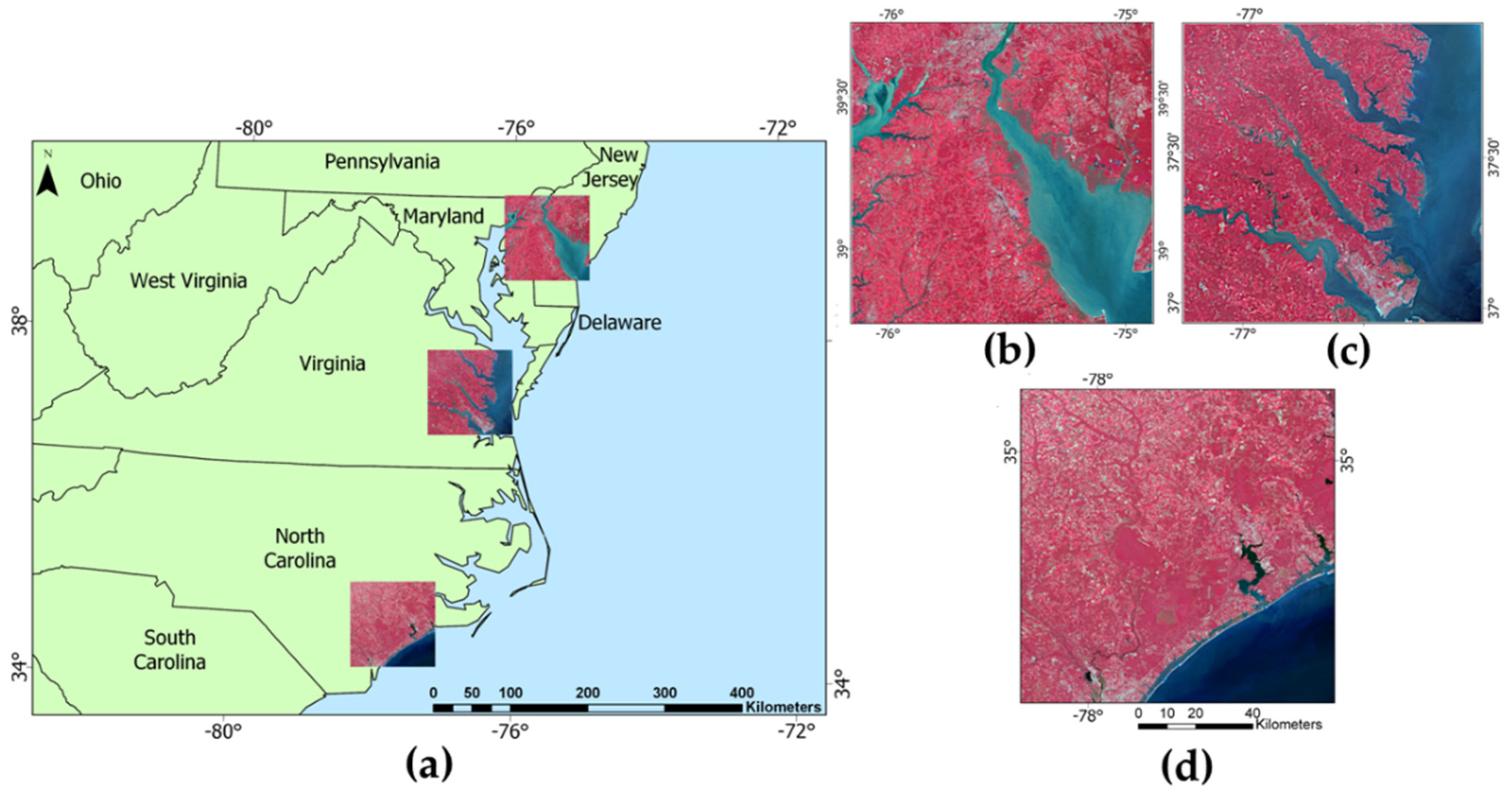
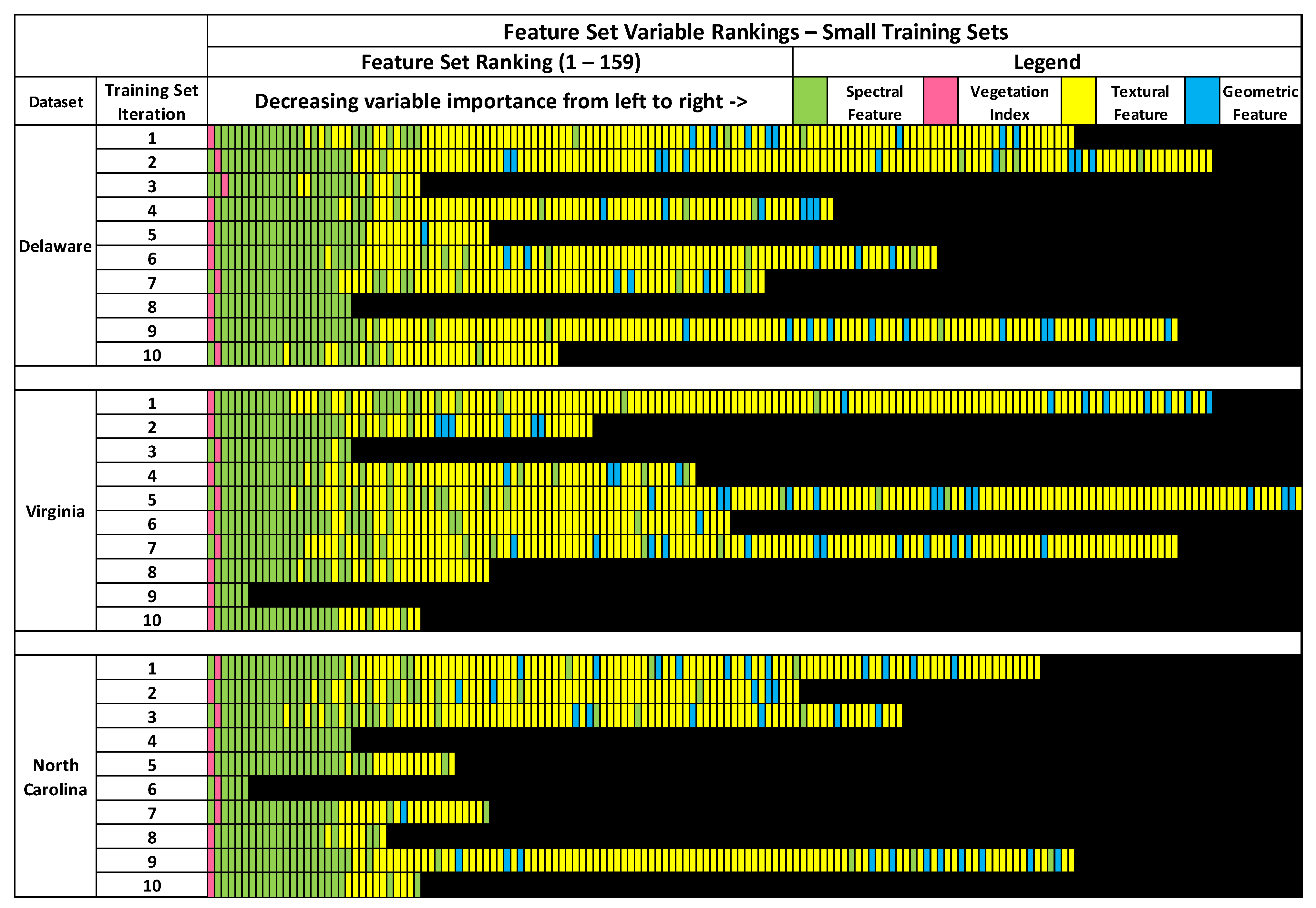
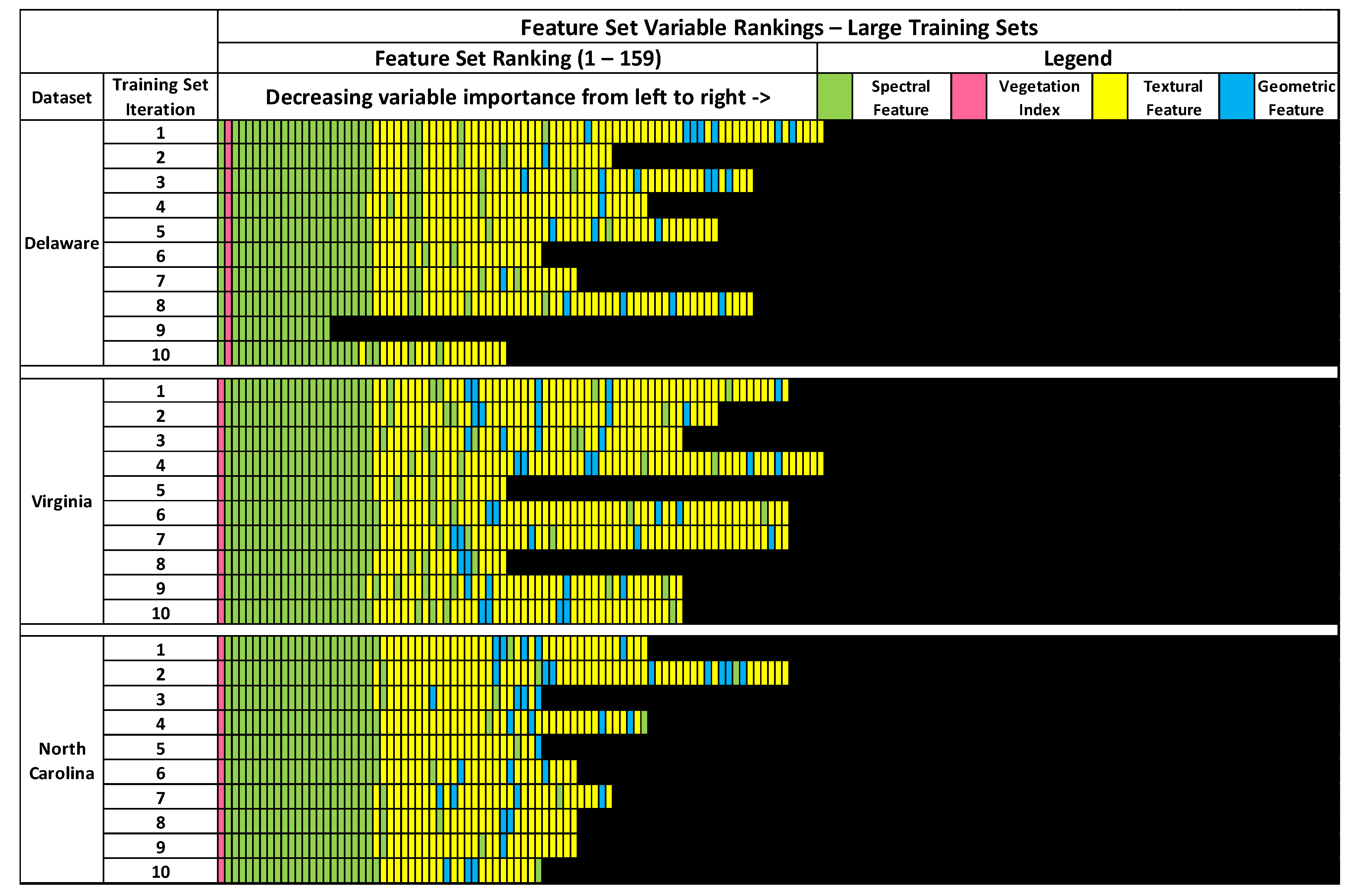
3.1. Feature Selection Results
As seen in
Figure 4, there was considerable variation in the feature selection results from the RFE process conducted on the small sample sets. Not only were there variations between the optimal feature sets of the sample sets acquired from different images, but also between sample sets acquired from the same image. Notably, none of the RFE-derived feature sets on the small training sets were identical. The number of optimal features between feature selection results of the Delaware small dataset ranged between 21 to 146. The feature sets of the small North Carolina dataset had the smallest range, between 6 to 126, while the small Virginia datasets had the largest range of features, between 6 to 159. Although there was minimal overlap in training samples between the small training sets, this wide variation in RFE results on training sets acquired from the same image was surprising.
The small Delaware feature sets also had the highest number of features that were included on all feature sets, at 21. Only six features were listed on all small Virginia and small North Carolina feature sets. Three features were found to be listed across all small training set feature sets: NDVI, max diff., and brightness. All feature sets contained at least one spectral variable. Textural features were included on 26 out of 30 (9 out 10 for Delaware, 9 out of 10 for Virginia, 8 out of 10 for North Carolina) of the small training set feature sets, while 18 out of 30 (7 out of 10 for Delaware, 6 out of 10 for Virginia, 5 out of 10 for North Carolina) small training set feature sets contained at least one geometric feature. Five feature sets were entirely comprised of spectral variables and did not include any geometric or textural features. Interestingly, only 32 out of 159 features were included in two-thirds or more of the small training set feature sets across all 3 images.
