Estimation of National Forest Aboveground Biomass from Multi-Source Remotely Sensed Dataset with Machine Learning Algorithms in China
Abstract
1. Introduction
2. Materials and Methods
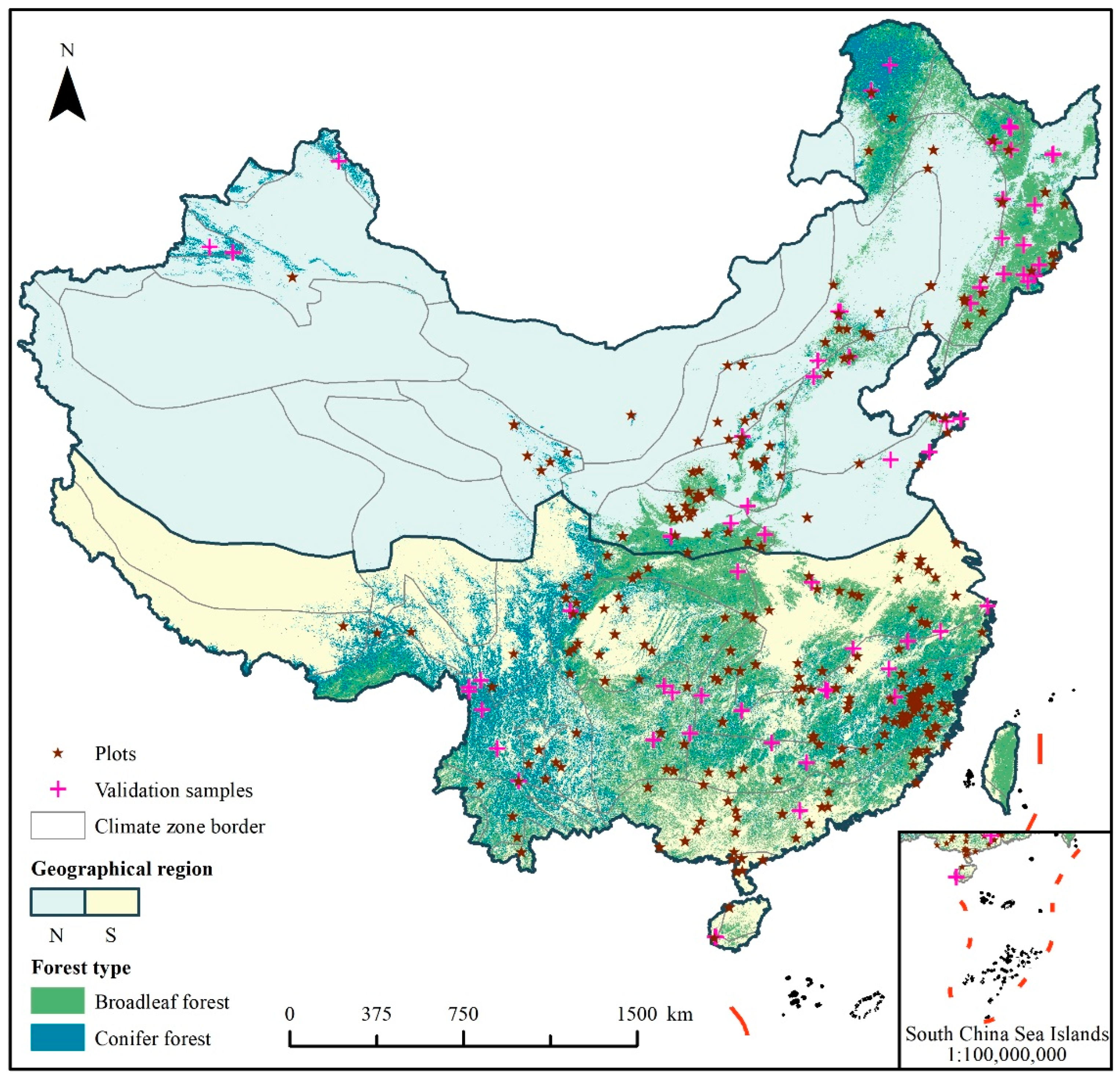
2.1. Forest Inventory Data and Allometric Equation
2.2. Remote Sensing Data Collection
2.2.1. Spaceborne LiDAR
2.2.2. MODIS Dataset
2.2.3. NPP and Climate Factors
2.2.4. Topography
2.3. Forest AGB Estimation and Uncertainty Determination
2.3.1. Model Design
2.3.2. Forest AGB Estimation
2.3.3. Uncertainty Determination
2.4. Accuracy Assessment
3. Results
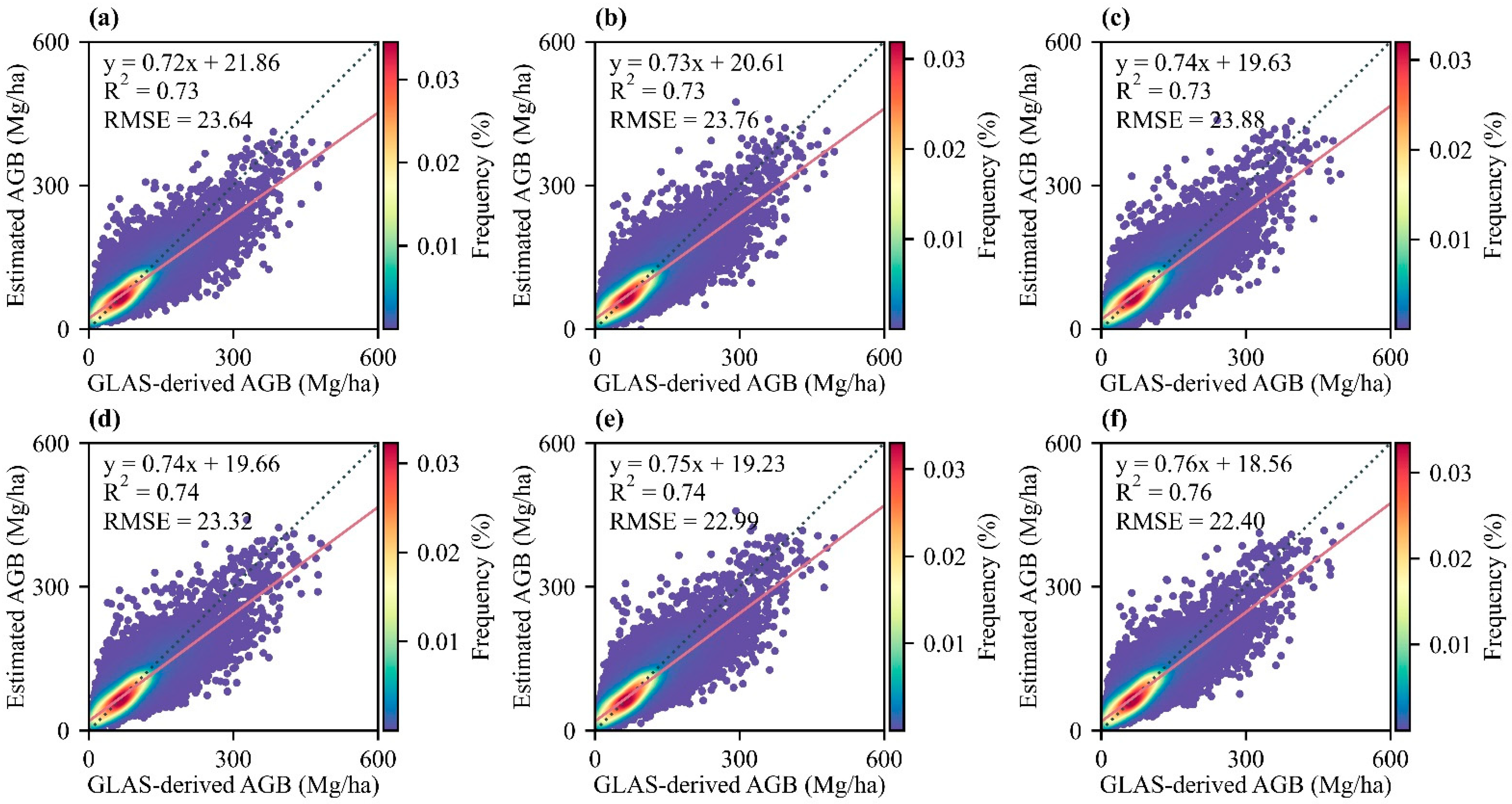
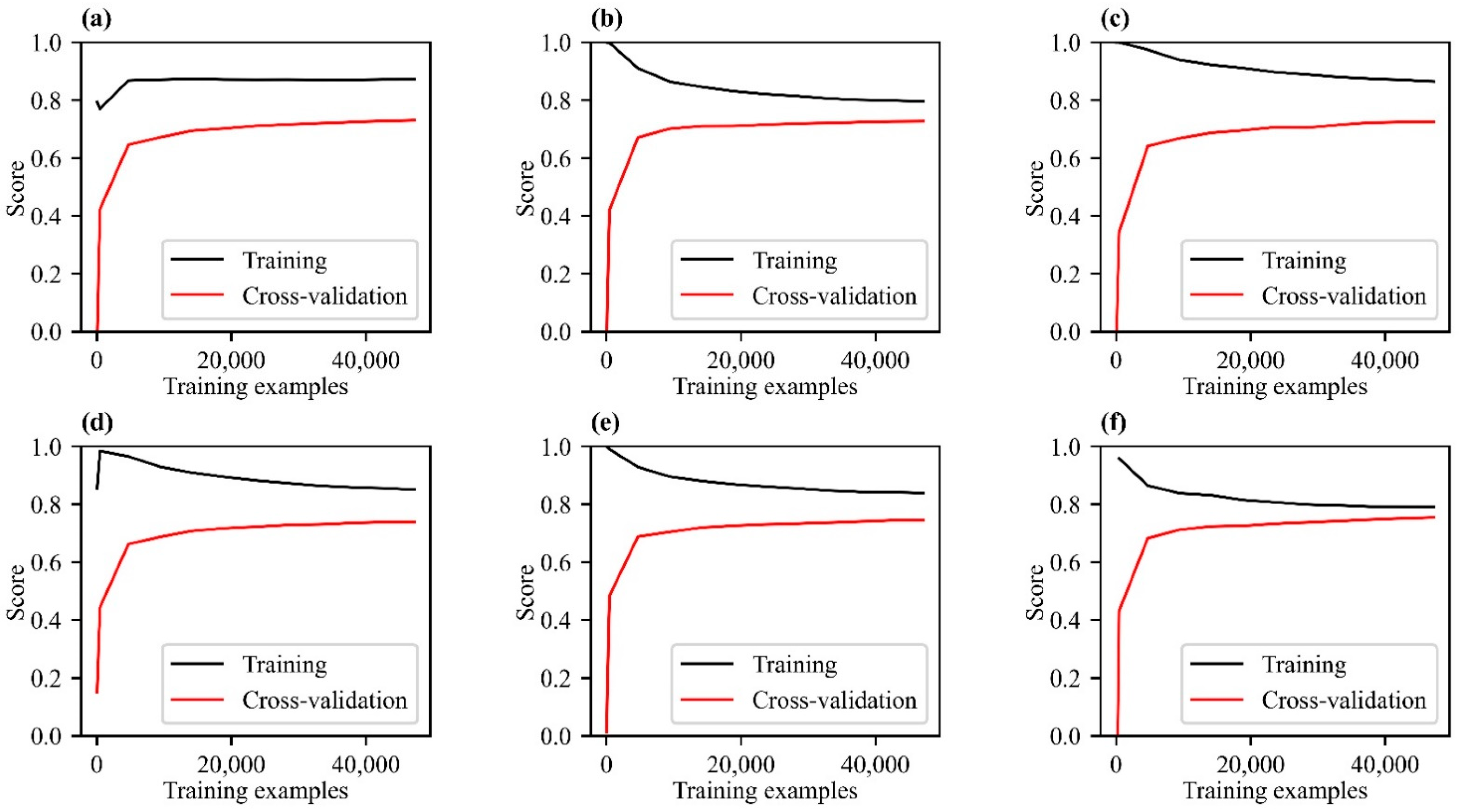
3.1. Model Comparison and Accuracy Verification
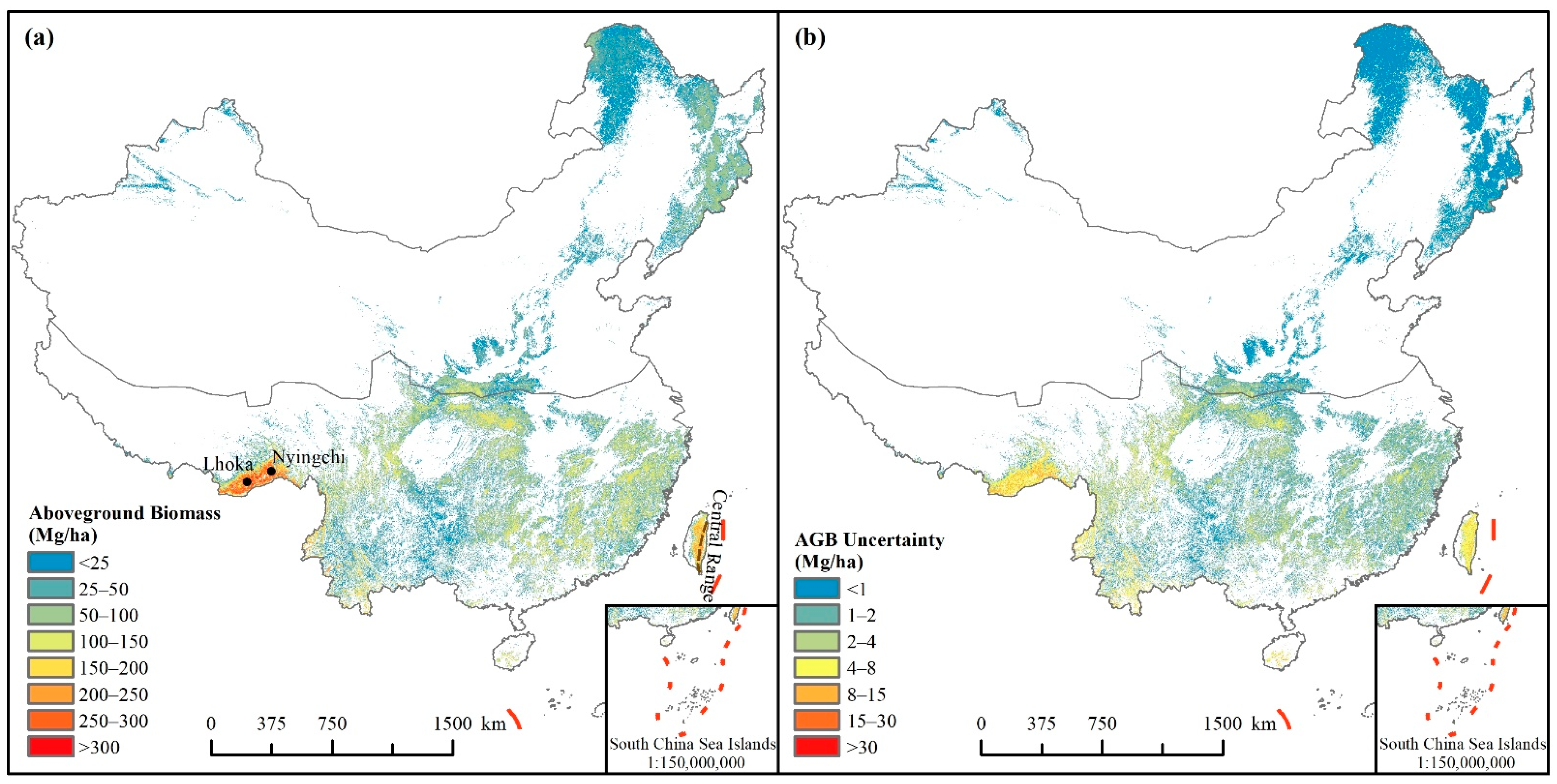
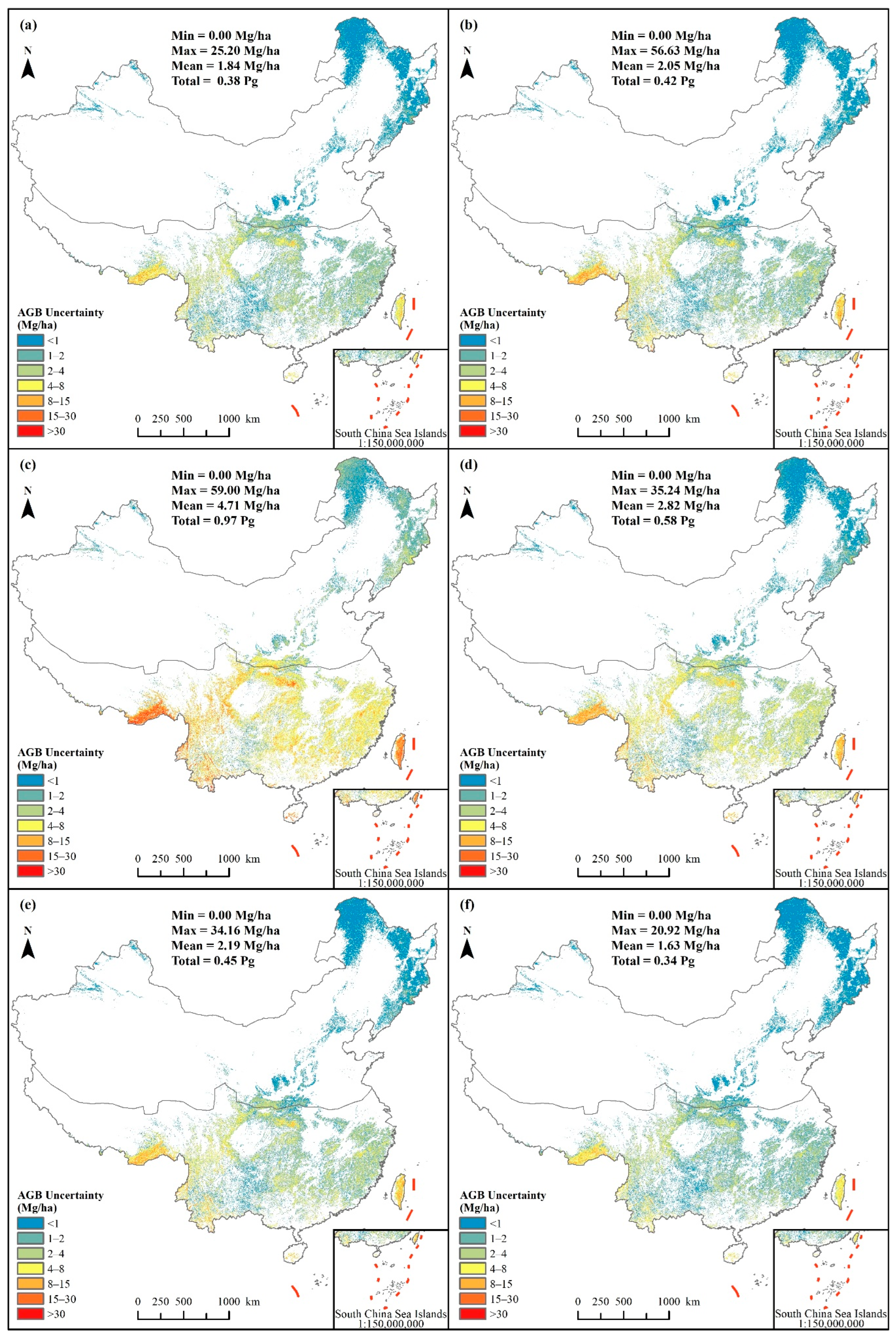
3.2. Spatially Continuous Forest AGB Map and Uncertainty Analysis
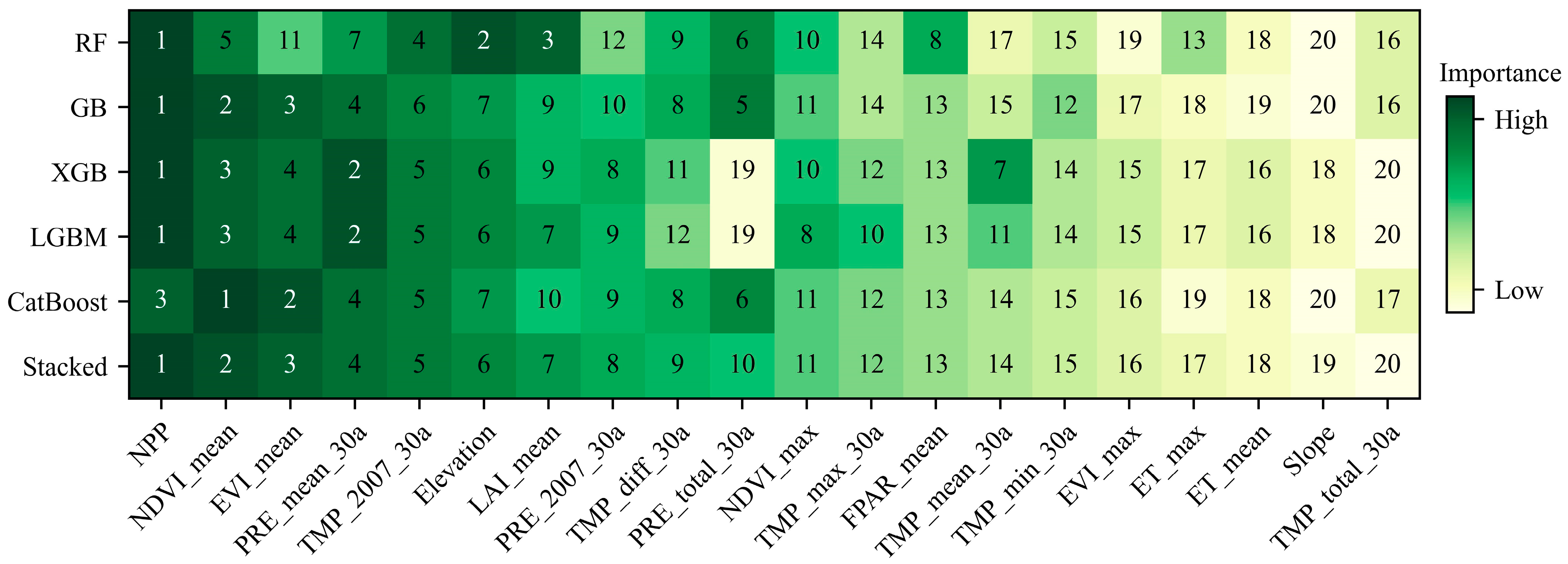
3.3. Feature Importance
4. Discussion
4.1. Comparison and Uncertainties
4.2. Feature Contribution
4.3. Limitations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO. Global Forest Resources Assessment 2020; FAO: Quebec City, QC, Canada, 2020. [Google Scholar]
- Houghton, R.A.; Hall, F.; Goetz, S.J. Importance of biomass in the global carbon cycle. J. Geophys. Res. Biogeosciences 2009, 114, G00E03. [Google Scholar] [CrossRef]
- Houghton, R.A.; Nassikas, A.A. Negative emissions from stopping deforestation and forest degradation, globally. Glob. Chang. Biol. 2018, 24, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Friedlingstein, P.; O’Sullivan, M.; Jones, M.W.; Andrew, R.M.; Hauck, J.; Olsen, A.; Peters, G.P.; Peters, W.; Pongratz, J.; Sitch, S.; et al. Global Carbon Budget 2020. Earth Syst. Sci. Data 2020, 12, 3269–3340. [Google Scholar] [CrossRef]
- Griscom, B.W.; Adams, J.; Ellis, P.W.; Houghton, R.A.; Lomax, G.; Miteva, D.A.; Schlesinger, W.H.; Shoch, D.; Siikamäki, J.V.; Smith, P.; et al. Natural climate solutions. Proc. Natl. Acad. Sci. USA 2017, 114, 11645–11650. [Google Scholar] [CrossRef]
- Feng, Y.; Zeng, Z.; Searchinger, T.D.; Ziegler, A.D.; Wu, J.; Wang, D.; He, X.; Elsen, P.R.; Ciais, P.; Xu, R.; et al. Doubling of annual forest carbon loss over the tropics during the early twenty-first century. Nat. Sustain. 2022, 5, 444–451. [Google Scholar] [CrossRef]
- Gatti, L.V.; Gloor, M.; Miller, J.B.; Doughty, C.E.; Malhi, Y.; Domingues, L.G.; Basso, L.S.; Martinewski, A.; Correia, C.S.C.; Borges, V.F.; et al. Drought sensitivity of Amazonian carbon balance revealed by atmospheric measurements. Nature 2014, 506, 76–80. [Google Scholar] [CrossRef] [PubMed]
- IPCC. 2006 IPCC Guidelines for National Greenhouse Gas Inventories; Institute for Global Environmental Strategies: Hayama, Japan, 2006. [Google Scholar]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Chirici, G.; Barbati, A.; Corona, P.; Marchetti, M.; Travaglini, D.; Maselli, F.; Bertini, R. Non-parametric and parametric methods using satellite images for estimating growing stock volume in alpine and Mediterranean forest ecosystems. Remote Sens. Environ. 2008, 112, 2686–2700. [Google Scholar] [CrossRef]
