. Agric. 2019, 163, 104859. [Google Scholar] [CrossRef]
- Fernandez-Beltran, R.; Baidar, T.; Kang, J.; Pla, F. Rice-yield prediction with multi-temporal Sentinel-2 Data and 3D CNN: A case study in Nepal. Remote Sens. 2021, 13, 1391. [Google Scholar] [CrossRef]
- Chen, Y.; Lee, W.S.; Gan, H.; Peres, N.; Fraisse, C.; Zhang, Y.; He, Y. Strawberry Yield Prediction Based on a Deep Neural Network Using High-Resolution Aerial Orthoimages. Remote Sens. 2019, 11, 1584. [Google Scholar] [CrossRef]
- Tian, H.; Wang, P.; Tansey, K.; Zhang, J.; Zhang, S.; Li, H. An LSTM neural network for improving wheat yield estimates by integrating remote sensing data and meteorological data in the Guanzhong Plain, PR China. Agric. For. Meteorol. 2021, 310, 108629. [Google Scholar] [CrossRef]
- Tian, H.; Wang, P.; Tansey, K.; Han, D.; Zhang, J.; Zhang, S.; Li, H. A deep learning framework under attention mechanism for wheat yield estimation using remotely sensed indices in the Guanzhong Plain, PR China. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102375. [Google Scholar] [CrossRef]
- Baghdadi, N.; Zribi, M. Optical Remote Sensing of Land Surface: Techniques and Methods; Elsevier: Oxford, UK, 2016. [Google Scholar]
- Eliu, P. A survey of remote-sensing big data. Front. Environ. Sci. 2015, 3, 45. [Google Scholar] [CrossRef]
- Kirkaya, A. Smart farming—Precision agriculture technologies and practices. J. Sci. Perspect. 2020, 4, 123–136. [Google Scholar] [CrossRef]
- Gómez, D.; Salvador, P.; Sanz, J.; Casanova, J.L. Potato yield prediction using machine learning techniques and Sentinel 2 data. Remote Sens. 2019, 11, 1745. [Google Scholar] [CrossRef]
- Kobayashi, N.; Tani, H.; Wang, X.; Sonobe, R. Crop classification using spectral indices derived from Sentinel-2A imagery. J. Inf. Telecommun. 2020, 4, 67–90. [Google Scholar] [CrossRef]
- Thomas, G.; Taylor, J.; Wood, G. Mapping yield potential with remote sensing. In Proceedings of the First European Conference on Precision Agriculture, Warwick University Conference Centre, Coventry, UK, 7–10 September 1997. [Google Scholar]
- Liang, S.; Wang, J. Advanced Remote Sensing: Terrestrial Information Extraction and Applications; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Liang, S.; Wang, J. Chapter 3—Compositing, Smoothing, and Gap-Filling Techniques; Academic Press: Cambridge, MA, USA, 2020; pp. 107–130. [Google Scholar]
- Fang, H.; Liang, S. Leaf area index models. In Reference Module in Earth Systems and Environmental Sciences; Elsevier: Burlington, MA, USA, 2014. [Google Scholar]
- Santos, G.O.; Rosalen, D.L.; de Faria, R.T. Use of active optical sensor in the characteristics analysis of the fertigated brachiaria with treated sewage. Eng. Agrícola 2017, 37, 1213–1221. [Google Scholar] [CrossRef]
- Zhao, B.; Duan, A.; Ata-Ul-Karim, S.T.; Liu, Z.; Chen, Z.; Gong, Z.; Zhang, J.; Xiao, J.; Liu, Z.; Qin, A.; et al. Exploring new spectral bands and vegetation indices for estimating nitrogen nutrition index of summer maize. Eur. J. Agron. 2018, 93, 113–125. [Google Scholar] [CrossRef]
