1. Introduction
Seagrasses are important coastal and marine habitats in temperate and tropical regions [
1], providing a broad range of ecological functions and ecosystem services (ES) [
2,
3]. Due to over-exploitation, physical modification, nutrient and sediment pollution, the introduction of non-native species and global climate change, these important habitats are declining worldwide [
4,
5] at even faster rates than tropical forests [
6]. This decline, partly explained by global scale phenomena [
7], is greatly enhanced by the accumulation of local threats, exposing them as one of the main causes of seagrasses regression [
8].
Among the three species of seagrasses present in the Canarian Archipelago (Spain),
Cymodocea nodosa (Ucria) Ascherson,
Zostera noltei Hornemann and
Halophila decipiens Ostenfeld [
9],
C. nodosa is by far the best-established species, sustaining the most important marine ecosystem in sandy bottoms and being considered as a suitable bioindicator of ecosystem conservation status [
10]. In the archipelago, local threats menacing this species are mainly related to human coastal activities [
4,
11].
Although the distribution of
C. nodosa has been already studied in the Archipelago [
11,
12,
13,
14,
15,
16] these one-time cartography attempts present limitations in terms of spatial coverage. Some areas were not covered due to technical infeasibility, and temporal discrepancies could be found in
C. nodosa’s historic distribution datasets, with a few years’ time difference. These discrepancies, together with the high seasonal variability of the species [
17], make it particularly difficult to map its actual distribution accurately.
Previous attempts to model the distribution of
C. nodosa can also be found in the literature [
18], although at global scales, with coarser resolutions and failing to capture the intrinsic specificities of
C. nodosa in the archipelago. Species distribution models (SDM) rely on presence records (known localities where the species can be found) and a series of predictor variables describing the environmental conditions of the study area to find their statistical relation and predict species distributions in any given geographic area [
19]. As an alternative, some modeling techniques rely on absence data as well, depicting localities where the species is known to be absent. Nevertheless, due to the nature itself of these data, true absences are commonly unavailable [
20], and the generation of pseudo-absence is presented as an alternative [
21]. Pseudo-absence data allow the use of standard presence/absence analysis methods [
22] while improving model performance [
23]. There is no clear consensus on the best approach [
24]. Two common approaches for pseudo-absence generation are the random selection from a background dataset or the limiting to areas with different environmental characteristics [
25]. Another concern when selecting pseudo-absences is the prevalence of the species in the study area, or the ratio between presence and absence records, as the optimal ratio of presence data to pseudo-absence data is debated [
23,
26]. Among other alternative techniques for species distribution modeling, we can find ensemble models. This technique has recently gained popularity in SDM [
27,
28,
29,
30,
31,
32] and can improve model robustness [
33,
34] and predictive capabilities [
35].
Seagrasses produce ES of great socio-economic importance, providing food sources and playing climate regulation and coastal protection roles, as well as positively affecting tourist activities and helping climate change mitigation [
36,
37]. Nevertheless, they are often overlooked in comparison with other terrestrial ecosystems [
38], with an even more pronounced difference when considering ES mapping [
39].
Evaluations/analyses of ES supply related to seagrass have been carried out previously in the archipelago [
4,
40]. Following the general trend, these kinds of studies focused more on mainland Europe rather than on Outermost Regions of Europe (ORs), such as the Canary Islands. A general lack of spatially explicit assessments of island ecosystems and their services can be found, mainly related to a lack of data and research efforts [
41]. This particular bias leads to overlooking the socio-economic importance of ES supply [
42].
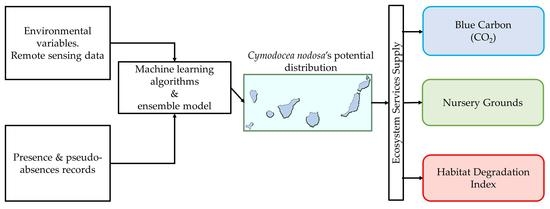
The main objective of this study was to explore the capabilities of different algorithms and modeling procedures to produce the potential distribution of C. nodosa in the Canarian Archipelago. The proposed methodology is presented as an alternative to cope with the lack of standardized cartographies of the coastal ecosystems and their ES supply, while relying on Remote Sensing (RS) products and images as an alternative to data-scarce scenarios. Finally, we propose using this potential distribution to estimate the ES supply and its economic valuation, producing spatially explicit assessments of the ecological and socioeconomic consequences of the impacts of human coastal activities, affecting this species distribution in the Canarian Archipelago.