- Piao, S. Forest biomass carbon stocks in China over the past 2 decades: Estimation based on integrated inventory and satellite data. J. Geophys. Res. 2005, 110, G01006. [Google Scholar] [CrossRef]
- Patenaude, G.; Milne, R.; Dawson, T.P. Synthesis of remote sensing approaches for forest carbon estimation: Reporting to the Kyoto Protocol. Environ. Sci. Policy 2005, 8, 161–178. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. Lidar Remote Sensing for Ecosystem Studies: Lidar, an emerging remote sensing technology that directly measures the three-dimensional distribution of plant canopies, can accurately estimate vegetation structural attributes and should be of particular interest to forest, landscape, and global ecologists. BioScience 2002, 52, 19–30. [Google Scholar] [CrossRef]
- Asner, G.P.; Powell, G.V.N.; Mascaro, J.; Knapp, D.E.; Clark, J.K.; Jacobson, J.; Kennedy-Bowdoin, T.; Balaji, A.; Paez-Acosta, G.; Victoria, E.; et al. High-resolution forest carbon stocks and emissions in the Amazon. Proc. Natl. Acad. Sci. USA 2010, 107, 16738–16742. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Spies, T.A. An evaluation of alternate remote sensing products for forest inventory, monitoring, and mapping of Douglas-fir forests in western Oregon. Can. J. For. Res. 2001, 31, 78–87. [Google Scholar] [CrossRef]
- Silva, C.A.; Duncanson, L.; Hancock, S.; Neuenschwander, A.; Thomas, N.; Hofton, M.; Fatoyinbo, L.; Simard, M.; Marshak, C.Z.; Armston, J.; et al. Fusing simulated GEDI, ICESat-2 and NISAR data for regional aboveground biomass mapping. Remote Sens. Environ. 2021, 253, 112234. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.A.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Beer, C.; Cartus, O.; Schmullius, C.; Shvidenko, A.; McCallum, I.; Wegmüller, U.; Wiesmann, A. Retrieval of growing stock volume in boreal forest using hyper-temporal series of Envisat ASAR ScanSAR backscatter measurements. Remote Sens. Environ. 2011, 115, 490–507. [Google Scholar] [CrossRef]
- Abbas, S.; Wong, M.S.; Wu, J.; Shahzad, N.; Muhammad Irteza, S. Approaches of Satellite Remote Sensing for the Assessment of Above-Ground Biomass across Tropical Forests: Pan-tropical to National Scales. Remote Sens. 2020, 12, 3351. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.M. Machine Learning; McGraw-HillK: New York, NY, USA, 1997; Volume 1. [Google Scholar]
- Sun, S.L.; Cao, Z.H.; Zhu, H.; Zhao, J. A Survey of Optimization Methods from a Machine Learning Perspective. IEEE Trans. Cybern. 2020, 50, 3668–3681. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Cheng, G.; Han, J.W. A survey on object detection in optical remote sensing images. ISPRS J. Photogramm. Remote Sens. 2016, 117, 11–28. [Google Scholar] [CrossRef]