- Junior, C.K.; Guimarães, A.M.; Caires, E.F. Use of active canopy sensors to discriminate wheat response to nitrogen fertilization under no-tillage. Eng. Agrícola 2016, 36, 886–894. [Google Scholar] [CrossRef][Green Version]
- Pantazi, X.-E.; Moshou, D.; Bravo, C. Active learning system for weed species recognition based on hyperspectral sensing. Biosyst. Eng. 2016, 146, 193–202. [Google Scholar] [CrossRef]
- Yao, Y.; Miao, Y.; Huang, S.; Gao, L.; Ma, X.; Zhao, G.; Jiang, R.; Chen, X.; Zhang, F.; Yu, K.; et al. Active canopy sensor-based precision N management strategy for rice. Agron. Sustain. Dev. 2012, 32, 925–933. [Google Scholar] [CrossRef]
- Mkhabela, M.; Bullock, P.; Raj, S.; Wang, S.; Yang, Y. Crop yield forecasting on the Canadian Prairies using MODIS NDVI data. Agric. For. Meteorol. 2011, 151, 385–393. [Google Scholar] [CrossRef]
- Wall, L.; Larocque, D.; Léger, P. The early explanatory power of NDVI in crop yield modelling. Int. J. Remote Sens. 2008, 29, 2211–2225. [Google Scholar] [CrossRef]
- Bolton, D.K.; Friedl, M.A. Forecasting crop yield using remotely sensed vegetation indices and crop phenology metrics. Agric. For. Meteorol. 2013, 173, 74–84. [Google Scholar] [CrossRef]
- Basnyat, B.M.P.; Lafond, G.P.; Moulin, A.; Pelcat, Y. Optimal time for remote sensing to relate to crop grain yield on the Canadian prairies. Can. J. Plant Sci. 2004, 84, 97–103. [Google Scholar] [CrossRef]
- Ren, J.; Chen, Z.; Zhou, Q.; Tang, H. Regional yield estimation for winter wheat with MODIS-NDVI data in Shandong, China. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 403–413. [Google Scholar] [CrossRef]
- Salazar, L.; Kogan, F.; Roytman, L. Use of remote sensing data for estimation of winter wheat yield in the United States. Int. J. Remote Sens. 2007, 28, 3795–3811. [Google Scholar] [CrossRef]
- Hack, H.; Bleiholder, H.; Buhr, L.; Meier, U.; Schnock-Fricke, U.; Weber, E.; Witzenberger, A. A uniform code for phenological growth stages of mono-and dicotyledonous plants - Extended BBCH scale, general. Nachr. Des. Dtsch. Pflan-Zenschutzd. 1992, 44, 265–270. [Google Scholar]
- Knoblauch, C.; Watson, C.; Berendonk, C.; Becker, R.; Wrage-Mönnig, N.; Wichern, F. Relationship between remote sensing data, plant biomass and soil nitrogen dynamics in intensively managed grasslands under controlled conditions. Sensors 2017, 17, 1483. [Google Scholar] [CrossRef]
- Marti, J.; Bort, J.; Slafer, G.A.; Araus, J.L. Can wheat yield be assessed by early measurements of normalized difference vege-tation index? Ann. Appl. Biol. 2007, 150, 253–257. [Google Scholar] [CrossRef]
- Ali, A.; Martelli, R.; Lupia, F.; Barbanti, L. Assessing multiple years’ spatial variability of crop yields using satellite vegetation indices. Remote Sens. 2019, 11, 2384. [Google Scholar] [CrossRef]
- Domínguez, J.; Kumhálová, J.; Novák, P. Assessment of the relationship between spectral indices from satellite remote sensing and winter oilseed rape yield. Agron. Res. 2017, 15, 055–068. [Google Scholar]
- Panek, E.; Gozdowski, D. Analysis of relationship between cereal yield and NDVI for selected regions of Central Europe based on MODIS satellite data. Remote Sens. Appl. Soc. Environ. 2019, 17, 100286. [Google Scholar] [CrossRef]
