Biomass Calculations of Individual Trees Based on Unmanned Aerial Vehicle Multispectral Imagery and Laser Scanning Combined with Terrestrial Laser Scanning in Complex Stands
Abstract
1. Introduction
2. Materials and Methods
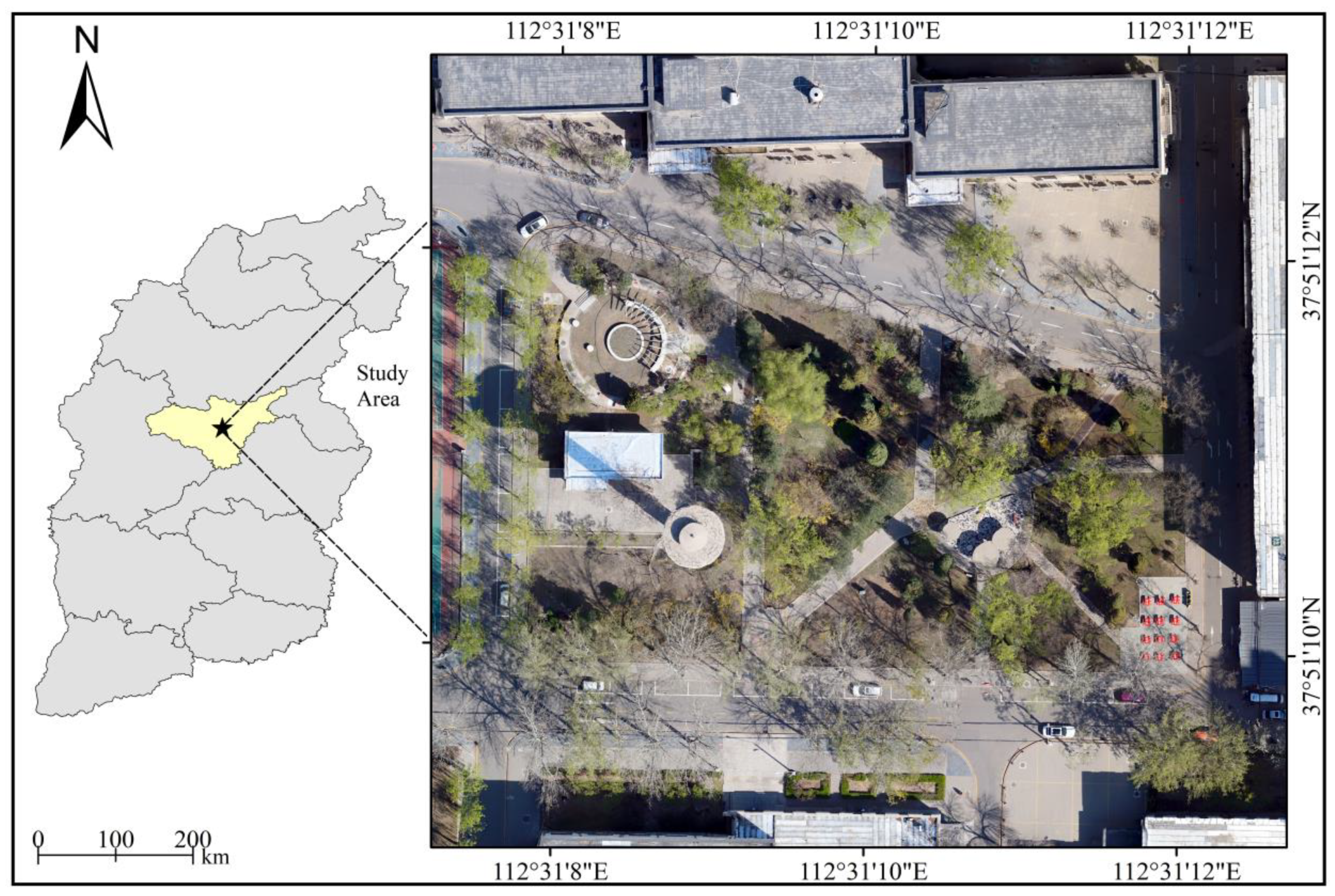
2.1. Study Site
2.2. Field Data Collection
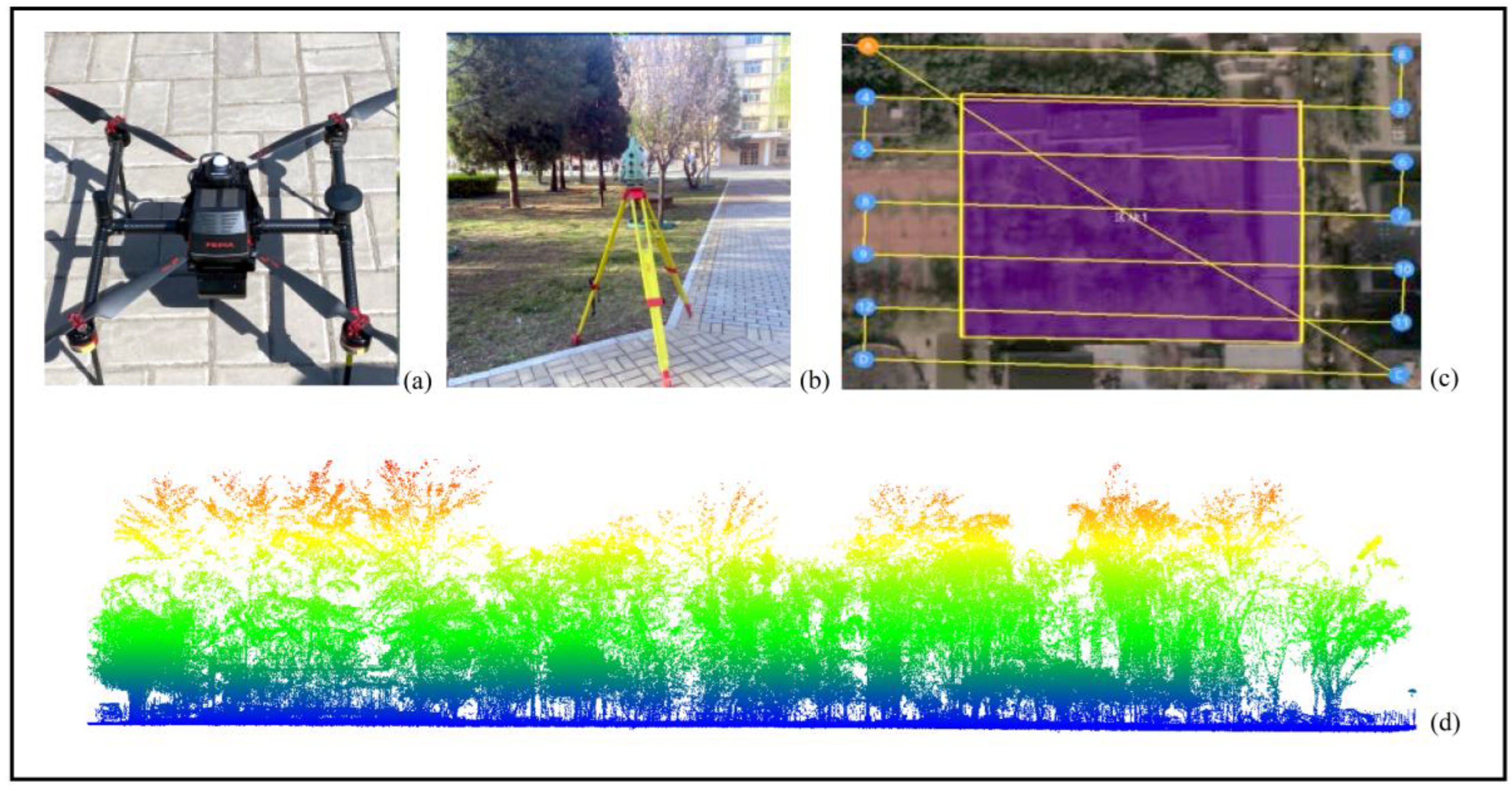
2.2.1. Manual Data Collection
2.2.2. LIDAR Remote Sensing Data
- (1)
- Terrestrial Laser Scanning (TLS) Data
- (2)
- Unmanned aerial vehicle laser scanning (UAV-LS) data
2.2.3. Optical Remote Sensing Data
- (1)
- Orthophoto Data
- (2)
- Multispectral Data
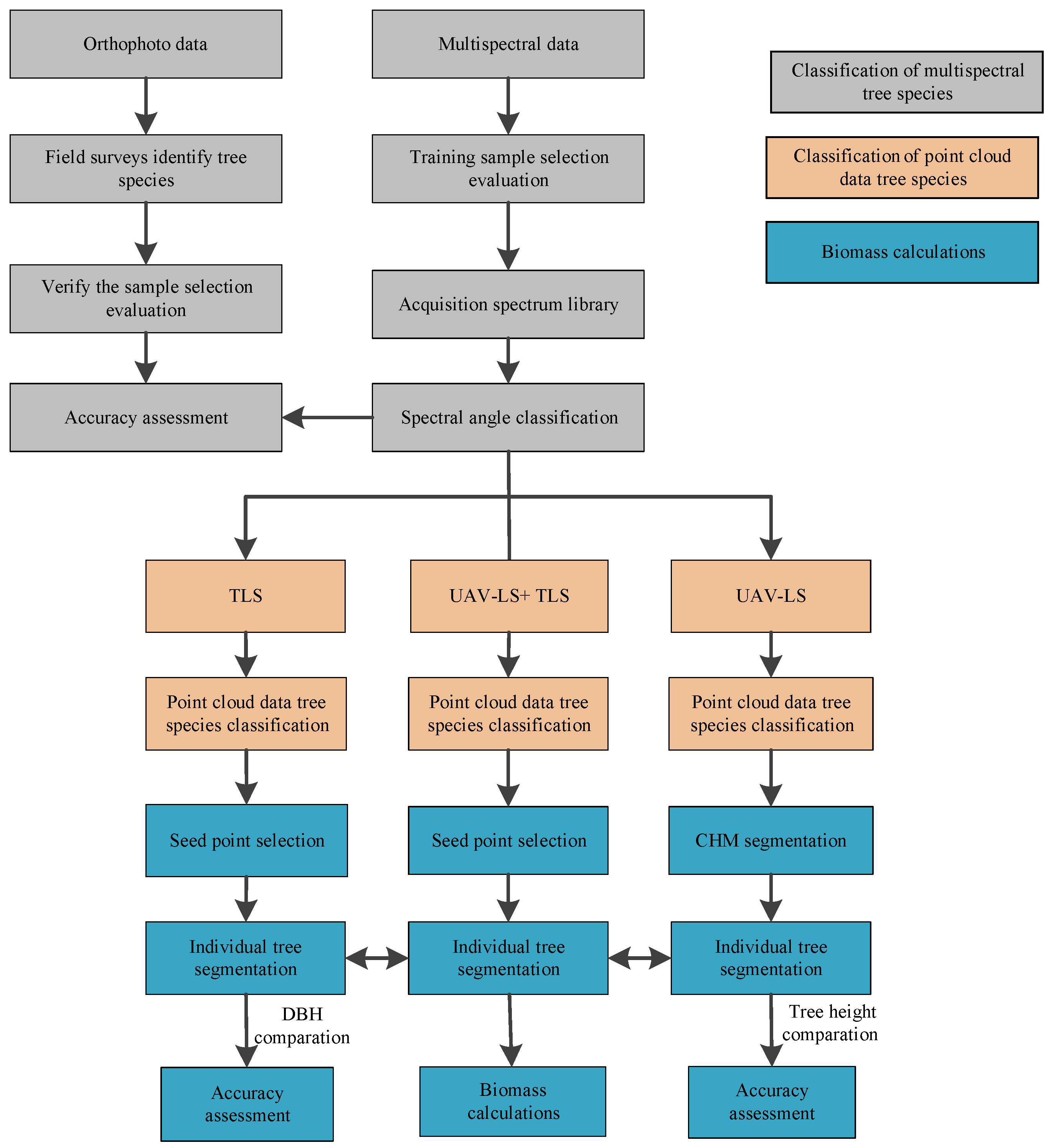
2.3. Research Methods
2.3.1. Tree Species Classification in the Study Area
- (1)
- A multinomial distribution algorithm introduced by Mosimani and James is adopted [40]:where is the total number of samples; is the percentage of the tree category in the total tree species; is the confidence of ( is the number of classification categories, and is the expected significance level) with a chi-square test value having a degree of freedom of 1; and is the expected classification error ratio percentage of the class. The minimum reasonable number of samples is then calculated.
- (2)
- Gong and Howarth studied different sampling strategies and believed that the highest classification accuracy can be obtained by collecting a pixel sample every N pixels [41]. Here, simple random sampling with an unbiased estimation of the population parameters is used.
- (3)
- The spectral angle classification (SAM) algorithm is used to compare and classify the unknown spectral lines with the sample spectrum in N-dimensional space [42]. By comparing the angle between the reference spectrum vector and each pixel vector, it is found that a greater angle causes the pixels to be more similar to the reference spectrum.