- Ali, I.; Greifeneder, F.; Stamenkovic, J.; Neumann, M.; Notarnicola, C. Review of Machine Learning Approaches for Biomass and Soil Moisture Retrievals from Remote Sensing Data. Remote Sens. 2015, 7, 16398–16421. [Google Scholar] [CrossRef]
- Rex, F.E.; Silva, C.A.; Dalla Corte, A.P.; Klauberg, C.; Mohan, M.; Cardil, A.; Silva, V.S.d.; Almeida, D.R.A.d.; Garcia, M.; Broadbent, E.N.; et al. Comparison of Statistical Modelling Approaches for Estimating Tropical Forest Aboveground Biomass Stock and Reporting Their Changes in Low-Intensity Logging Areas Using Multi-Temporal LiDAR Data. Remote Sens. 2020, 12, 1498. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 3149–3157. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased Boosting with Categorical Features. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 3–8 December 2018; pp. 6639–6649. [Google Scholar]
- Luo, M.; Wang, Y.; Xie, Y.; Zhou, L.; Qiao, J.; Qiu, S.; Sun, Y. Combination of Feature Selection and CatBoost for Prediction: The First Application to the Estimation of Aboveground Biomass. Forests 2021, 12, 216. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Li, C.; Liu, Z. Forest aboveground biomass estimation using Landsat 8 and Sentinel-1A data with machine learning algorithms. Sci. Rep. 2020, 10, 9952. [Google Scholar] [CrossRef]
- Ahmad, A.; Gilani, H.; Ahmad, S.R. Forest Aboveground Biomass Estimation and Mapping through High-Resolution Optical Satellite Imagery—A Literature Review. Forests 2021, 12, 914. [Google Scholar] [CrossRef]
- Mendes-Moreira, J.; Soares, C.; Jorge, A.M.; Sousa, J.F.D. Ensemble approaches for regression. ACM Comput. Surv. 2012, 45, 1–40. [Google Scholar] [CrossRef]
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Liang, S.; Li, X.; Liu, J. A stacking ensemble algorithm for improving the biases of forest aboveground biomass estimations from multiple remotely sensed datasets. GIScience Remote Sens. 2022, 59, 234–249. [Google Scholar] [CrossRef]
- Chen, C.; Park, T.; Wang, X.; Piao, S.; Xu, B.; Chaturvedi, R.K.; Fuchs, R.; Brovkin, V.; Ciais, P.; Fensholt, R.; et al. China and India lead in greening of the world through land-use management. Nat. Sustain. 2019, 2, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, X.; Zhang, X.; Lu, F. Biomass and Its Allocation of Forest Ecosystems in China; Chinese Forestry Publishing House Press: Beijing, China, 2013. [Google Scholar]
- Zhang, X.; Liu, L.; Chen, X.; Gao, Y.; Xie, S.; Mi, J. GLC_FCS30: Global land-cover product with fine classification system at 30 m using time-series Landsat imagery. Earth Syst. Sci. Data 2021, 13, 2753–2776. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Tan, Z.; Chen, Q. Mapping of the north-south demarcation zone in China based on GIS. J. Lanzhou Univ. Nat. Sci. 2012, 48, 28–33. [Google Scholar]