Regarding the feature selection results of the large training sets, variation between RFE-derived feature sets of training sets within the same image dataset, and between different image datasets was less than variations observed in the RFE-derived feature sets of the small training sets. This was expected, as the large training sets contain a considerable amount of overlap in terms of training image objects, at least between training sets of the same image dataset. Despite the high number of shared observations or training data overlap between the large training sample sets, there was still a notable amount of variation between training sets.
As seen in
Figure 5, feature sets varied between training sets of the same image dataset, and between image datasets. Similar to the feature sets of the small training sets, no two RFE-derived feature sets of the large training sets were identical, which is surprising given the high overlap in training data between training sets acquired from the same image. However, the ranges of feature set size between different large training set feature sets were much smaller than the ranges of the small training set feature sets. The range of feature set sizes for the Delaware large training sets was between 16 to 86, while large training sets from the Virginia ranged between 41 to 86 features. The feature sets of the large North Carolina training sets had the smallest range of 46 to 81 features. Feature importance rankings were also more consistent between large training sets of the same image dataset, which is unsurprising given the overlap in image object samples between training sets. Most spectral features were consistently ranked higher in importance than textural or geometric features across all feature sets. All feature sets also included at least one spectral variable. All feature sets, except for one of the large Delaware training sets, included textural features. At least 1 geometric feature was included on 7 out of 10 of the large Delaware training sets, 9 out of 10 of the large Virginia training sets, and all of the large North Carolina feature sets.
The consistency of features across all training sets was also higher in the large training sets than the small training sets. In total, 16 features were included on all 10 large Delaware feature sets, while 38 features were included on all large Virginia feature sets. The large North Carolina feature sets had the most commonality, with 40 features that were included in all 10 datasets, including several textural and geometric features. Across all 30 training sets from all three image datasets, 16 spectral features were found on all datasets. Additionally, 47 out of 159 features were included on two-thirds or more of all large training set feature sets, a higher value than observed on the small training set feature sets. While the increase in feature set consistency between training sets acquired from the same image is likely due to the high overlap between the training sets, the increase in feature set consistency between the large training sets acquired from different images is surprising, and may suggest that larger training sets may be more useful for identifying image features that are consistently beneficial across multiple image datasets.