- (4)
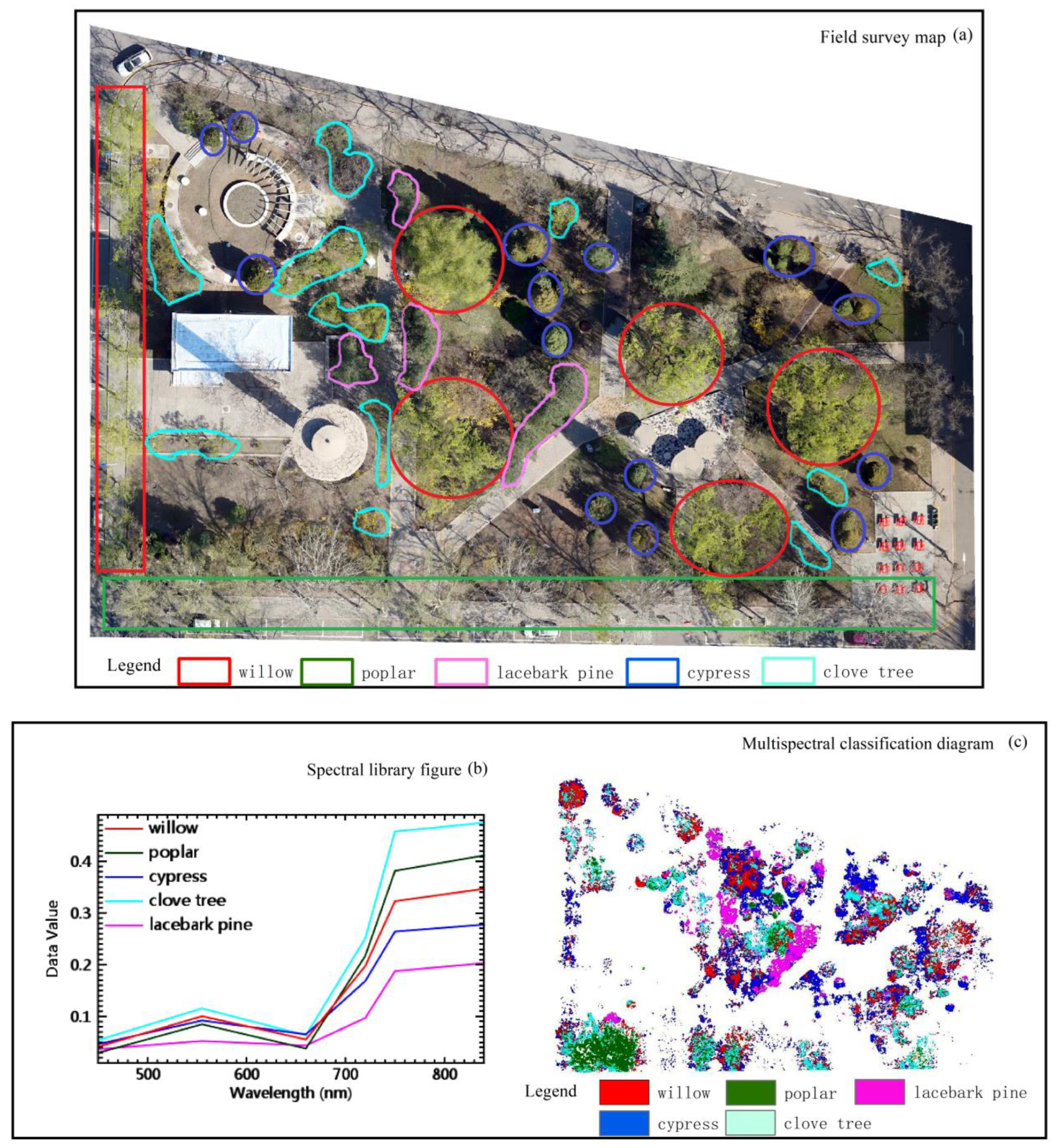
- According to the field investigation of tree species, the tree spectra are extracted from the image pure pixels to establish the spectrum library, which is used to sort the tree species in the study area, as represented in Figure 4.
2.3.2. Individual Tree Biomass Calculation
LIDAR Point Cloud Data Tree Species Classification
- (1)
- Fusion of UAV-LS and TLS Data
- (2)
- Point cloud data for tree species classification
Calculation of Individual Tree Biomass of Classified Tree Species
- (1)
- The noise mainly includes high-position gross error and low-position gross error. The algorithm searches adjacent points within a specified neighborhood; calculates the average distance from the point to the adjacent points; calculates the median, mean, and standard deviation of the average distances; and removes noise by selecting appropriate parameters.
- (2)
- The improved progressive triangulation filtering algorithm (iPTD) is used to classify ground points [45]. A sparse triangulation is generated through seed points and is later encrypted by the layer through iterative processing until all ground points are classified.
- (3)
- The digital terrain model (DTM) is removed from the digital surface model (DSM) to obtain the canopy height model (CHM) [46]. Through the upstream ridge segmentation algorithm, the high points of the CHM are regarded as peaks, and the low points are regarded as valleys. The water areas are filled, and barriers are built as the water edges as determined from segmentation. In experiments, when performing CHM segmentation, a tree is often identified as several trees based on the algorithm alone, resulting in multi-division so that there are more trees after CHM segmentation than actual trees. Consequently, we need to combine CHM segmentation and manual operations to classify trees. The data classification was conducted using Cloud Compare and LIDAR360 software. The individual tree segmentation of the seed points is used to define the parameter variables such as the tree height and DBH, as shown in Figure 7.
- (4)
- The volume calculations of the study area adopt the individual tree binary volume model volume , where , , and are model parameters; is the DBH; and is the tree height.
3. Results
3.1. Evaluation of Tree Species Identification Based on Multispectral Data
3.1.1. Evaluation of Samples before Classification
3.1.2. Evaluation of Results after Classification
3.2. Biomass Based on Classified Tree Species
3.2.1. Comparison between DBH and Tree Height
- (1)
- A comparison between the DBH and tree height of various species is shown in Figure 8.
- (2)
- A comparison between DBH and tree height of all tree species is shown in Figure 9.
3.2.2. Biomass Calculations for Each Tree Species
4. Discussion and Conclusions
4.1. Multispectral Identification of Tree Species
4.2. Individual Tree Parameter Segmentation and Comparison
4.3. Conclusion
- (1)
- In the study area, the multispectral tree species identification shows that the extracted spectra can accurately identify Lacebark pine, and the identification of other tree species is slightly lower than that of Lacebark pine. Thus, the reflectance spectrum of Lacebark pine can be applied to the identification of Lacebark pine species.
- (2)
- The comparison of DBH after TLS and UAV-LS+TLS and the comparison of tree height after UAV-LS and UAV-LS+TLS under appropriate conditions show that the DBH parameters obtained by TLS and tree height parameters obtained by UAV-LS have good availability.