4. Discussion
A distribution model of C. nodosa meadows was produced, being one of the first attempts to apply this methodology in the Canarian Archipelago. Based on the modeled potential distribution of this species, we assessed the ES provision while characterizing the potential impacts of human coastal activities.
On the one hand, the proposed methodology focused on the Canarian Archipelago, and better captured the species particularities in this geographic region than previous modeling attempts, on much broader scales [
18]. On the other hand, the modeled distribution allowed us to overcome the intrinsic limitations of the previous cartographies of
C. nodosa in terms of technical infeasibility and temporal discrepancies due to the species’ high seasonal variability [
17,
87].
Only presence records were available for the study, with no true absence records, and we relied on the generation of pseudo-absence data as an alternative. This kind of data may improve model performance [
23], while on the other hand allowing the use of standard analysis methods for presence/absence data [
19,
88]. The generation of pseudo-absence data is a critical process, as it dramatically affects the model’s performance [
26]. An unbalanced design with more pseudo-absences than presence records has been found to affect the performance of some models positively, and others negatively [
24]. Some studies suggest an optimal prevalence of 0.5 [
89], while others lower that ratio down to 0.1 [
90]. We considered different types of pseudo-absences and prevalence ratios to cope with this lack of a clear consensus, and the final model selection criteria were based entirely on model performance. For the present modeling procedure, an equilibrated ratio between presences and pseudo-absences was found to yield better results. In one out of three cases, 0.25 was the preferred prevalence, while the rest of the models were run with a prevalence of 1, feeding the narrative of a lack of consensus in this matter. In contrast to some studies highlighting worse performances for models run with aleatory pseudo-absences [