3.2. Classification Results—Small Training Sets
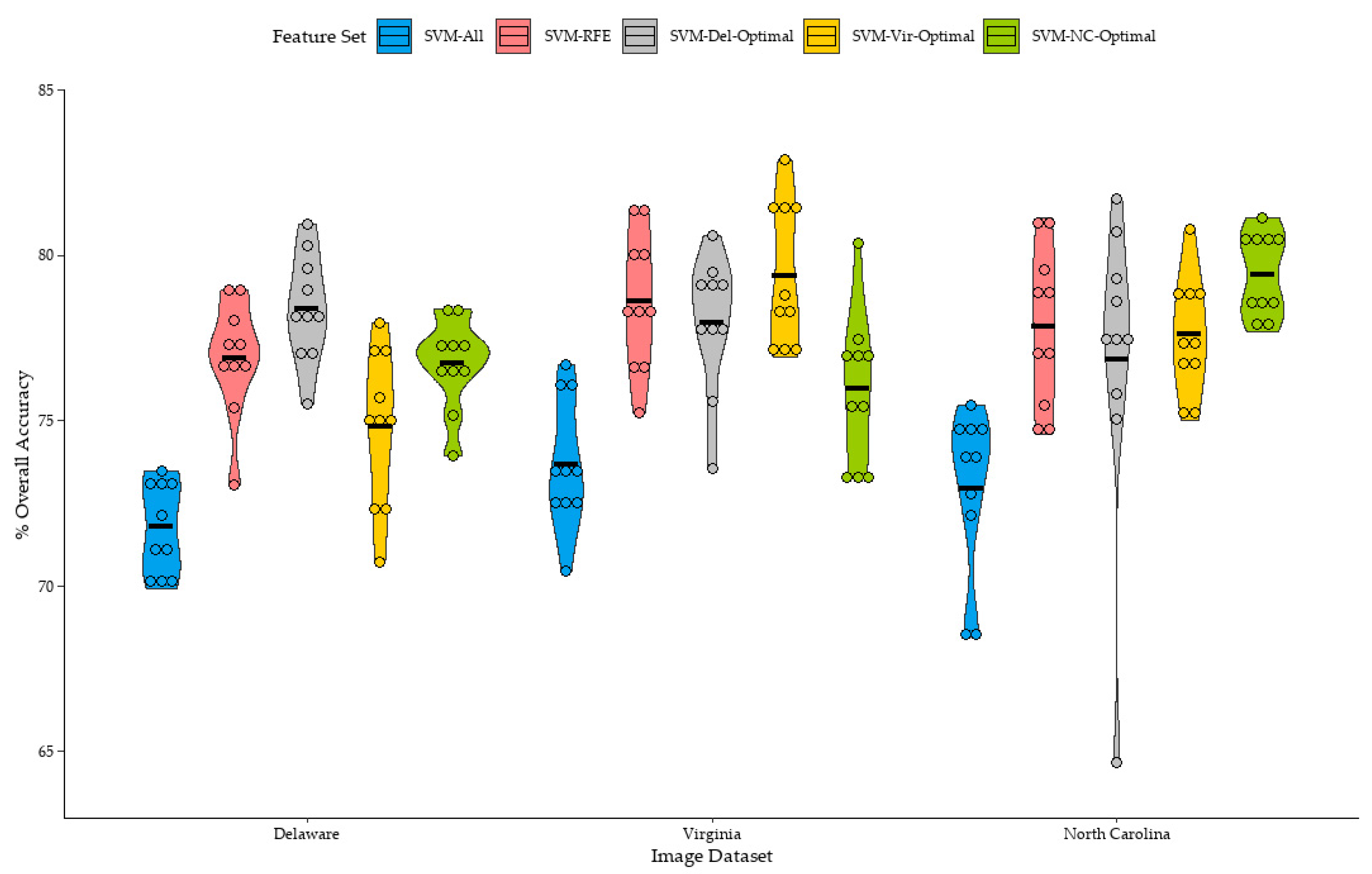
Figure 6 summarizes the distribution of overall accuracies of the classifications trained from the small sample sets. Classifications trained from the full sample set (SVM-All) produced the lowest mean overall accuracy for the Delaware, Virginia, and North Carolina datasets at 71.8%, 73.7%, and 72.9%, respectively. Classification accuracies for all training sets were consistently improved when the RFE process was individually applied to each training set (SVM-RFE). SVM-RFE mean classification accuracies improved by 5.1%, 4.9%, and 4.9% over SVM-All for classifications of the Delaware, Virginia, and North Carolina datasets, respectively. In general, feature selection was beneficial for improving classification accuracy for all classifications trained from the small sample sets.
Classifications trained using the feature set acquired from best SVM-RFE classification of the Delaware dataset (SVM-Del-Optimal) produced the highest overall mean accuracy of the Delaware dataset, at 78.3%, a 1.4% increase over the mean accuracy of the SVM-RFE classifications. While this is a relatively small increase, this result was unexpected, as the SVM-RFE process is thought to identify a feature set which optimizes the accuracy of each individual training set. It is interesting that a single feature set derived from an RFE process applied to a different training set was able to improve overall accuracy for some classifications. It should also be mentioned that, while the difference in means was small, the difference in means was found to be statistically significant.