- (3)
- In complex stands, multispectral techniques can be used to identify tree species, and LIDAR technology can be used to perform individual tree parameter calculations. Accurately combining the two data, it is feasible to identify tree species in complex forest stands and calculate the biomass of individual trees.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hember, R.; Kurz, W. Low tree-growth elasticity of forest biomass indicated by an individual-based model. Forests 2018, 9, 21. [Google Scholar] [CrossRef]
- Anjin, C.; Yongmin, K.; Yongil, K.; Yangdam, E. Estimation of individual tree biomass from airborne lidar data using tree height and crown diameter. Disaster Adv. 2012, 5, 360–365. [Google Scholar]
- Krofcheck, D.; Litvak, M.; Lippitt, C.; Neuenschwander, A. Woody biomass estimation in a southwestern US. Juniper savanna using lidar-derived clumped tree segmentation and existing allometries. Remote Sens. 2016, 8, 453. [Google Scholar] [CrossRef]
- Zeng, W.; Fu, L.; Xu, M.; Wang, X.; Chen, Z.; Yao, S. Developing individual tree-based models for estimating aboveground biomass of five key coniferous species in China. J. For. Res. 2018, 29, 1251–1261. [Google Scholar] [CrossRef]
- Kitahara, F.; Mizoue, N.; Yoshida, S. Effects of training for in experienced survey or son data quality of tree diameter and height measurements. Silva Fenn. 2010, 44, 657–667. [Google Scholar] [CrossRef]
- Hunter, M.; Keller, M.; Victoria, D.; Morton, D. Tree height and tropical forest biomass estimation. Biogeosciences 2013, 10, 8385–8399. [Google Scholar] [CrossRef]
- Larjavaara, M.; Muller-Landau, H.C. Measuring tree height: A quantitative comparison of two common field methods in a moist tropical forest. Methods Ecol. Evol. 2013, 4, 793–801. [Google Scholar] [CrossRef]
- Sterenczak, K.; Mielcarek, M.; Wertz, B.; Bronisz, K.; Zajączkowski, G.; Jagodziński, A.M.; Ochał, W.; Skorupski, M. Factors influencing the accuracy of ground-based tree-height measurements for major European tree species. J. Environ. Manag. 2019, 231, 1284–1292. [Google Scholar] [CrossRef]
- Laasasenaho, J. Taper curve and volume functions for pine, Spruce and Birch. Commun. Inst. Forest Fenn. 1982, 108, 1–74. [Google Scholar]
- Repola, J. Biomass equations for birch in Finland. Silva Fenn. 2008, 42, 605–624. [Google Scholar] [CrossRef]
- Repola, J. Biomass equations for scots pine and Norway spruce in Finland. Silva Fenn. 2009, 43, 625–647. [Google Scholar] [CrossRef]
- Koch, H.; Heyder, U.; Weinacker, H. Detection of individual tree crowns in airborne lidar data. Photogramm. Eng. Remote Sens. 2006, 72, 357–363. [Google Scholar] [CrossRef]
- Bohlin, J.; Wallerman, J.; Fransson, E.S. Forest variable estimation using photogrammetric matching of digital aerial images in combination with a high-resolution DEM. Scand. J. For. Res. 2012, 27, 692–699. [Google Scholar] [CrossRef]
- White, J.; Coops, N.; Wulder, M.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote. Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Olofsson, K.; Holmgren, J. Single tree stem profile detection using terrestrial laser scanner data, flatness saliency features and curvature properties. Forests 2016, 7, 207. [Google Scholar] [CrossRef]
- Gobakken, T.; Bollandsås, O.; Næsset, E. Comparing biophysical forest characteristics estimated from photo grammatic matching of aerial images and air born elaser scanning data. Scand. J. For. Res. 2015, 30, 73–86. [Google Scholar] [CrossRef]
- Vastaranta, M.; Wulder, M.; White, J.; Pekkarinen, A.; Tuominen, S.; Ginzler, C. Airborne laser scanning and digital stereo imagery measures of forest structure: Comparative results and implications to forest mapping and inventory update. Can. J. Remote Sens. 2013, 39, 382–395. [Google Scholar] [CrossRef]