91], the majority of the selected models identified as statistically significant were constructed with randomly selected pseudo-absences. Nevertheless, these results are completely in concordance with findings in [
24], where better results were obtained when randomly selected pseudo-absences were used with regression modeling techniques. On the other hand, authors found better results when applying environmental-based criteria for pseudo-absence selection with classification techniques, such is the case for the selected RF model in this study, being the only model run with EPA pseudoabsence data.
The environmental variables correlation is another critical step in species distribution modeling, as it negatively affects the model’s performance. VIF analysis was used in addition to pairwise correlation, as it proved to highlight correlation sometimes missed by pairwise analyses [
92,
93]. The approach followed, combined with the construction of different sets of environmental variables, allowed us to minimize the information loss, a method recognized as capable of improving the model’s performance [
71] but still not commonly used in species distribution models [
94].
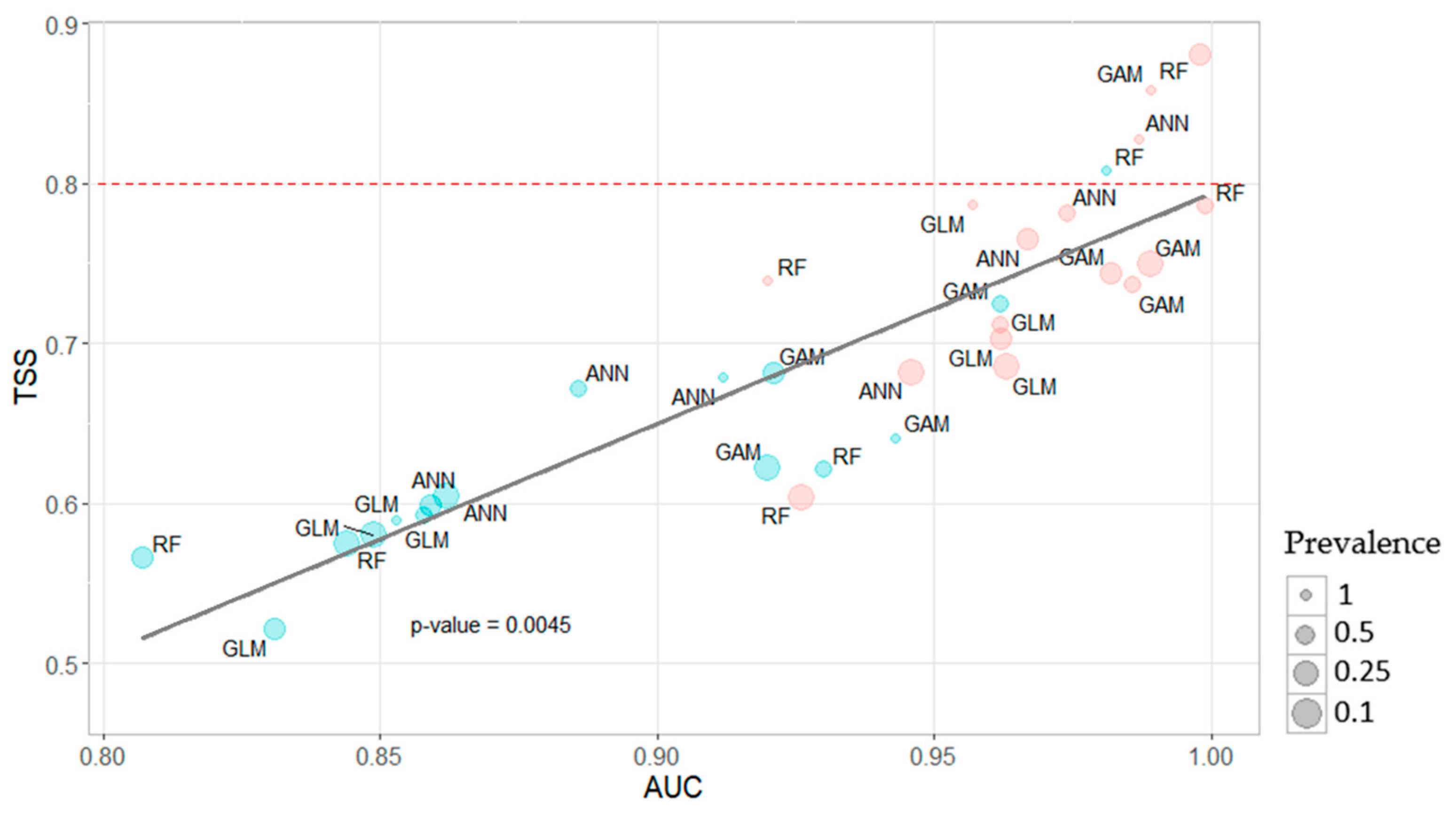
In general, evaluation metrics for MaxEnt models were very robust, and the selected model outperformed all the other candidates, showing very consistent values of AUC, AIC, and omission rates. The rest of the models based on pseudo-absence data presented robust AUC, specificity, and sensitivity, pointing to a balanced trade-off between commission and omission errors, while TSS was generally lower.
Traditional model evaluation approaches using metrics such as AUC have been criticized for not being a strong metric on its own [