Similarly, mean overall accuracies for the Virginia and North Carolina datasets were highest when the SVM-Vir-Optimal and SVM-NC-Optimal feature sets were applied to classifications of the Virginia, and North Carolina datasets, respectively. Mean overall accuracy of the SVM-Vir-Optimal classifications of the Virginia dataset was 0.7% higher than SVM-RFE classifications of the Virginia Dataset, although the difference in means between SVM-Vir-Optimal and SVM-RFE was found to be not statistically significant. However, while the difference in means was minimal, SVM-Vir-Optimal classifications provided higher overall accuracies over SVM-RFE classifications in six out of the ten replications.
For the North Carolina dataset, SVM-NC-Optimal classifications had a mean accuracy of 79.4%, 1.6% higher than the mean accuracy of the SVM-RFE classifications. The difference in means were also found to be statistically significant. Notably, one classification of the North Carolina dataset trained using the SVM-NC-Optimal feature set (
Table 6) had an overall accuracy of 80.4%, an improvement of 4.9% overall accuracy over the corresponding SVM-RFE classification using the same training set (
Table 7).
As indicated in
Table 6 and
Table 7, an example of improved performance of a classification trained using the NC-Optimal feature set was partially due to the RFE-SVM classifications’ relatively low user’s and producer’s accuracies of the water and wetlands classes. When the NC-Optimal feature set was used for this training set, user’s and producer’s accuracies of these classes were greatly improved, as well as the user’s and producer’s accuracies of the exposed soil, forest, and grassland classes. As these three classes comprise much of the composition of the test set, reduced classification error of these classes resulted in a substantial improvement in overall accuracy.
It should be mentioned that while the Del-Optimal, Virginia-Optimal, and NC-Optimal feature sets slightly improved the mean overall accuracy of the classifications of their respective image datasets, not all classifications saw an increase in overall accuracy. For example, two out of ten of the Delaware classifications saw a decrease in overall accuracy when the Del-Optimal feature set was used. When the Virginia-Optimal feature set was used for classifications of the Virginia dataset, one of the ten classifications saw a decrease in overall accuracy. Four out of ten SVM-NC-Optimal classifications saw a decrease in overall accuracy when compared to the SVM-RFE classifications of the North Carolina dataset. This suggests that a single optimal feature set may not be universally beneficial for improving classification accuracy for all training sample sets.
The SVM-Del-Optimal, SVM-Vir-Optimal, and SVM-NC-Optimal feature sets seemed to be beneficial to improving the mean accuracy of classification models of the same image dataset. However, when the optimal feature sets were transferred between images, mean classification accuracy tended to slightly decrease below the mean accuracy of the SVM-RFE classifications of each image dataset.
For example, the mean overall accuracy of the SVM-Del-Optimal and SVM-NC-Optimal classifications of the Virginia dataset were 77.9% and 75.9%, respectively, both lower than the mean overall accuracy of SVM-RFE and SVM-Vir-Optimal classifications of the Virginia dataset, at 78.6%, and 79.3%, respectively, although the difference in means between SVM-Del-Optimal and SVM-RFE classifications were found not to be significant. Classifications of the Delaware and North Carolina dataset followed a similar pattern. SVM-Vir-Optimal and SVM-NC-Optimal mean overall classification accuracies were lower than SVM-RFE or SVM-Del-Optimal when applied to classify the Delaware dataset. SVM-Vir-Optimal and SVM-Del-Optimal mean overall classification accuracies were also lower than SVM-RFE and SVM-NC-Optimal when applied to classify the North Carolina dataset.
Some mean classification accuracies were relatively close. For example, for the classifications of the Delaware dataset, the mean classification accuracy of SVM-RFE was 76.9% and SVM-NC-Optimal at 76.7%; the difference in means was found to be not statistically significant. While the mean accuracy of SVM-NC-Optimal was lower, performance was relatively similar to SVM-RFE for the Delaware dataset, but still lower than the mean overall accuracy of SVM-Del-Optimal at 78.4%. However, there were specific classifications which saw large differences in overall accuracy when trained from different feature sets. For example, an SVM-Del-Optimal classification of the North Carolina dataset saw a decrease in overall accuracy of 10.8% when compared to the corresponding SVM-RFE classification.
In general, for the classification’s trained from the small sample sets, optimal feature sets tended to slightly improve overall classification accuracy within the same image, but generally provided slightly lower overall accuracy when transferred to classifications of other images. However, these results were inconsistent between individual classifications, as some classifications saw a decrease in overall accuracy. Furthermore, the increase in mean overall accuracy over the SVM-RFE classifications for each dataset was relatively small. It should also be mentioned that overall accuracies of classifications which used an RFE-derived feature set, whether individually optimized (SVM-RFE), or one of the optimal feature sets, either from the same image or other images (SVM-Del-Optimal, SVM-Vir-Optimal, or SVM-NC-Optimal), were still typically higher than classifications trained using the full feature set (SVM-All).