- White, J.; Wulder, M.; Vastaranta, M.; Coops, N.; Pitt, D.; Woods, M. The utility of image-based point clouds for forest inventory: A comparison with airborne laser scanning. Forests 2013, 4, 518–536. [Google Scholar] [CrossRef]
- Moe, K.; Owari, T.; Furuya, N.; Hiroshima, T. Comparing individual tree height information derived from field surveys, lidar and UAV-dap for high-value timber species in northern Japan. Forests 2020, 11, 223. [Google Scholar] [CrossRef]
- Olli, N.; Eija, H.; Sakari, T.; Niko, V.; Teemu, H.; Xiaowei, Y. Individual tree detection and classification with UAV-based photogrammetric point clouds and hyperspectral imaging. Remote Sens. 2017, 9, 185. [Google Scholar]
- Reason, M.; Iain, W.; France, G.; Karen, A. Structure from motion (sfm) photogrammetry with drone data: A low cost method for monitoring greenhouse gas emissions from forests in developing countries. Forests 2017, 8, 68. [Google Scholar]
- Cao, L.; Liu, H.; Fu, X.; Zhang, Z.; Ruan, H. Comparison of UAV lidar and digital aerial photogrammetry point clouds for estimating forest structural attributes in subtropical planted forests. Forests 2019, 10, 145. [Google Scholar] [CrossRef]
- He, R.; Zhao, Y.; Xiao, W.; Yang, X.; Ding, B.; Chen, C. Monitoring the potential spontaneous combustion in a coal waste dump after reclamation through UAV RGB imagery-based on alfalfa above ground biomass (AGB). Land Degrad. Dev 2022. In press. [Google Scholar]
- Ginzler, C.; Waser, L. Progress in remote sensing for forestry applications. Schweiz. Z. Forstwes. 2017, 168, 118–126. [Google Scholar] [CrossRef]
- Torresan, B.; Carotenuto, F.; Berton, A.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L.; et al. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sen. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Lim, K.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. LiDAR remote sensing of forest structure. Prog. Phys. Geogr. 2003, 27, 88–106. [Google Scholar] [CrossRef]
- Yong, P.; Li, Z.Y.; Chen, E.X.; Sun, G.Q. Lidar remote sensing technology and its application in forestry. Sci. Silvae Sin. 2005, 41, 129. [Google Scholar]
- Li, M.; Im, J.; Quackenbush, L.J.; Liu, T. Forest biomass and carbon stock quantification using Airborne LiDAR Data: A case study over Huntington wildlife forest in the Adirondack park. IEEE J.-Stars. 2017, 7, 3143–3156. [Google Scholar] [CrossRef]
- Chen, R.; Li, C.; Zhu, Y.; Lei, L.; Zhang, C. Extraction of crown information from individual fruit tree by UAV LiDAR. Nongye Gongcheng Xuebao Trans. Chin. Soc. Agric. Eng. 2020, 36, 50–59. [Google Scholar]
- Geng, L.; Li, M.; Fan, W.; Wang, B.; Forestry, S. Individual tree structure parameters and effective crown of the stand extraction base on Airborn LiDAR Data. Sci. Silvae Sin. 2018, 54, 62–72. [Google Scholar]
- Brede, B.; Calders, K.; Lau, A.; Raumonen, P.; Kooistra, L. Non-destructive tree volume estimation through quantitative structure modelling: Comparing UAV laser scanning with terrestrial LIDAR. Remote Sens. Environ. 2019, 233, 111355. [Google Scholar] [CrossRef]
- Douss, R.; Farah, I.R. Extraction of individual trees based on Canopy Height Model to monitor the state of the forest. Trees For. People 2022, 8, 100257. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A new method for segmenting individual trees from LiDAR point cloud. Photogram Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef]
- Xu, D.; Wang, H.; Xu, W. LiDAR applications to estimate forest biomass at individual tree scale: Opportunities, challenges and future perspectives. Forests 2021, 12, 550. [Google Scholar] [CrossRef]