- Vallentin, C.; Harfenmeister, K.; Itzerott, S.; Kleinschmit, B.; Conrad, C.; Spengler, D. Suitability of satellite remote sensing data for yield estimation in northeast Germany. Precis. Agric. 2021, 23, 52–82. [Google Scholar] [CrossRef]
- Prey, L.; Hu, Y.; Schmidhalter, U. High-throughput field phenotyping traits of grain yield formation and nitrogen use efficiency: Optimizing the selection of vegetation indices and growth stages. Front. Plant Sci. 2020, 10, 1672. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, Q.; Ren, H.; Sun, Y.; Li, M.; Zhang, T.; Ren, S. Optimal hyperspectral characteristics determination for winter wheat yield prediction. Remote Sens. 2018, 10, 2015. [Google Scholar] [CrossRef]
- Kayad, A.G.; Al-Gaadi, K.A.; Tola, E.; Madugundu, R.; Zeyada, A.M.; Kalaitzidis, C. Assessing the spatial variability of alfalfa yield using satellite imagery and ground-based data. PLoS ONE 2016, 11, e0157166. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, K.; Shibasaki, R. Estimating crop yields with deep learning and remotely sensed data. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 858–861. [Google Scholar]
- You, J.; Li, X.; Low, M.; Lobell, D.; Ermon, S. Deep Gaussian process for crop yield prediction based on remote sensing data. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–10 February 2017; pp. 4559–4565. [Google Scholar]
- Fernandes, J.L.; Ebecken, N.F.F.; Esquerdo, J. Sugarcane yield prediction in Brazil using NDVI time series and neural networks ensemble. Int. J. Remote Sens. 2017, 38, 4631–4644. [Google Scholar] [CrossRef]
- Haghverdi, A.; Washington-Allen, R.A.; Leib, B.G. Prediction of cotton lint yield from phenology of crop indices using artificial neural networks. Comput. Electron. Agric. 2018, 152, 186–197. [Google Scholar] [CrossRef]
- Wang, X.; Huang, J.; Feng, Q.; Yin, D. Winter wheat yield prediction at county level and uncertainty analysis in main wheat-producing regions of China with deep learning approaches. Remote Sens. 2020, 12, 1744. [Google Scholar] [CrossRef]
- de Freitas Cunha, R.L.; Silva, B. Estimating crop yields with remote sensing and deep learning. In Proceedings of the 2020 IEEE Latin American GRSS & ISPRS Remote Sensing Conference (LAGIRS), Santiago, Chile, 22–26 March 2020; pp. 273–278. [Google Scholar]
- Gavahi, K.; Abbaszadeh, P.; Moradkhani, H. DeepYield: A combined convolutional neural network with long short-term memory for crop yield forecasting. Expert Syst. Appl. 2021, 184, 115511. [Google Scholar] [CrossRef]
- Qiao, M.; He, X.; Cheng, X.; Li, P.; Luo, H.; Zhang, L.; Tian, Z. Crop yield prediction from multi-spectral, multi-temporal re-motely sensed imagery using recurrent 3D convolutional neural networks. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102436. [Google Scholar] [CrossRef]
- Abbaszadeh, P.; Gavahi, K.; Alipour, A.; Deb, P.; Moradkhani, H. Bayesian multi-modeling of deep neural nets for probabilistic crop yield prediction. Agric. For. Meteorol. 2021, 314, 108773. [Google Scholar] [CrossRef]