- Jiang, J.; Jiang, D.; Lin, Y. Monsoon Area and Precipitation over China for 1961–2009. Chin. J. Atmos. Sci. 2015, 39, 722–730. [Google Scholar]
- Huang, H.; Liu, C.; Wang, X.; Zhou, X.; Gong, P. Integration of multi-resource remotely sensed data and allometric models for forest aboveground biomass estimation in China. Remote Sens. Environ. 2019, 221, 225–234. [Google Scholar] [CrossRef]
- Simard, M.; Pinto, N.; Fisher, J.B.; Baccini, A. Mapping forest canopy height globally with spaceborne lidar. J. Geophys. Res. 2011, 116, G04021. [Google Scholar] [CrossRef]
- Su, Y.; Guo, Q.; Xue, B.; Hu, T.; Alvarez, O.; Tao, S.; Fang, J. Spatial distribution of forest aboveground biomass in China: Estimation through combination of spaceborne lidar, optical imagery, and forest inventory data. Remote Sens. Environ. 2016, 173, 187–199. [Google Scholar] [CrossRef]
- Green, J.K.; Keenan, T.F. The limits of forest carbon sequestration. Science 2022, 376, 692–693. [Google Scholar] [CrossRef]
- Richardson, A.D.; Carbone, M.S.; Keenan, T.F.; Czimczik, C.I.; Hollinger, D.Y.; Murakami, P.; Schaberg, P.G.; Xu, X. Seasonal dynamics and age of stemwood nonstructural carbohydrates in temperate forest trees. New Phytol. 2013, 197, 850–861. [Google Scholar] [CrossRef]
- Richardson, A.D.; Anderson, R.S.; Arain, M.A.; Barr, A.G.; Bohrer, G.; Chen, G.; Chen, J.M.; Ciais, P.; Davis, K.J.; Desai, A.R.; et al. Terrestrial biosphere models need better representation of vegetation phenology: Results from the North American Carbon Program Site Synthesis. Glob. Chang. Biol. 2012, 18, 566–584. [Google Scholar] [CrossRef]
- Zheng, Y.; Shen, R.; Wang, Y.; Li, X.; Liu, S.; Liang, S.; Chen, J.M.; Ju, W.; Zhang, L.; Yuan, W. Improved estimate of global gross primary production for reproducing its long-term variation, 1982–2017. Earth Syst. Sci. Data 2020, 12, 2725–2746. [Google Scholar] [CrossRef]
- Peng, S.; Ding, Y.; Liu, W.; Li, Z. 1 km monthly temperature and precipitation dataset for China from 1901 to 2017. Earth Syst. Sci. Data 2019, 11, 1931–1946. [Google Scholar] [CrossRef]
- Farr, T.G.; Rosen, P.A.; Caro, E.; Crippen, R.; Duren, R.; Hensley, S.; Kobrick, M.; Paller, M.; Rodriguez, E.; Roth, L.; et al. The Shuttle Radar Topography Mission. Rev. Geophys. 2007, 45, RG2004. [Google Scholar] [CrossRef]
- Sexton, J.O.; Song, X.-P.; Feng, M.; Noojipady, P.; Anand, A.; Huang, C.; Kim, D.-H.; Collins, K.M.; Channan, S.; DiMiceli, C.; et al. Global, 30-m resolution continuous fields of tree cover: Landsat-based rescaling of MODIS vegetation continuous fields with lidar-based estimates of error. Int. J. Digit. Earth 2013, 6, 427–448. [Google Scholar] [CrossRef]
- Asner, G.P.; Hughes, R.F.; Mascaro, J.; Uowolo, A.L.; Knapp, D.E.; Jacobson, J.; Kennedy-Bowdoin, T.; Clark, J.K. High-resolution carbon mapping on the million-hectare Island of Hawaii. Front. Ecol. Environ. 2011, 9, 434–439. [Google Scholar] [CrossRef]