- Huadong, X.; Chen, W.; Liu, H. Single-wood DBH and tree height extraction using terrestrial laser scanning. J. Forest Environ. 2019, 39, 524–529. [Google Scholar]
- Ling, Z.; Jie, Z. Obtaining three-dimensional forest canopy structure using TLS. In Proceedings of the SPIE—The International Society for Optical Engineering, San Diego, CA, USA, 10 August 2008; Volume 70, pp. 7–10. [Google Scholar]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sen. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, A.; Soederman, U. Species identification of individual trees by combining high resolution LiDAR data with multi: Pectoral images. Int. J. Remote Sens. 2008, 29, 1537–1552. [Google Scholar] [CrossRef]
- Lian, X.; Hu, H.; Cai, Y.; Ma, H. Engineering Application and Practice of 3D Laser Scanning Technology; Surveying and Mapping Press: Beijing, China, 2017. [Google Scholar]
- Mosimann, J.E. On the compound multinomial distribution, the multivariate β-distribution, and correlations among proportions. Biometrika 1962, 49, 65–82. [Google Scholar]
- Gong, P.; Howarth, P. Performance analyses of probabilistic relaxation methods for land-cover classification. Remote Sens. Environ. 1989, 30, 33–42. [Google Scholar] [CrossRef]
- Park, B.; Windham, W.; Lawrence, K.; Smith, D. Contaminant classification of poultry hyperspectral imagery using a spectral angle mapper Algorithm. Biosyst. Eng. 2007, 96, 323–333. [Google Scholar] [CrossRef]
- Bolkas, D.; Chiampi, J.; Chapman, J.; Pavill, V. Creating a virtual reality environment with a fusion of sUAS and TLS point-clouds. Int. J. Image Data Fusion 2020, 11, 136–161. [Google Scholar] [CrossRef]
- Lin, X.; Xie, W. A segment-based filtering method for mobile laser scanning point cloud. Int. J. Image Data Fusion 2022, 13, 136–154. [Google Scholar] [CrossRef]
- Wang, H.; Wang, S.; Chen, Q.; Jin, W.; Sun, M. An improved filter of progressive TIN densification for LiDAR point cloud data. Wuhan Univ. J. Nat. Sci. 2015, 20, 362–368. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surovy, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Li, H.; Lei, Y. Estimation and Evaluation of Forest Biomass Carbon Storage in China; China Forestry Press: Beijing, China, 2010. [Google Scholar]
- Zhu, W.; Lin, W. Remote Sensing Digital Image Processing; Higher Education Press: Beijing, China, 2015. [Google Scholar]
- Roberts, C.; Mcnamee, R. Assessing the reliability of ordered categorical scales using kappa-type statistics. Stat. Methods Med. Res. 2005, 14, 493–514. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, M. Weighted kappa statistic for clustered matched-pair ordinal data. Comput. Stat. Data Anal. 2015, 82, 1–18. [Google Scholar] [CrossRef]
- Xiao, W.; Ren, H.; Sui, T.; Zhang, H.; Zhao, Y.; Hu, Z. A drone and field-based investigation of the land degradation and soil erosion at opencast coal mine dumps after 5 years’ evolution of natural processes. Int. J. Coal Sci. Technol. 2022, 9, 42. [Google Scholar] [CrossRef]
- Bo, H.; Bei, Z.; Song, Y. Urban land-use mapping using a deep convolutional neural network with high spatial resolution multispectral remote sensing imagery. Remote Sens. Environ. 2018, 214, 73–86. [Google Scholar]
- Hauser, L.; Timmermans, J.; Windt, N. Explaining discrepancies between spectral and in-situ plant diversity in multispectral satellite earth observation. Remote Sens Environ. 2021, 265, 112684. [Google Scholar] [CrossRef]
- Levick, S.; Whiteside, T.; Loewensteiner, D. Leveraging TLS as a calibration and validation tool for MLS and ULS mapping of savanna structure and biomass at landscape-scales. Remote Sens. 2021, 13, 257. [Google Scholar] [CrossRef]