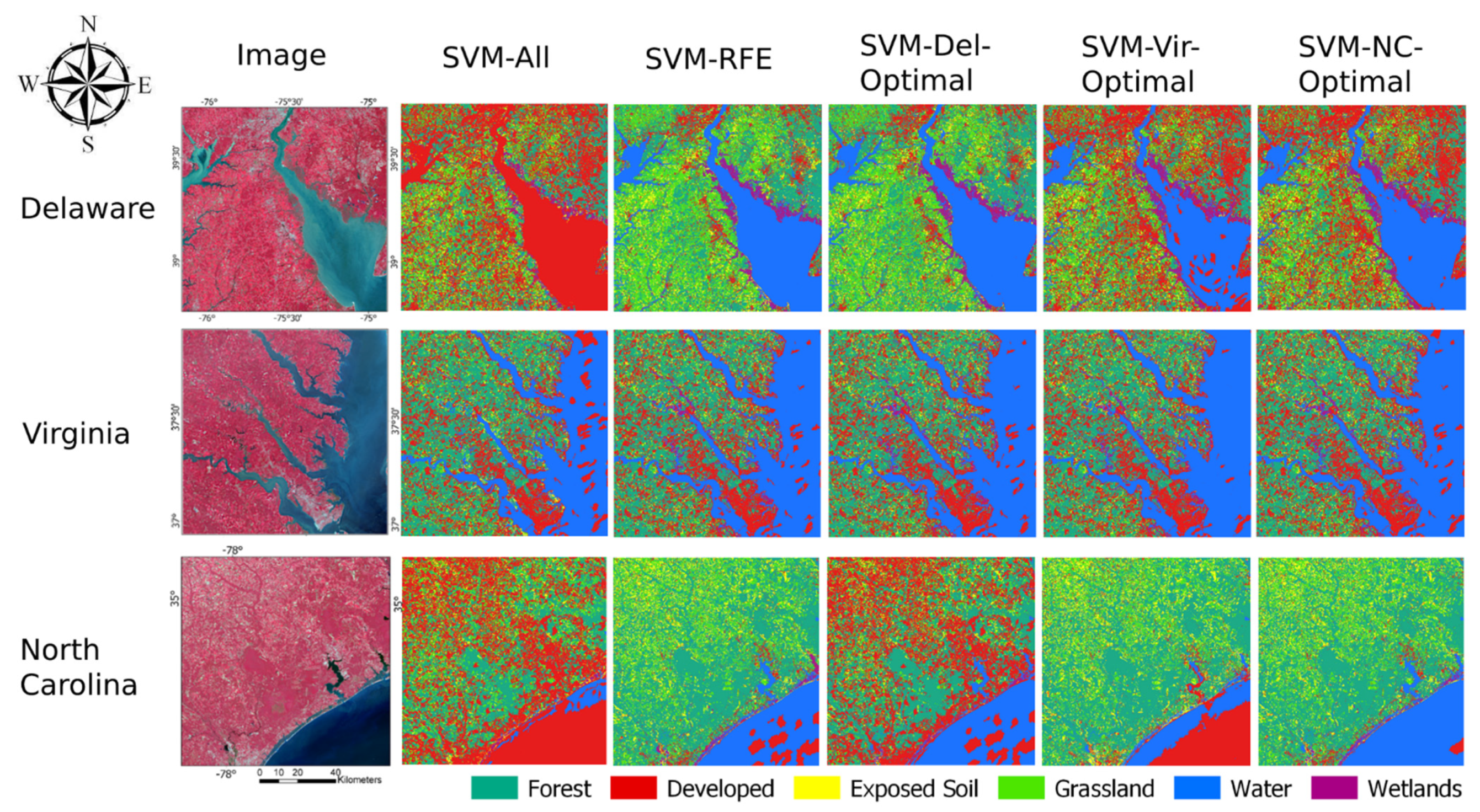
Figure 7 provides an example visual representation of classifications of all three image datasets using different feature sets. There was notable classification error in all three image datasets between the water and developed classes, especially in the SVM-All classifications which used the full feature set.
3.3. Classification Results—Large Training Sets
The distribution of overall accuracy of classifications trained from the large training sets are shown in
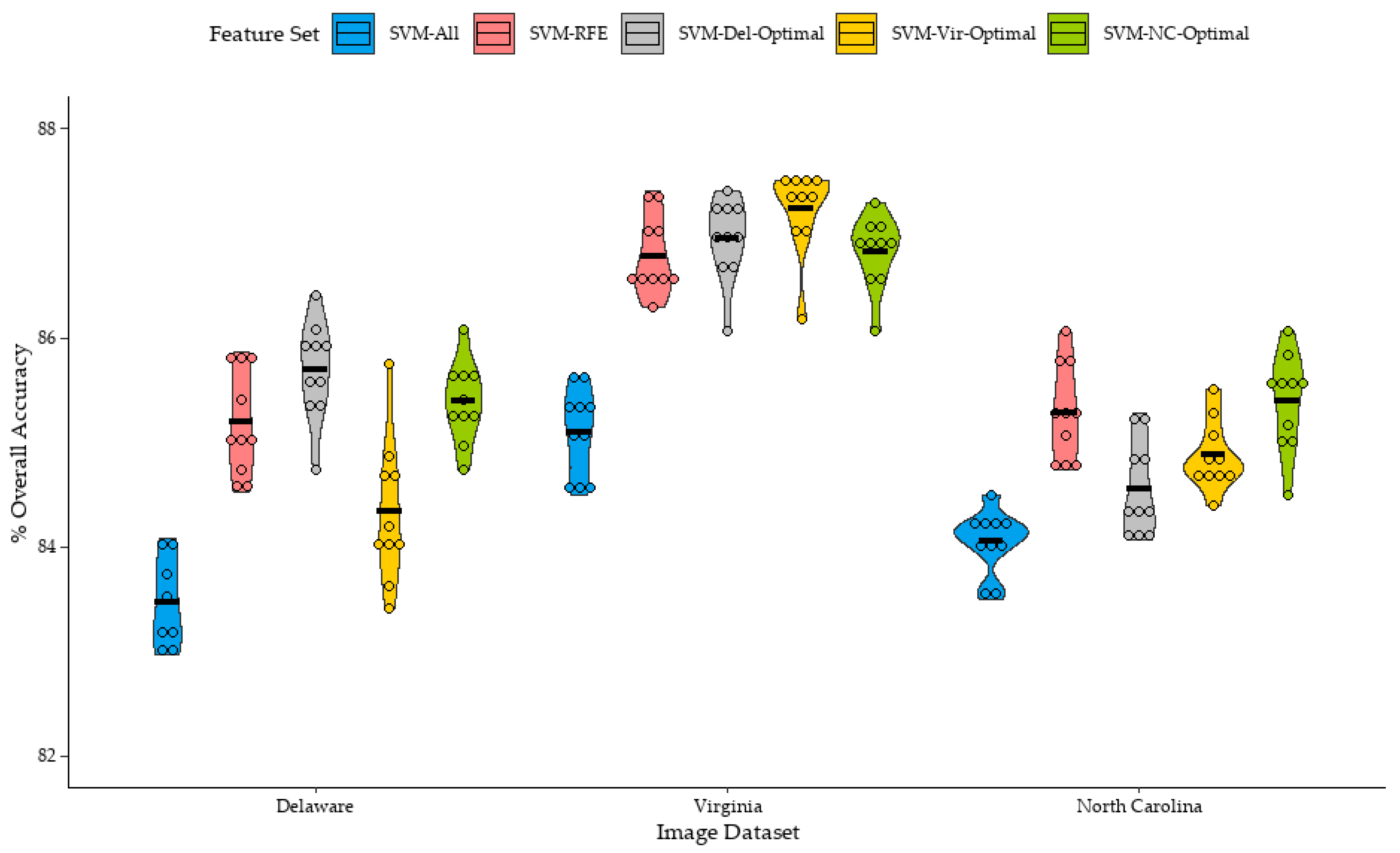
Figure 8. Similar to the results of the small training set classifications, classifications which were trained from the full feature set (SVM-All) generally provided the lowest mean overall accuracies for the Delaware, Virginia, and North Carolina datasets, at 83.1%, 85.1%, and 84.0%, respectively. Feature selection was beneficial for improving classification accuracy, as the mean overall accuracy of the SVM-RFE classifications were 85.2%, 86.8%, and 85.3% for the Delaware, Virginia, and North Carolina datasets, respectively. It should be mentioned that improvement in classification accuracy using RFE-optimized feature sets over classifications using the full feature set was relatively smaller in the large training set classifications, compared to the small training set classifications. In congruence with observations from other studies employing the SVM classifier, this suggests that SVM may be sensitive to the Hughes effect, or the “curse of dimensionality” [
50,
51].
While the application of the Del-Optimal feature set to the large training set classifications of the Delaware dataset improved mean classification accuracy by 0.5% over SVM-RFE, the differences in means was not statistically significant. The largest improvement in classification accuracy of an SVM-Del-Optimal classification over an SVM-RFE