- Chen, Q.; Vaglio Laurin, G.; Valentini, R. Uncertainty of remotely sensed aboveground biomass over an African tropical forest: Propagating errors from trees to plots to pixels. Remote Sens. Environ. 2015, 160, 134–143. [Google Scholar] [CrossRef]
- Zhu, J.; Hu, H.; Tao, S.; Chi, X.; Li, P.; Jiang, L.; Ji, C.; Zhu, J.; Tang, Z.; Pan, Y.; et al. Carbon stocks and changes of dead organic matter in China’s forests. Nat. Commun. 2017, 8, 151. [Google Scholar] [CrossRef]
- Chang, Z.; Hobeichi, S.; Wang, Y.-P.; Tang, X.; Abramowitz, G.; Chen, Y.; Cao, N.; Yu, M.; Huang, H.; Zhou, G.; et al. New Forest Aboveground Biomass Maps of China Integrating Multiple Datasets. Remote Sens. 2021, 13, 2892. [Google Scholar] [CrossRef]
- Tang, X.; Zhao, X.; Bai, Y.; Tang, Z.; Wang, W.; Zhao, Y.; Wan, H.; Xie, Z.; Shi, X.; Wu, B.; et al. Carbon pools in China’s terrestrial ecosystems: New estimates based on an intensive field survey. Proc. Natl. Acad. Sci. USA 2018, 115, 4021–4026. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Ghosh, S.M.; Behera, M.D. Aboveground biomass estimation using multi-sensor data synergy and machine learning algorithms in a dense tropical forest. Appl. Geogr. 2018, 96, 29–40. [Google Scholar] [CrossRef]
- Hu, T.; Su, Y.; Xue, B.; Liu, J.; Zhao, X.; Fang, J.; Guo, Q. Mapping Global Forest Aboveground Biomass with Spaceborne LiDAR, Optical Imagery, and Forest Inventory Data. Remote Sens. 2016, 8, 565. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.; Huang, J.; Guo, Z.; Ni, W.; Fu, A. National Forest Aboveground Biomass Mapping from ICESat/GLAS Data and MODIS Imagery in China. Remote Sens. 2015, 7, 5534–5564. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.Y.; Guindon, L.; Villemaire, P.; Guo, X.J.; Stinson, G.; Bergeron, T.; Magnussen, S.; Hall, R.J. Mapping attributes of Canada’s forests at moderate resolution through kNN and MODIS imagery. Can. J. For. Res. 2014, 44, 521–532. [Google Scholar] [CrossRef]
- Moradi, F.; Darvishsefat, A.A.; Pourrahmati, M.R.; Deljouei, A.; Borz, S.A. Estimating Aboveground Biomass in Dense Hyrcanian Forests by the Use of Sentinel-2 Data. Forests 2022, 13, 104. [Google Scholar] [CrossRef]
- Sexton, J.O.; Noojipady, P.; Song, X.-P.; Feng, M.; Song, D.-X.; Kim, D.-H.; Anand, A.; Huang, C.; Channan, S.; Pimm, S.L.; et al. Conservation policy and the measurement of forests. Nat. Clim. Chang. 2016, 6, 192–196. [Google Scholar] [CrossRef]
- Li, Y.; Sulla-Menashe, D.; Motesharrei, S.; Song, X.-P.; Kalnay, E.; Ying, Q.; Li, S.; Ma, Z. Inconsistent estimates of forest cover change in China between 2000 and 2013 from multiple datasets: Differences in parameters, spatial resolution, and definitions. Sci. Rep. 2017, 7, 8748. [Google Scholar] [CrossRef]