- Brede, B.; Lau, A.; Bartholomeus, H.M. Comparing RIEGL RiCOPTER UAV LiDAR derived canopy height and DBH with terrestrial LiDAR. Sensors 2017, 17, 2371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Z.; Wang, Y.; Zheng, Z.; Zheng, S.; Zhao, D.; Bai, Y. UAV-based individual shrub aboveground biomass estimation calibrated against terrestrial LiDAR in a shrub-encroached grassland. Int. J. Appl. Earth Obs. Geoinf. 2021, 101, 102358. [Google Scholar] [CrossRef]
- Ren, H.; Zhao, Y.; Wu Xiao, W.; Zhang, J.; Chen, C.; Ding, B.; Yang, X. Vegetation growth status as an early warning indicator for the spontaneous combustion disaster of coal waste dump after reclamation: An unmanned aerial vehicle remote sensing approach. J. Environ. Manag. 2022, 317, 115502. [Google Scholar] [CrossRef] [PubMed]
- Donev, P.; Wang, H.; Qin, S.; Meng, P.; Lu, J. Estimating the forest above-ground biomass based on extracted LiDAR metrics and predicted diameter at breast height. Chin. J. Surv. Mapp. Engl. Ed. 2021, 4, 13–24. [Google Scholar]
- Wang, Y.; Pyrl, J.; Liang, X. In situ biomass estimation at tree and plot levels: What did data record and what did algorithms derive from terrestrial and aerial point clouds in boreal forest. Remote Sens. Environ. 2019, 232, 111309. [Google Scholar] [CrossRef]
- Li, Q.; Wong, F.; Fung, T. Mapping multi-layered mangroves from multispectral, hyperspectral, and LiDAR data. Remote Sens Environ. 2021, 258, 112403. [Google Scholar] [CrossRef]
- Jl, A.; Hong, W.; Qin, S.; Cao, L.; Pu, R.; Li, G.; Sun, J. Estimation of aboveground biomass of Robinia pseudoacacia forest in the Yellow River Delta based on UAV and Backpack LiDAR point clouds. Int. J. Appl. Earth. Obs. Geoinf. 2020, 86, 102014. [Google Scholar]
- Kukkonen, M.; Maltamo, M.; Korhonen, L. Comparison of multispectral airborne laser scanning and stereo matching of aerial images as a single sensor solution to forest inventories by tree species. Remote Sens. Environ. 2019, 231, 111208. [Google Scholar] [CrossRef]
- Shukhrat, S.; Michael, S.; Shaun, R.; Tommaso, J.; Justin, B.; Ilhom, A.; Kara, Y. Multi-platform LiDAR approach for detecting coarse woody debris in a landscape with varied ground cover. Int. J. Remote Sens. 2021, 42, 9316–9342. [Google Scholar]
- Michele, D.; Lorenzo, B.; Damiano, G. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar]
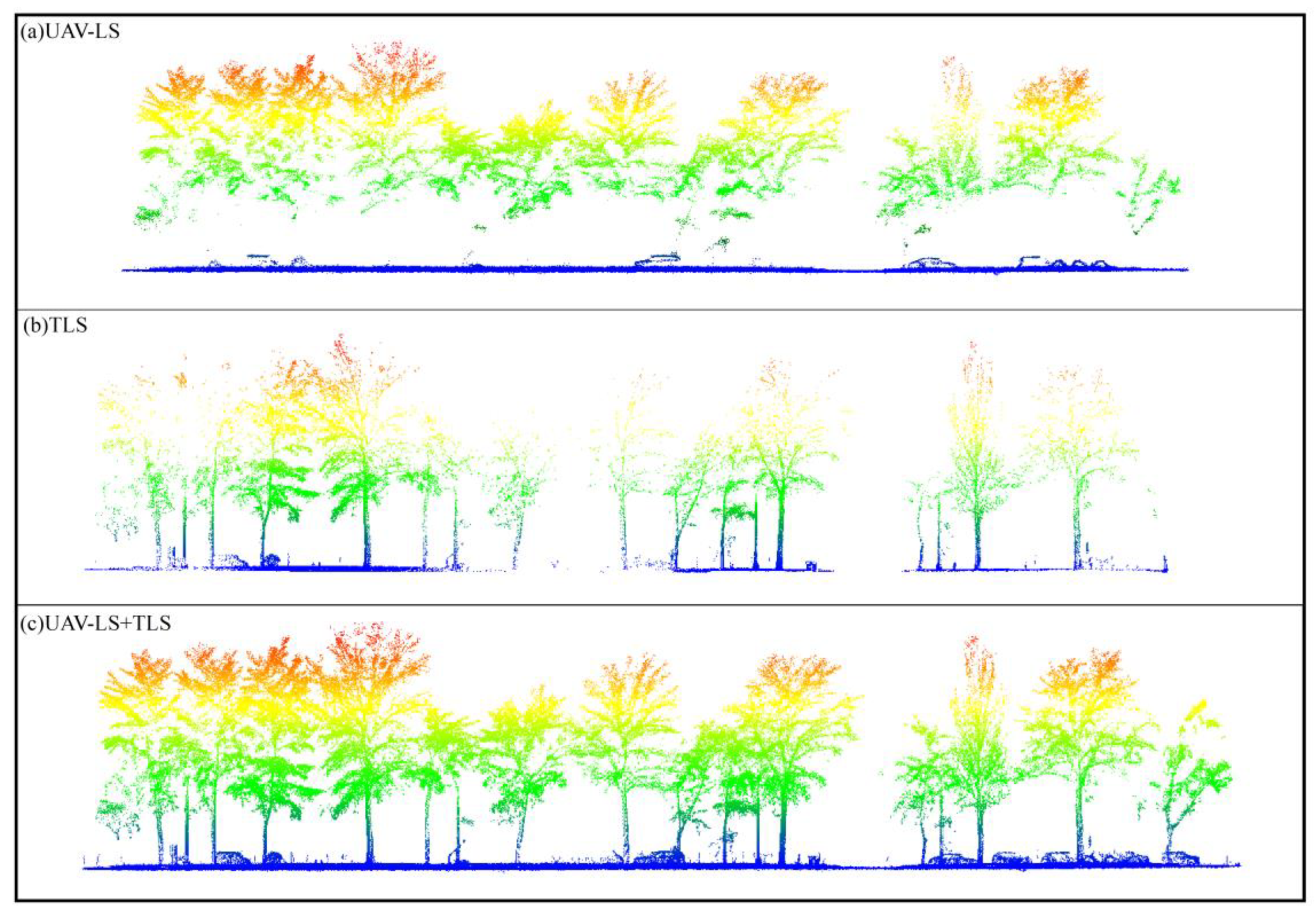
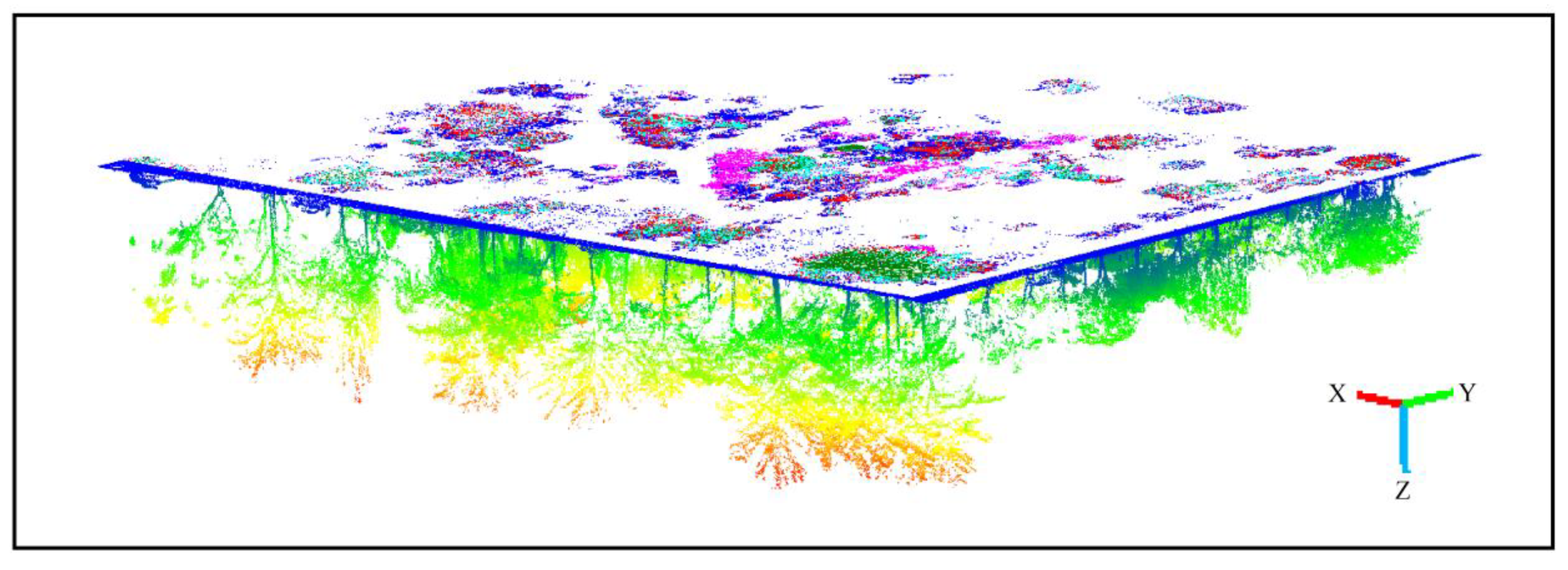


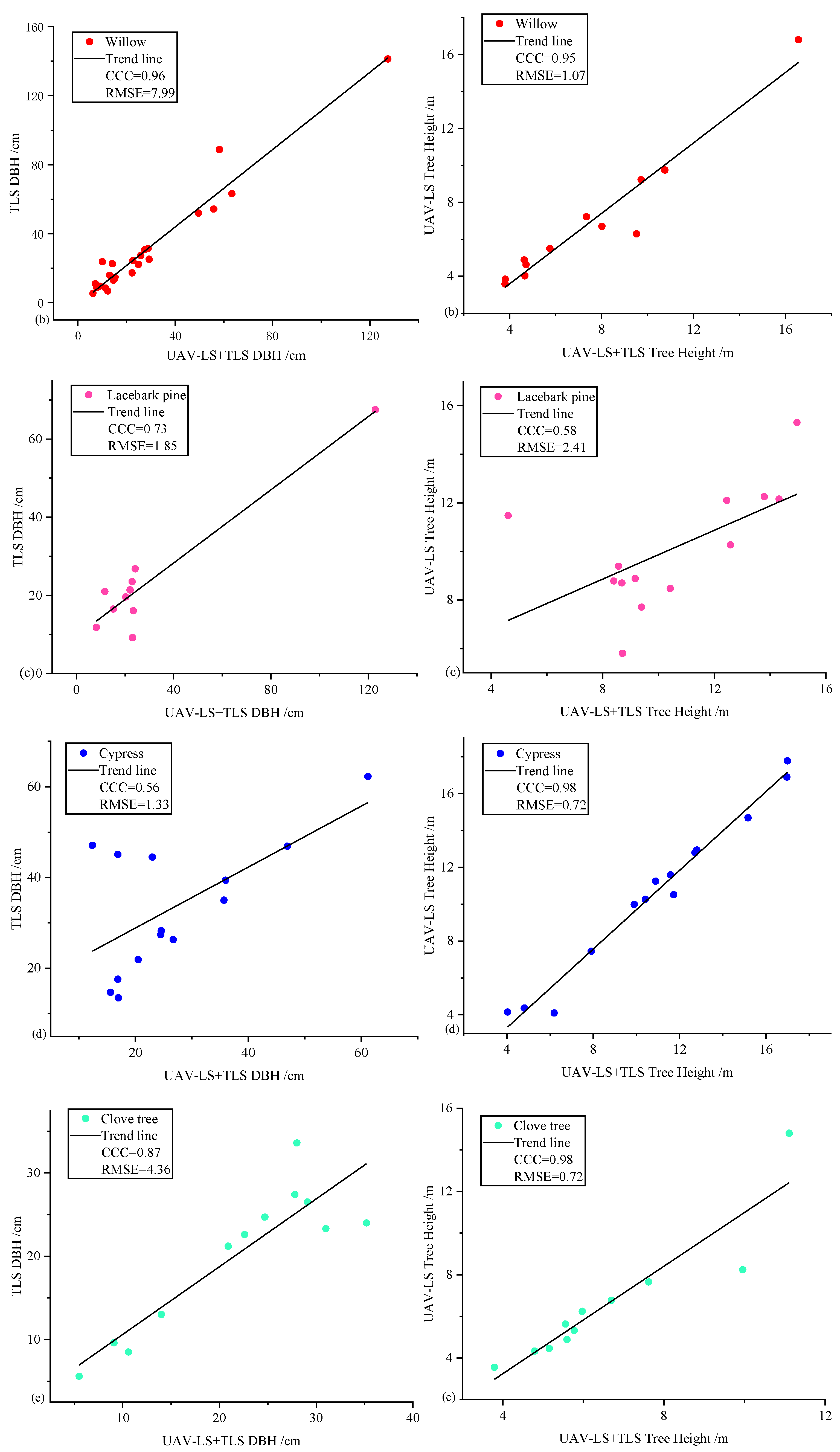
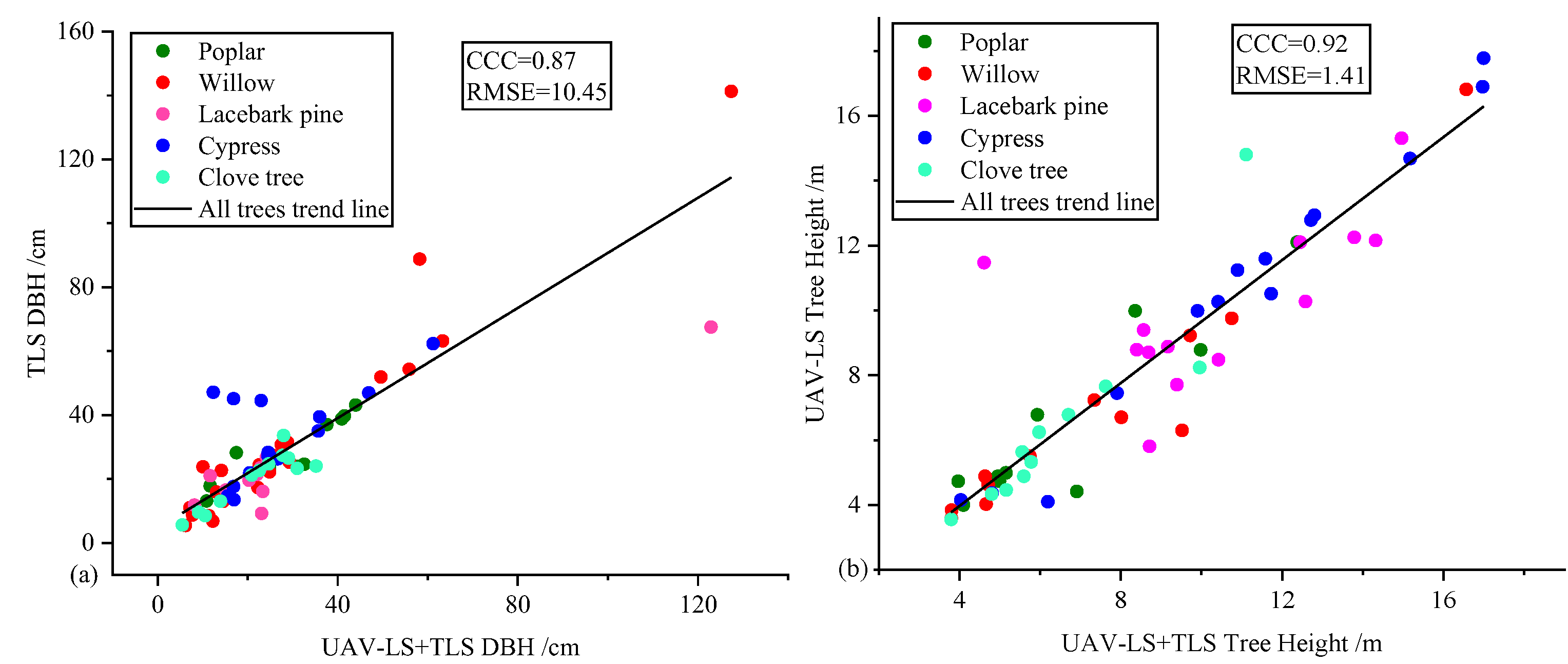
| Tree Species | Trees | Morphological Characteristics |
|---|---|---|
| Willow | 17 | Switch branches are slender, soft, and drooping; bark tissue is thick and longitudinally split; and the center of the old trunk is rotten and hollow. |
| Poplar | 12 | Bark grayish brown, fissured at the lower part; and sprouts thin, round, smooth, or slightly tomentose. |
| Clove tree | 42 | The trunk is forked; the crown is conical; and the bark is smooth, yellowish brown. |
| Lacebark pine | 15 | There is a large trunk; the branches are slender, obliquely spread, tower-shaped or umbrella-shaped crown; and winter buds are reddish-brown, oval, without resin. |
| Cypress | 29 | The bark is dark gray, and young trees often have branches that extend diagonally to form a spire-shaped canopy. |
| MS50 | ||
|---|---|---|
| Range | Prism (GPR1,GPH1P) | 1.5 to 3500 m |
| No prism/any surface | 1.5 m to >1000 m | |
| Reflector (60 mm × 60 mm) | 250 m | |
| Accuracy/ Measurement | single time (prism) | 0.6 mm + 1 ppm/ typically 2.4 |
| Single (any surface) | 2 mm + 2 ppm/typically 3 s | |
| Spot size | At 50m | 8 mm × 20 mm |