classification of the Delaware datasets was 1.6%. However, similar to the small training sets, not all classifications saw an improvement in overall accuracy, as two classifications saw a decrease in overall accuracy of 0.3% and 0.6% when the SVM-Del-Optimal feature set was applied to the classifier.
The transference of the Del-Optimal feature set to large training set classifications of the Virginia and North Carolina datasets yielded mixed results. When the Del-Optimal feature set was applied to classifications of the North Carolina dataset, mean overall accuracy decreased by 0.7% compared to the mean accuracy of the SVM-RFE classifications, with the difference in means being found to be statistically significant. Nine out of ten classifications saw a decrease in accuracy; however, this decrease in accuracy was relatively small, with the largest decrease in overall accuracy at 1.8%. However, when the Del-Optimal feature set was applied to classifications of the Virginia dataset, mean overall accuracy slightly increased by 0.2%, and overall accuracy was comparable to SVM-RFE classifications of the Virginia dataset.
The Vir-Optimal feature set provided slightly higher mean overall accuracy for large sample set classifications of the Virginia dataset, at 86.9%, an increase of 0.5% over the SVM-RFE classification of the Virginia dataset, with the difference in means being statistically significant, while providing slightly lower mean overall accuracies for classifications of the Delaware and North Carolina datasets at 84.3%, and 84.9%, a decrease of 0.8% and 0.4%, respectively. Similar to the Del-Optimal dataset, not all Vir-Optimal datasets were beneficial for classifications of the Virginia dataset, as one classification saw a decrease in overall accuracy compared to the SVM-RFE classification, with the largest increase in overall accuracy being 1.0%.