- Santoro, M.; Cartus, O.; Carvalhais, N.; Rozendaal, D.M.A.; Avitabile, V.; Araza, A.; de Bruin, S.; Herold, M.; Quegan, S.; Rodríguez-Veiga, P.; et al. The global forest above-ground biomass pool for 2010 estimated from high-resolution satellite observations. Earth Syst. Sci. Data 2021, 13, 3927–3950. [Google Scholar] [CrossRef]
- Anderegg, W.R.L.; Trugman, A.T.; Badgley, G.; Anderson, C.M.; Bartuska, A.; Ciais, P.; Cullenward, D.; Field, C.B.; Freeman, J.; Goetz, S.J.; et al. Climate-driven risks to the climate mitigation potential of forests. Science 2020, 368, eaaz7005. [Google Scholar] [CrossRef]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.C.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Neuville, R.; Bates, J.S.; Jonard, F. Estimating Forest Structure from UAV-Mounted LiDAR Point Cloud Using Machine Learning. Remote Sens. 2021, 13, 352. [Google Scholar] [CrossRef]
- Dalla Corte, A.P.; Rex, F.E.; Almeida, D.R.A.d.; Sanquetta, C.R.; Silva, C.A.; Moura, M.M.; Wilkinson, B.; Zambrano, A.M.A.; Cunha Neto, E.M.d.; Veras, H.F.P.; et al. Measuring Individual Tree Diameter and Height Using GatorEye High-Density UAV-Lidar in an Integrated Crop-Livestock-Forest System. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef]


| Region | Forest Type | a | b | RMSE | R2 | Number of Samples |
|---|---|---|---|---|---|---|
| N | Broadleaf | 5.291 | 1.093 | 15.37 | 0.75 | 63 |
| Conifer | 7.022 | 1.047 | 20.73 | 0.88 | 63 | |
| S | Broadleaf | 2.47 | 1.476 | 26.32 | 0.86 | 184 |
| Conifer | 6.849 | 1.123 | 48.26 | 0.65 | 60 |
| Models | R2 | RMSE |
|---|---|---|
| Random Forest (RF) | 0.74 | 23.36 |
| Gradient Boosting (GB) | 0.70 | 25.06 |
| Extreme Gradient Boosting (XGB) | 0.73 | 23.84 |
| Light Gradient Boosting Machine (LGBM) | 0.73 | 23.70 |
| Categorical Boosting (CatBoost) | 0.74 | 23.00 |
| Linear Regression (LR) * | 0.61 | 52.71 |
| k-Nearest Neighbor (KNN) * | 0.45 | 33.91 |
| Multilayer Perceptron (MLP) * | 0.18 | 69.32 |
| Ridge Regression (RR) * | 0.42 | 77.78 |
| Support Vector Regression (SVR) * | 0.29 | 38.52 |
| Feature Name | Source |
|---|---|
| NDVI_mean | Normalized difference vegetation index based on MOD13Q1 using mean synthesis |
| NDVI_max | Normalized difference vegetation index based on MOD13Q1 using maximum synthesis |
| EVI_mean | Enhanced vegetation index based on MOD13Q1 using mean synthesis |
| EVI_max | Enhanced vegetation index based on MOD13Q1 using maximum synthesis |
| LAI_mean | Leaf area index based on MOD13Q1 using mean synthesis |
| LAI_max | Leaf area index based on MOD13Q1 using maximum synthesis |
| FPAR_mean | Fraction of photosynthetically active radiation based on MOD13Q1 using mean synthesis |
| FPAR_max | Fraction of photosynthetically active radiation based on MOD13Q1 using maximum synthesis |
| ET_max | Evapotranspiration based on MOD13Q1 using mean synthesis |
| ET_mean | Evapotranspiration based on MOD13Q1 using maximum synthesis |
| NPP | Net primary productivity from GLASS |
| PRE_mean_30a | Average precipitation of 30 years |