| Measurement technology | System analysis technology based on phase principle | Coaxial, red visible light |
| FEIMA D-LIDAR 2000 Module | |||
|---|---|---|---|
| Laser Type | RIEGL mini VUX-1UAV | Channels | 1 |
| Dot Frequency | 100 kpts/s | Measurement Range | 250 m |
| Range Accuracy | ±1 cm | Echo Number | 5 (Max.) |
| Scanning Speed | 10~100 Hz | Echo Intensity | 16 bit |
| Wavelength | 905 nm (Class 1) | Laser Divergence Angle | 1.6 × 0.5 mrad |
| Horizontal Field of View | 360° | Resolution-horizontal | 0.05~0.5° |
| FEIMA robotics D-CAM2000 Aerial Survey Module | |||
|---|---|---|---|
| Camera Type | SONY ILCE-6000 (α6000) | Sensor Size | 23.5 × 15.6 mm |
| Effective Size | (6000 × 4000) 2400 million | Lens | 20 mm fixed focus |
| Gimbal | 2-axis | ||
| FEIMA Robotics D-MSPC2000 Multi-Spectral Module | |||
|---|---|---|---|
| Sensor parameters | CMOS: 1/3″ global shutter | Effective pixels | 1.2 million |
| Resolution | 1280 × 960 | Sensor size | 4.8 mm × 3.6 mm |
| Focal length | 5.2 mm | Field of view | HFOV: 49.6°, VFOV: 38° |
| Aperture | F/2.2 | Quantization bits | 12 bit |
| Shooting speed | 1 time/s | Ground resolution | GSD: 8.65 cm/pix, AGL: 120 m |
| Training Samples | Poplar | Cypress | Clove Tree | Lacebark Pine |
| Willow | −1.877 | −1.939 | −1.979 | −1.999 |
| Poplar | −1.999 | −1.945 | −1.999 | |
| Cypress | −1.999 | −1.998 | ||
| Clove tree | −1.999 | |||
| Validation Samples | Poplar | Cypress | Clove Tree | Lacebark Pine |
| Willow | −1.874 | −1.981 | −1.967 | −1.999 |
| Poplar | −1.968 | −1.910 | −1.998 | |
| Cypress | −1.997 | −1.869 | ||
| Clove tree | −1.999 |
| Overall Accuracy (40/78) 51.28% | Kappa Coefficient 0.42 | ||||
|---|---|---|---|---|---|
| Tree | Willow | Poplar | Cypress | Clove Tree | Lacebark Pine |
| Commission (Percent) | 58.33 | 0.00 | 28.57 | 53.85 | 10.00 |
| Omission (Percent) | 79.17 | 28.57 | 50.00 | 50.00 | 18.18 |
| Tree Species | Volume/m3 | |||
|---|---|---|---|---|
| Willow | 13.99 | 12,251.55 | 3182.22 | 15,433.77 |
| Poplar | 10.29 | 9105.79 | 2365.14 | 11,470.93 |
| Clove tree | 6.56 | 5738.49 | 1490.52 | 7229.01 |
| Cypress | 12.73 | 11,257.70 | 3881.98 | 15,139.68 |
| Lacebark pine | 1.42 | 1256.47 | 326.36 | 1582.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, X.; Zhang, H.; Xiao, W.; Lei, Y.; Ge, L.; Qin, K.; He, Y.; Dong, Q.; Li, L.; Han, Y.; et al. Biomass Calculations of Individual Trees Based on Unmanned Aerial Vehicle Multispectral Imagery and Laser Scanning Combined with Terrestrial Laser Scanning in Complex Stands. Remote Sens. 2022, 14, 4715. https://doi.org/10.3390/rs14194715
Lian X, Zhang H, Xiao W, Lei Y, Ge L, Qin K, He Y, Dong Q, Li L, Han Y, et al. Biomass Calculations of Individual Trees Based on Unmanned Aerial Vehicle Multispectral Imagery and Laser Scanning Combined with Terrestrial Laser Scanning in Complex Stands. Remote Sensing. 2022; 14(19):4715. https://doi.org/10.3390/rs14194715
Chicago/Turabian StyleLian, Xugang, Hailang Zhang, Wu Xiao, Yunping Lei, Linlin Ge, Kai Qin, Yuanwen He, Quanyi Dong, Longfei Li, Yu Han, and et al. 2022. "Biomass Calculations of Individual Trees Based on Unmanned Aerial Vehicle Multispectral Imagery and Laser Scanning Combined with Terrestrial Laser Scanning in Complex Stands" Remote Sensing 14, no. 19: 4715. https://doi.org/10.3390/rs14194715
APA StyleLian, X., Zhang, H., Xiao, W., Lei, Y., Ge, L., Qin, K., He, Y., Dong, Q., Li, L., Han, Y., Fan, H., Li, Y., Shi, L., & Chang, J. (2022). Biomass Calculations of Individual Trees Based on Unmanned Aerial Vehicle Multispectral Imagery and Laser Scanning Combined with Terrestrial Laser Scanning in Complex Stands. Remote Sensing, 14(19), 4715. https://doi.org/10.3390/rs14194715