- Mu, H.; Zhou, L.; Dang, X.; Yuan, B. Winter wheat yield estimation from multitemporal remote sensing images based on convolutional neural networks. In Proceedings of the 2019 10th International Workshop on the Analysis of Multitemporal Remote Sensing Images (MultiTemp), Shanghai, China, 5–7 August 2019; pp. 1–4. [Google Scholar]
- Terliksiz, A.S.; Altylar, D.T. Use of deep neural networks for crop yield prediction: A case study of soybean yield in Lauderdale County, Alabama, USA. In Proceedings of the 2019 8th International Conference on Agro-Geoinformatics (Agro-Geoinformatics), Istanbul, Turkey, 16–19 July 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Wolanin, A.; Mateo-García, G.; Camps-Valls, G.; Gómez-Chova, L.; Meroni, M.; Duveiller, G.; Liangzhi, Y.; Guanter, L. Esti-mating and understanding crop yields with explainable deep learning in the Indian wheat belt. Environ. Res. Lett. 2020, 15, 024019. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Z.; Luo, Y.; Zhang, L.; Zhang, J.; Li, Z.; Tao, F. Wheat yield predictions at a county and field scale with deep learning, machine learning, and google earth engine. Eur. J. Agron. 2020, 123, 126204. [Google Scholar] [CrossRef]
- Sun, J.; Di, L.; Sun, Z.; Shen, Y.; Lai, Z. County-level soybean yield prediction using deep CNN-LSTM model. Sensors 2019, 19, 4363. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rai, S.; Krishnan, N.C. Wheat crop yield prediction using deep LSTM model. arXiv 2020, arXiv:2011.01498 2020. [Google Scholar]
- Ghazaryan, G.; Skakun, S.; König, S.; Rezaei, E.E.; Siebert, S.; Dubovyk, O. Crop yield estimation using multi-source satellite image series and deep learning. In Proceedings of the 2020 IEEE International Geoscience and Remote Sensing Symposium, Online, 26 September–2 October 2020; pp. 5163–5166. [Google Scholar]
- Gastli, M.S.; Nassar, L.; Karray, F. Satellite images and deep learning tools for crop yield prediction and price forecasting. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Jeong, S.; Ko, J.; Yeom, J.M. Predicting rice yield at pixel scale through synthetic use of crop and deep learning models with satellite data in South and North Korea. Sci. Total Environ. 2021, 802, 149726. [Google Scholar] [CrossRef]
- Dang, C.; Liu, Y.; Yue, H.; Qian, J.; Zhu, R. Autumn crop yield prediction using data-driven approaches: Support vector machines, random forest, and deep neural network methods. Can. J. Remote Sens. 2020, 47, 162–181. [Google Scholar] [CrossRef]
- Gao, J.; Li, P.; Chen, Z.; Zhang, J. A survey on deep learning for multimodal data fusion. Neural Comput. 2020, 32, 829–864. [Google Scholar] [CrossRef]
- Khaki, S.; Pham, H.; Wang, L. Simultaneous corn and soybean yield prediction from remote sensing data using deep transfer learning. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Kaneko, A.; Kennedy, T.; Mei, L.; Sintek, C.; Burke, M.; Ermon, S.; Lobell, D. Deep learning for crop yield prediction in Africa. In Proceedings of the International Conference on Machine Learning AI for Social Good Workshop, Long Beach, CA, USA, 10–15 June 2019; pp. 1–5. [Google Scholar]