Curiously, the NC-Optimal feature set provided slightly improved mean overall accuracy of the Delaware, Virginia, and North Carolina datasets and were largely comparable to the mean overall accuracies of each image dataset’s SVM-RFE classifications. However, similar to the Del-Optimal and Vir-Optimal feature sets, results tended to vary, with some classifications seeing an improvement in overall accuracy while others saw a decrease. Notably, the NC-Optimal feature set had the smallest increase in mean overall accuracy for the image dataset it was derived from, at 0.1%, at least when compared to the mean overall accuracy of the SVM-RFE classifications of the North Carolina dataset. It should be mentioned that the difference in means between SVM-RFE and SVM-NC-Optimal classifications of the North Carolina dataset were not statistically significant, which is expected given the extremely small difference in mean overall accuracy of the 10 replications.
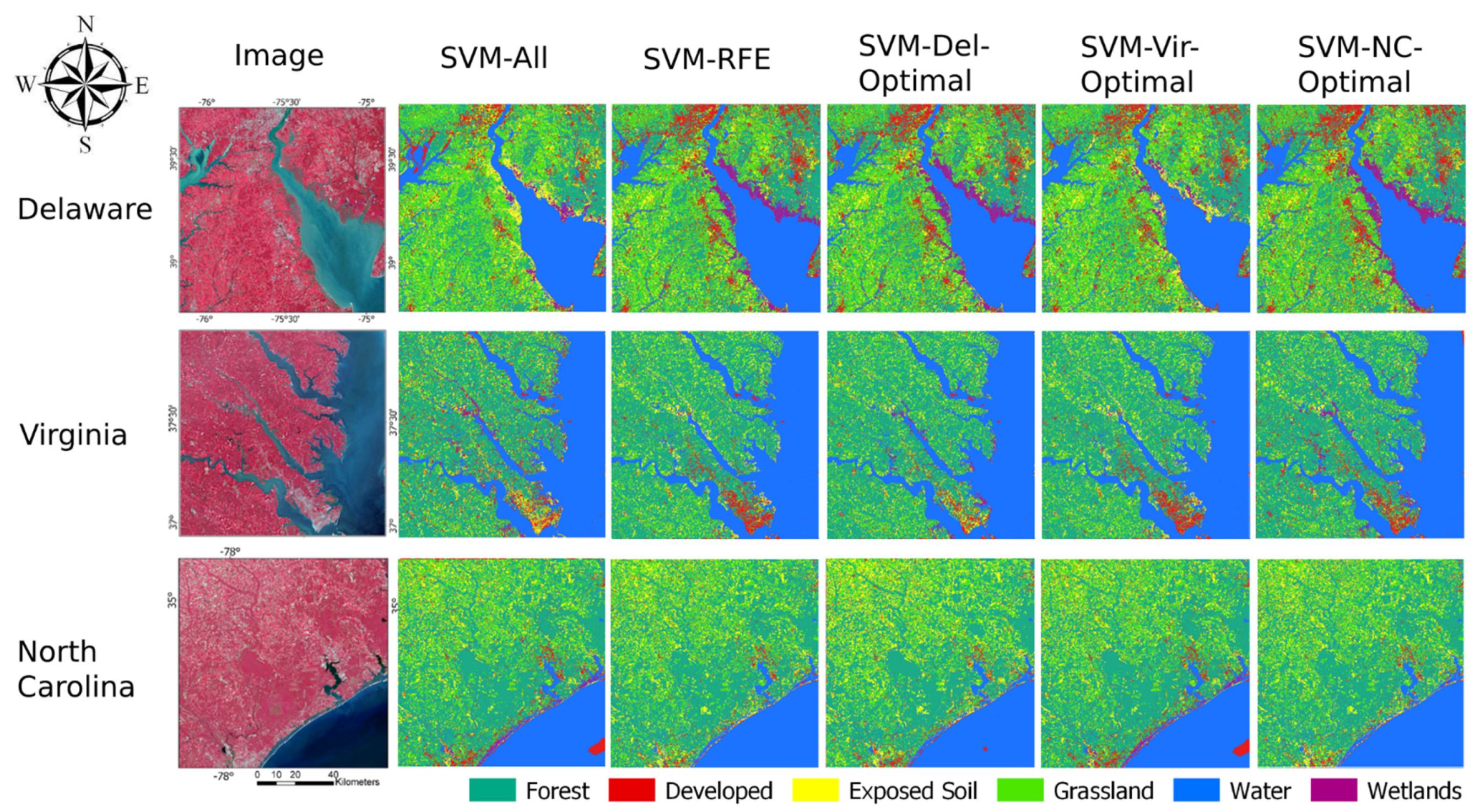
A visual example set of classifications trained from the large training sets is depicted in
Figure 9. In general, the highest performing classifications of each sample set looked visually good. However, there were visual differences between several classifications, some of which highlight errors within some of the maps. In the Delaware dataset, there was notable classification error between the exposed soil and wetlands classes in some coastal areas in the SVM-All and SVM-Vir-Optimal classifications. In this example, there was also notable classification error of the developed and exposed soil classes of the Virginia classification in the SVM-All and SVM-Del-Optimal classifications. The North Carolina classifications were visually similar, except for the SVM-All and SVM-Vir-Optimal classifications, which contained a high amount of error between the wetlands and exposed soil classes on the beachfront coastal area.