| PRE_total_30a | Total precipitation of 30 years |
| PRE_2007_30a | The anomalies of the average precipitation in 2007 from the 30-year average precipitation |
| TMP_max_30a | Maximum temperature of 30 years |
| TMP_mean_30a | Average temperature from of 30 years |
| TMP_min_30a | Minimum temperature of 30 years |
| TMP_diff_30a | Temperature range of 30 years |
| TMP_total_30a | Total temperature of 30 years |
| TMP_2007_30a | The anomalies of the average temperature in 2007 from the 30-year average temperature |
| Elevation | Surface elevation extracted from SRTM |
| Slope | Surface slope extracted from SRTM |
| Feature Name | Permutation Importance/% | ||||
|---|---|---|---|---|---|
| RF | GB | XGB | LGBM | CatBoost | |
| Slope | 1.90 | 0.07 | 1.49 | 0.82 | 1.06 |
| Elevation | 13.97 | 5.54 | 8.27 | 5.98 | 6.62 |
| LAI_mean | 12.79 | 1.98 | 4.44 | 2.55 | 3.07 |
| NPP | 65.06 | 37.33 | 39.26 | 37.26 | 29.61 |
| FPAR_mean | 7.18 | 0.67 | 4.16 | 1.58 | 3.37 |
| LAI_max * | 0.68 | 0.20 | 0.39 | 0.18 | 0.30 |
| ET_mean | 2.11 | 0.14 | 1.83 | 0.99 | 1.58 |
| FPAR_max * | 0.77 | 0.51 | 0.62 | 0.39 | 0.65 |
| ET_max | 4.23 | 0.77 | 1.64 | 1.05 | 1.30 |
| NDVI_mean | 12.15 | 12.26 | 29.25 | 21.50 | 30.39 |
| EVI_mean | 6.45 | 5.62 | 20.66 | 13.22 | 27.66 |
| NDVI_max | 6.63 | 2.73 | 4.34 | 3.28 | 3.47 |
| EVI_max | 2.09 | 0.32 | 1.77 | 0.80 | 1.60 |
| PRE_mean_30a | 8.13 | 4.76 | 37.23 | 24.98 | 13.23 |
| PRE_total_30a | 8.49 | 7.51 | 0 | 0 | 11.66 |
| PRE_2007_30a | 5.78 | 1.70 | 5.36 | 2.64 | 5.12 |
| TMP_diff_30a | 7.09 | 0.94 | 5.90 | 1.97 | 6.25 |
| TMP_max_30a | 3.57 | 1.01 | 3.39 | 1.86 | 3.44 |
| TMP_mean_30a | 1.91 | 0.48 | 6.36 | 2.13 | 2.49 |
| TMP_min_30a | 2.69 | 0.35 | 3.08 | 1.03 | 1.81 |
| TMP_total_30a | 1.89 | 0.56 | 0 | 0 | 1.62 |
| TMP_2007_30a | 12.13 | 5.61 | 12.34 | 7.18 | 12.59 |
| Base Learners | R2 | RMSE |
|---|---|---|
| RF + CatBoost | 0.75 | 22.75 |
| RF + CatBoost + LGBM | 0.75 | 22.75 |
| RF + CatBoost + XGB | 0.75 | 22.74 |
| RF + CatBoost + LGBM + XGB | 0.75 | 22.74 |
| RF + CatBoost + LGBM + XGB + GB | 0.76 | 22.70 |
| Model | Parameters | |
|---|---|---|
| RF | n_estimations | 100 |
| max_depth | 16 | |
| min_samples_leaf | 6 | |
| Others | default | |
| GB | n_estimations | 600 |
| Others | default | |
| XGB | All | default |
| LGBM | n_estimations | 300 |
| max_depth | 11 | |
| Others | default | |
| CatBoost | All | default |
| Source | Approach | Method | Study Area | Year | Object | Average (Mg/ha) | Total (Pg) |
|---|---|---|---|---|---|---|---|
| FRA 2020 1 [1] | Forest inventory | Biomass expansion factor | China | 2010 | Forest aboveground biomass | 55.13 | 11.18 |
| Piao et al., 2005 [11] | Remote sensing | Regression analysis (Linear Regression) | China | 1997–1999 | Forest biomass | 93.68 | 11.98 |
| Beaudoin et al., 2014 [61] | Remote sensing | Machine learning (KNN) | Canada | 2001 | Forest aboveground biomass | 61.32 | 17.6 |