95], so we considered a wide range of evaluation metrics, aiming for scrupulous filtering of candidate models, highlighting the role of dAIC as a strong metric [
96,
97], as well as omission rate [
98]. Two out of four selected models used RF algorithm, in concordance with recent findings [
99,
100], feeding the narrative that RF models presented especially good performances when applied to seagrass distribution models [
53].
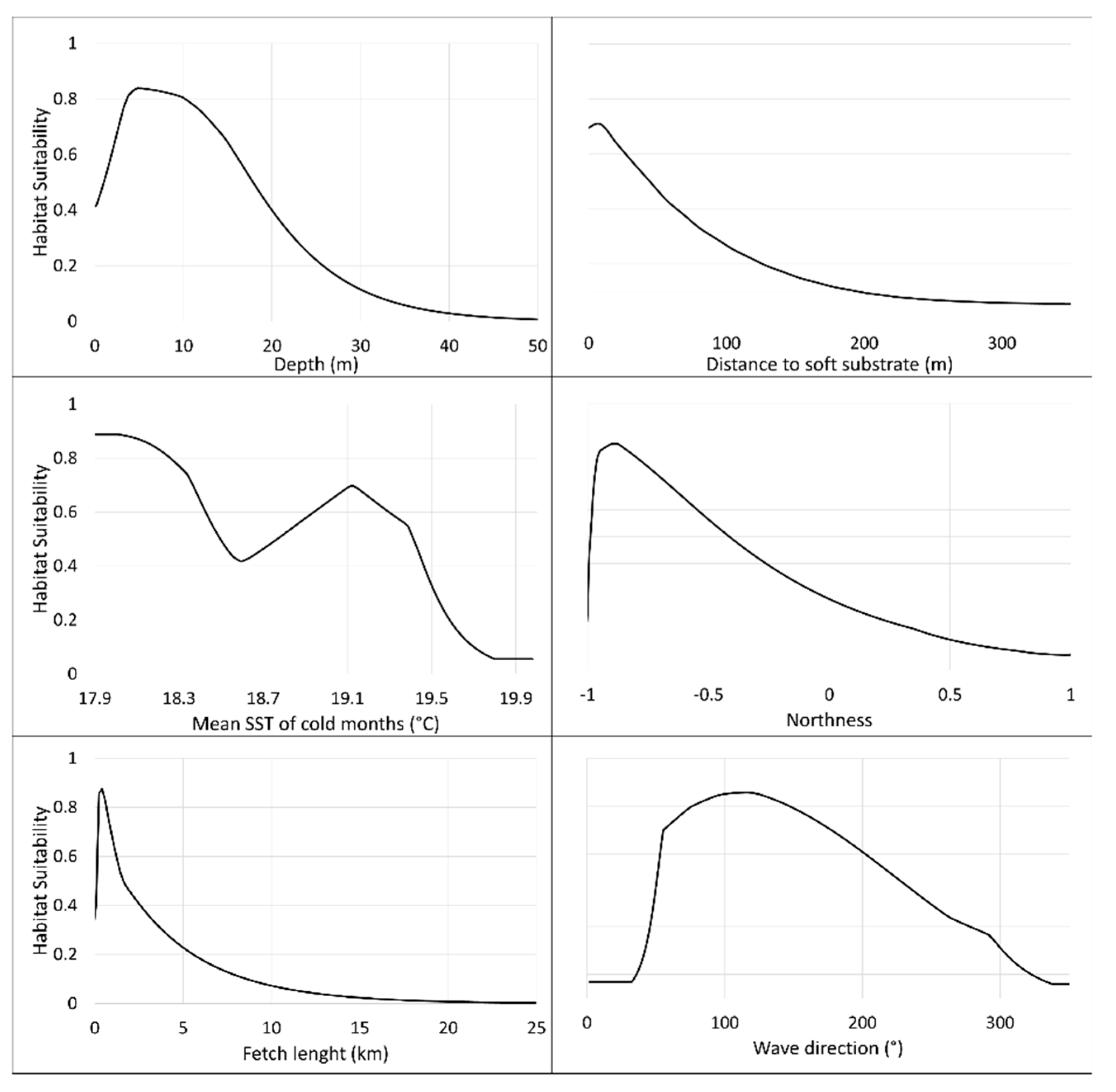
The results regarding predictors’ importance are robust and consistent in all selected models. In all cases, the same six environmental variables held more than 90% of models’ predictive capabilities (Depth, distance to the soft substrate, mean SST of cold months, northness, fetch length, and wave direction), although variable importance was generally more equilibrated for models run with pseudo-absence data.
Depth and fetch length shaped the potential distribution area for the species to shallow waters, relatively sheltered, while northness and wave direction delimited the potential suitable areas to the south-eastern coasts of the islands. The western region of the archipelago presents lower suitability for the species, as island geological age, following an east-to-west direction, is directly related to substrate availability. Variations in SST values also follow an east-to-west increasing trend, with differences between western and eastern regions up to 5 °C [
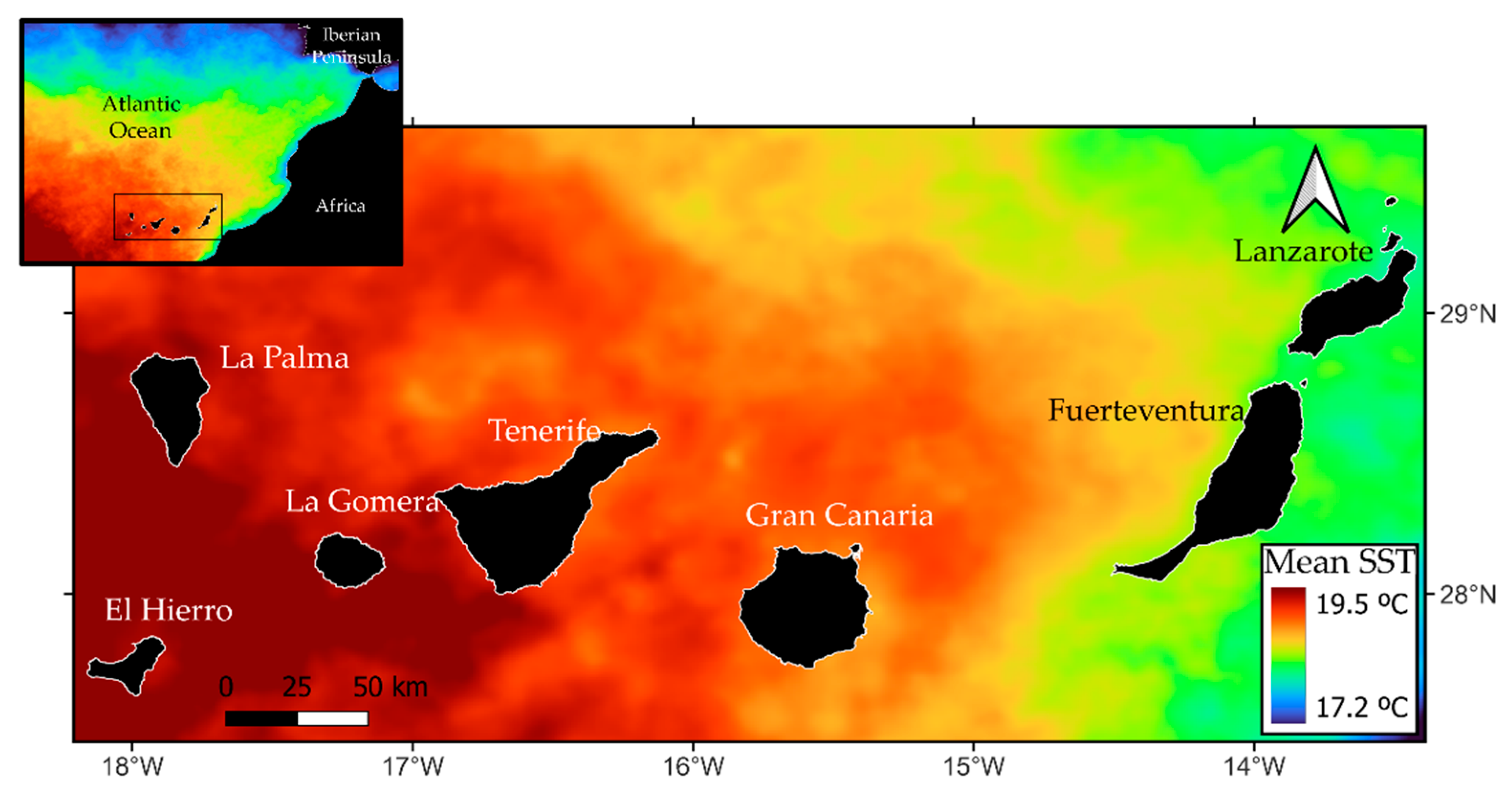
101], mainly due to the influence of the upwelling of the African coast [
45], which may cause differences in the longitudinal distribution of some species. The first peak of habitat suitability around lower values of SST could be explained by the proximity of the eastern islands to the aforementioned upwelling of the African coast. These islands present the lowest values of