- Jiang, H.; Hu, H.; Zhong, R.; Xu, J.; Xu, J.; Huang, J.; Wang, S.; Ying, Y.; Lin, T. A deep learning approach to conflating het-erogeneous geospatial data for corn yield estimation: A case study of the US corn belt at the county level. Glob. Chang. Biol. 2020, 26, 1754–1766. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Z.; Kang, Y.; Özdoğan, M. Corn yield prediction and uncertainty analysis based on remotely sensed variables using a Bayesian neural network approach. Remote Sens. Environ. 2021, 259, 112408. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Luo, Y.; Cao, J.; Xie, R.; Li, S. Integrating satellite-derived climatic and vegetation indices to predict smallholder maize yield using deep learning. Agric. For. Meteorol. 2021, 311, 108666. [Google Scholar] [CrossRef]
- Xie, Y.; Huang, J. Integration of a Crop Growth Model and Deep Learning Methods to Improve Satellite-Based Yield Estimation of Winter Wheat in Henan Province, China. Remote Sens. 2021, 13, 4372. [Google Scholar] [CrossRef]
- Jin, X.; Li, Z.; Feng, H.; Ren, Z.; Li, S. Deep neural network algorithm for estimating maize biomass based on simulated Sentinel 2A vegetation indices and leaf area index. Crop J. 2019, 8, 87–97. [Google Scholar] [CrossRef]
- Engen, M.; Sandø, E.; Sjølander, B.L.O.; Arenberg, S.; Gupta, R.; Goodwin, M. Farm-scale crop yield prediction from multi-temporal data using deep hybrid neural networks. Agronomy 2021, 11, 2576. [Google Scholar] [CrossRef]
- Xie, Y. Combining CERES-wheat model, Sentinel-2 data, and deep learning method for winter wheat yield estimation. Int. J. Remote Sens. 2022, 43, 630–648. [Google Scholar] [CrossRef]
- Nevavuori, P.; Narra, N.; Linna, P.; Lipping, T. Crop yield prediction using multitemporal UAV data and spatio-temporal deep learning models. Remote Sens. 2020, 12, 4000. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Han, J.; Zha, Y.; Zhu, P. Deep convolutional neural networks for rice grain yield estimation at the ripening stage using UAV-based remotely sensed images. Field Crop. Res. 2019, 235, 142–153. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Lin, L. Plot-scale rice grain yield estimation using UAV-based remotely sensed images via CNN with time-invariant deep features decomposition. In Proceedings of the 2019 IEEE International Geoscience and Remote Sensing Symposium, 28 July–2 August 2019; pp. 7180–7183. [Google Scholar] [CrossRef]
- Yang, W.; Nigon, T.; Hao, Z.; Dias Paiao, G.; Fernández, F.G.; Mulla, D.; Yang, C. Estimation of corn yield based on hyper-spectral imagery and convolutional neural network. Comput. Elect. Agric. 2021, 184, 106092. [Google Scholar] [CrossRef]
- Sagan, V.; Maimaitijiang, M.; Bhadra, S.; Maimaitiyiming, M.; Brown, D.R.; Sidike, P.; Fritschi, F.B. Field-scale crop yield pre-diction using multi-temporal WorldView-3 and PlanetScope satellite data and deep learning. ISPRS J. Photogramm. Remote Sens. 2021, 174, 265–281. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2019, 237, 111599. [Google Scholar] [CrossRef]
- Danilevicz, M.F.; Bayer, P.E.; Boussaid, F.; Bennamoun, M.; Edwards, D. Maize yield prediction at an early developmental stage using multispectral images and genotype data for preliminary hybrid selection. Remote Sens. 2021, 13, 3976. [Google Scholar] [CrossRef]