| Ghosh et al., 2018 [58] | Remote sensing | Machine learning (RF) 2 | Katerniaghat Wildlife Sanctuary, India | 2017 | Forest aboveground biomass | ||
| Luo et al., 2021 [31] | Remote sensing | Machine learning (CatBoost) 2 | Jilin province, China | 2014 | Forest aboveground biomass | 25.77 | |
| Moradi et al., 2022 [62] | Remote sensing | Machine learning (ANN) | Hyrcanian, Iran | 2016 | Forest aboveground biomass | 210 | |
| Saatchi et al., 2011 [17] | Remote sensing | Machine learning (Maximum Entropy) | Pan- tropical | 2000-2001 | Forest aboveground biomass | 157.04 (116.58) 3 | 386 (19.62) 3 |
| Santoro et al., 2021 [65] | Forest invention with remote sensing | Biomass expansion factor | Global | 2010 | Forest aboveground biomass | 108 (60) 3 | 521 (13.47) 3 |
| Hu et al., 2016 [59] | Remote sensing | Machine learning (RF) | Global | 2004 | Forest aboveground biomass | 210.09 (160.74) 3 | 532.75 (16.41) 3 |
| Chi et al., 2015 [49] | Remote sensing | Machine learning (RF) | China | 2006 | Forest aboveground biomass | 12.62 | |
| Su et al., 2016 [38] | Remote sensing | Machine learning (RF) | China | 2004 | Forest aboveground biomass | 120 | |
| Huang et al., 2019 [36] | Remote sensing | Machine learning (RF) | China | 2006 | Forest aboveground biomass | 69.87 | 10.88 |
| Chang et al., 2021 [55] | Remote sensing | Machine learning (RF) 2 | China | 2011-2015 | Forest aboveground biomass | 96.64 | 16.26 |
| This Study | Remote sensing | Machine learning (Stacking) | China | 2007 | Forest aboveground biomass | 53.16 | 11.00 |
| Model | R2 | Percentage of Decrease |
|---|---|---|
| RF | 0.62 | 15.07% |
| GB | 0.60 | 17.80% |
| XGB | 0.60 | 17.80% |
| LGBM | 0.62 | 16.21% |
| CatBoost | 0.63 | 14.86% |
| Stacked | 0.65 | 14.47% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Z.; Xia, X.; Huang, Y.; Lu, Y.; Guo, Z. Estimation of National Forest Aboveground Biomass from Multi-Source Remotely Sensed Dataset with Machine Learning Algorithms in China. Remote Sens. 2022, 14, 5487. https://doi.org/10.3390/rs14215487
Tang Z, Xia X, Huang Y, Lu Y, Guo Z. Estimation of National Forest Aboveground Biomass from Multi-Source Remotely Sensed Dataset with Machine Learning Algorithms in China. Remote Sensing. 2022; 14(21):5487. https://doi.org/10.3390/rs14215487
Chicago/Turabian StyleTang, Zhi, Xiaosheng Xia, Yonghua Huang, Yan Lu, and Zhongyang Guo. 2022. "Estimation of National Forest Aboveground Biomass from Multi-Source Remotely Sensed Dataset with Machine Learning Algorithms in China" Remote Sensing 14, no. 21: 5487. https://doi.org/10.3390/rs14215487
APA StyleTang, Z., Xia, X., Huang, Y., Lu, Y., & Guo, Z. (2022). Estimation of National Forest Aboveground Biomass from Multi-Source Remotely Sensed Dataset with Machine Learning Algorithms in China. Remote Sensing, 14(21), 5487. https://doi.org/10.3390/rs14215487