- Sun, J.; Lai, Z.; Di, L.; Sun, Z.; Tao, J.; Shen, Y. Multilevel deep learning network for county-level corn yield estimation in the U.S. corn belt. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 5048–5060. [Google Scholar] [CrossRef]
- Kross, A.; Znoj, E.; Callegari, D.; Kaur, G.; Sunohara, M.; Lapen, D.; McNairn, H. Using artificial neural networks and remotely sensed data to evaluate the relative importance of variables for prediction of within-field corn and soybean yields. Remote Sens. 2020, 12, 2230. [Google Scholar] [CrossRef]
- Barbosa, B.D.S.; Ferraz, G.A.e.S.; Costa, L.; Ampatzidis, Y.; Vijayakumar, V.; dos Santos, L.M. UAV-based coffee yield pre-diction utilizing feature selection and deep learning. Smart Agric. Technol. 2021, 1, 100010. [Google Scholar] [CrossRef]
- Bronstein, M.M.; Bronstein, A.M.; Michel, F.; Paragios, N. Data fusion through cross-modality metric learning using similarity-sensitive hashing. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010; pp. 3594–3601. [Google Scholar]
- Poria, S.; Cambria, E.; Bajpai, R.; Hussain, A. A review of affective computing: From unimodal analysis to multimodal fusion. Inf. Fusion 2017, 37, 98–125. [Google Scholar] [CrossRef]
- Khaki, S.; Wang, L. Crop yield prediction using deep neural networks. Front. Plant Sci. 2019, 10, 621. [Google Scholar] [CrossRef]
- Bramon, R.; Boada, I.; Bardera, A.; Rodriguez, J.; Feixas, M.; Puig, J.; Sbert, M. Multimodal data fusion based on mutual in-formation. IEEE Trans. Vis. Comput. Graph. 2011, 18, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, Aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef]
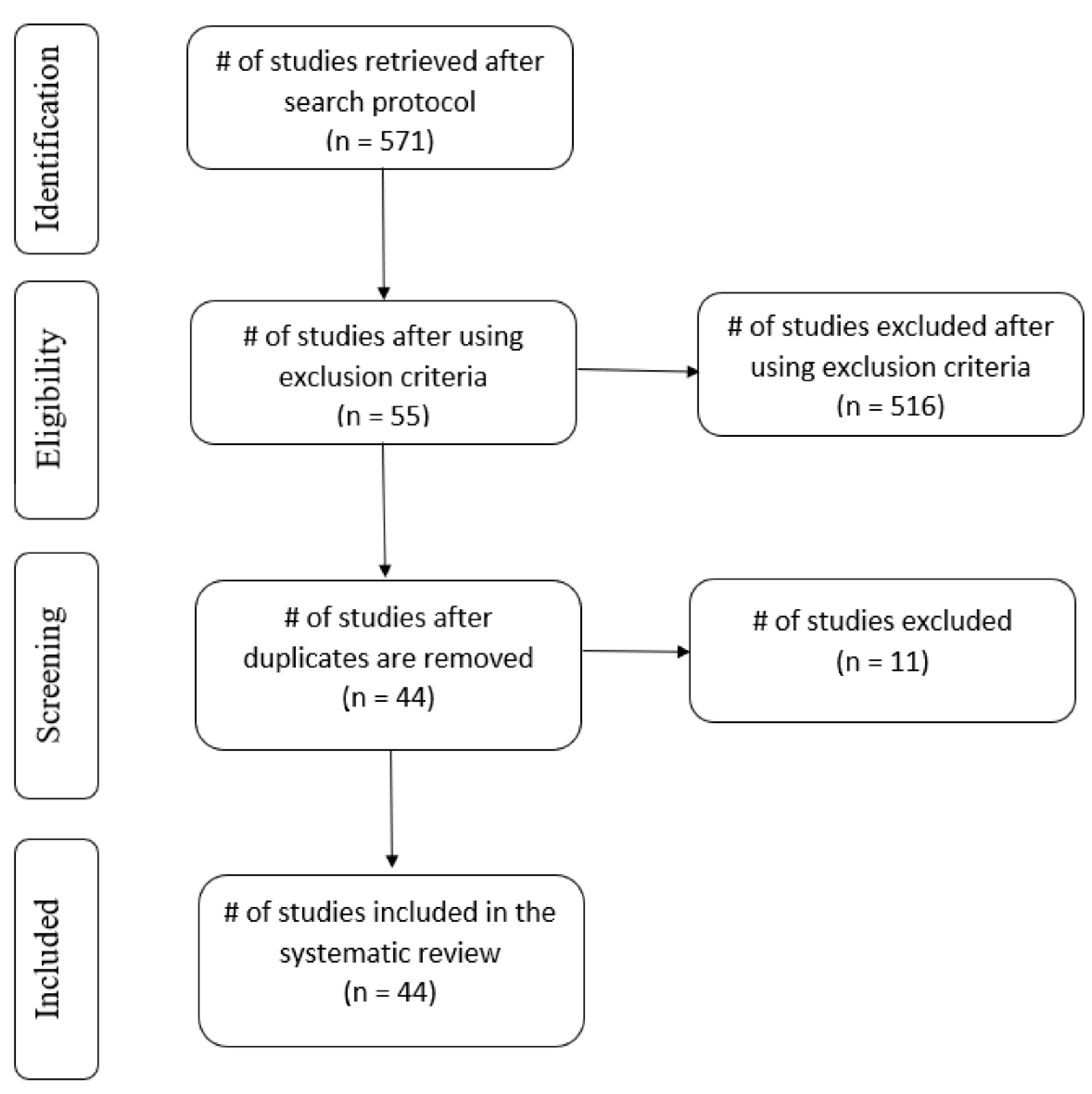

| Databases | # of Retrieved Articles after Search Protocol | # of Articles after Exclusion Criteria | # of Articles after Removing Repeated Articles |
|---|---|---|---|
| Scopus | 77 | 9 | 4 |
| Science Direct | 243 | 17 | 11 |
| IEEE Xplore | 20 | 7 | 7 |
| Google Scholar | 154 | 14 | 14 |
| MDPI | 13 | 4 | 4 |
| Web of Science | 64 | 4 | 4 |
| Total | 571 | 55 | 44 |
| Type of Remote Sensing | Model | Data and Features Used in Study | Authors |
|---|---|---|---|
| AVHRR | Bayesian neural networks (BNN) | Crop yield data | Johnson et al. [16] |
| LSTM | Yield data, surface reflectance, NDVI, air temperature, precipitation, air pressure, humidity | Wang et al. [60] | |
| ERA5 reanalysis | LSTM | Growing Degree Days (GDD), plant–harvest cycle, average yield data, maximum temperature, minimum temperature | Cunha and Silva [61] |
| Landsat 8 | ANN | NDVI, SR, NIR, GNDVI, GI, WI, and SBI | Haghverdi et al. [57] |
| MODIS | 3D-CNN | Surface reflectance, land surface temperature, and land cover type | Gavahi et al. [62], Qiao et al. [63], Abbaszadeh et al. [64] |
| CNN | NDVI, NDWI, NIR, precipitation, minimum, mean and maximum air temperatures | Kuwata and Shibasaki [56], Mu et al. [65], Terliksiz and Altýlar [66], Wolanin et al. [67], Cao et al. [68] | |
| CNN-LSTM | Surface reflectance, land surface temperature | You et al. [57], Sun et al. [69]. Sharma et al. [70], Ghazaryan et al. [71], Gastli et al. [72], Jeong et al. [73] | |
| DNN | NDVI, Absorbed Photosynthetically Active Radiation (APAR), land surface temperature | Dang et al. [74], Gao et al. [75] | |
| Deep forward neural network (DFNN) | Yield data, surface reflectance, land surface temperature, cropland data layers | Khaki et al. [76] | |
| LSTM | NDVI, EVI, land surface temperature | Tian et al. [23], Tian et al. [24], Kaneko. et al. [77], Jiang et al. [78], Ma et al. [79], Zhang et al. [80], Xie et al. [81] | |
| Neural networks ensemble | NDVI, Red, SR, NIR, GNDVI, GI, WI, and SBI | Fernandes et al. [58] | |
| Sentinel-2 | 3D-CNN | Crop yield, rice crop mask, B02-B08, B8A, B11, B12 and NDVI, climate data | Fernandez-Beltran et al. [21] |
| DNN | Precipitation, temperature, NIR, and SWIR | Jin et al. [82], Engen et al. [83] | |
| LSTM | Minimum and maximum temperature, integrated solar radiation, cumulative precipitation, soil texture, soil chemical parameters, hydrological properties | Xie et al. [84] | |
| UAV | CNN | EVI, GRVI, GNDVI, MSAVI, OSAVI, NDVI, SAVI, WDRVI | Nevavuori et al. [85], Yang et al. [86], Yang et al. [87], Yang et al. [88] |
| CNN-LSTM | RGB images, thermal time, crop yield data, cumulative temperature | Nevavuori et al. [85] | |
| DNN | NDVI, GNDVI, EVI, EVI2, WDRVI, SIPI, NRVI, VARI, TVI, OSAVI, MCARI, TCARI, NDWI, NDRE, RECI, GLCM | Sagan et al. [89] | |
| Faster R-CNN | Weather images | Chen et al. [22] | |
| Multimodal data fusion | Surface temperature, air temperature, humidity, normalized relative canopy temperature (NRCT), Vegetation Fraction (VF) | Maimaitijiang et al. [90] | |
| Spectral deep neural network (sp-DNN) | Crop yield, harvested yield, multispectral images, NIR, NDVI, NDVI-RE, NDRE, ENVI, CCCI, GNDVI, GLI, and OSAVI | Danilevica et al. [91] |
| Vegetation Indices | NDVI—Normalized Difference Vegetation Index, EVI—Enhanced Vegetation Index, GCI—Green Chlorophyll Index, NDWI—Normalized Difference Water Index, NIR—Near-Infrared, MODIS Surface Reflectance, MODIS Land Surface Temperature, GNDVI—Green Normalized Difference Vegetation Index, EVI2—Two-Band Enhanced Vegetation Index, WDRVI—Wide Dynamic Range Vegetation Index, SIPI—Structure Insensitive Pigment Index, NRVI—Normalized Ration Vegetation Index, VARI—Visible Atmospherically Resistant Index, TVI—Triangular Vegetation Index, OSAVI—Optimized Soil Adjusted Vegetation Index, MCARI—Modified Chlorophyll Absorption Ratio Index, TCARI—Transformed Chlorophyll Absorption Reflectance Index, NDRE—Normalized Difference Red-Edge Index, RECI—Red-Edge Chlorophyll Index, GLCM—Gray-Level Co-Occurrence Matrix, GI—Greenness Index, WI—Wetness Index, Red Band, SBI—Soil Brightness Index, SAVI—Soil-Adjusted Vegetation Index, MSAVI—Modified Soil-Adjusted Vegetation Index, SIF—Solar-Induced Chlorophyll Fluorescence, APAR—Absorbed Photosynthetically Active Radiation, PCI—Precipitation Condition Index, VHI—Vegetation Health Index, PAR—Photosynthetically Active Radiation, TVDI—Temperature Vegetation Dryness Index, VSWI—Vegetation Supply Water Index, PDI—Perpendicular Drought Index, RZSM—Root Zone Soil Moisture, NDMI—Normalized Difference Moisture Index, LAI—Leaf Area Index, ET—Total Evapotranspiration, LE—Average Latent Heat Flux, PET—Total Potential Evapotranspiration, PLE—Average Potential Latent Heat Flux, GPP—Gross Primary Productivity, PsnNet—Net Photosynthesis, CCCI—Canopy Chlorophyll Content Index, GLI—Green Leaf Index, NDVI-RE—Normalized Difference Vegetation Index Red Edge, NDRE-R—Normalized Difference Red Edge Red, VTCI—Vegetation temperature Condition Index, NRCT—Normalized Relative Canopy Temperature, VF—Vegetation Fraction, Sentinel—2—Bands B02 to B08, B8A, B11, B12, RDVI—Renormalized Difference Vegetation index, MTVI1—Modified Triangular Vegetation Index, TBWI—Three-Band Water Index, WDRVI—Wide Dynamic Range Vegetation Index, NDII—Normalized Difference Infrared Index, DCNI—Canopy Nitrogen Index |
| Meteorological Data/Weather Conditions | Precipitation, minimum temperature, mean temperature, maximum air temperature, temperature, weather, accumulated precipitation, cumulative temperature, average precipitation, average temperature, GDD—Growing Degree Days, KDD—Killing Degree Days, FDD—Frozen Degree Days, Surface Downward Shortwave Radiation Flux (SWdown), Water Vapor Pressure Deficit (VPD), air pressure, air-specific humidity, surface downward longwave radiation, wind speed, evapotranspiration, water stress indicator |
| Crop Yield Information (Excluding Crop Yield Data) | Growing phase as percentage of total thermal time, start of crop season, end of crop season, length of crop season, harvest cycle, country-level yield, field-level yield, biomass measure, fresh grain yield, dry grain yield, wheat crop fraction |
| Images | RGB images, hyperspectral images, moisture images |
| Soil Data | Clay content mass fraction, sand content mass fraction, water content, pH, bulk density, carbon content, silt content, coarse fragments, cation exchange capacity, pH in H2O, pH in KCL, Soil Available Water Holding Capacity (AWC), particle size distribution, total nitrogen |
| Others | Annual land cover, plant growth stage, micro-topographic fields, plant height, crop planting areas, thermal time, solar radiation, crop land data |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muruganantham, P.; Wibowo, S.; Grandhi, S.; Samrat, N.H.; Islam, N. A Systematic Literature Review on Crop Yield Prediction with Deep Learning and Remote Sensing. Remote Sens. 2022, 14, 1990. https://doi.org/10.3390/rs14091990
Muruganantham P, Wibowo S, Grandhi S, Samrat NH, Islam N. A Systematic Literature Review on Crop Yield Prediction with Deep Learning and Remote Sensing. Remote Sensing. 2022; 14(9):1990. https://doi.org/10.3390/rs14091990
Chicago/Turabian StyleMuruganantham, Priyanga, Santoso Wibowo, Srimannarayana Grandhi, Nahidul Hoque Samrat, and Nahina Islam. 2022. "A Systematic Literature Review on Crop Yield Prediction with Deep Learning and Remote Sensing" Remote Sensing 14, no. 9: 1990. https://doi.org/10.3390/rs14091990
APA StyleMuruganantham, P., Wibowo, S., Grandhi, S., Samrat, N. H., & Islam, N. (2022). A Systematic Literature Review on Crop Yield Prediction with Deep Learning and Remote Sensing. Remote Sensing, 14(9), 1990. https://doi.org/10.3390/rs14091990