SST. The second peak of habitat suitability starts around 19 °C and decreases when values of SST tend to increase, as these higher values are found in the western regions of the archipelago. These results show that the SST does not seem to be a key variable for the survival of the species in the Canary Islands, and contrast with findings in [
18], in which SST winter optimal values for the establishment of the species seem to be lower (around 14 °C). Although it has been suggested that the global loss of meadows has been partly caused by climate change [
102], this phenomenon has mainly concerned Mediterranean meadows formed by Posidonia and Zostera species. Caution is advised when comparing the results between models, as different study scales are known to lead to different variable importance [
73,
103]. There is still no evidence that it affects
C. nodosa in the same way, and it has even been suggested that
C. nodosa may even benefit from global warming due to its affinity for warm waters [
104]. Nevertheless, there are other mechanisms linked to climate change, rather than temperature, that may have a significant impact on
C. nodosa meadows, such as the increasing frequency and intensity of extreme weather events.
The consistency and robustness of the different models, regarding variables’ importance and predictive capabilities, allowed for the construction of an ensemble model. It is known to have the potential of improving model robustness over attempts using single algorithms [
33,
34], while enhancing our model’s prediction confidence [
35]. Even though the ensemble model was constructed with multiple base models, it performed as a single species distribution model.
A total of 14,047 ha were identified as potentially suitable for the species, in contrast with the 6709 ha in the Canary islands occupied by
C. nodosa meadows as stated in the Marine Phanerogams Atlas from Spain [
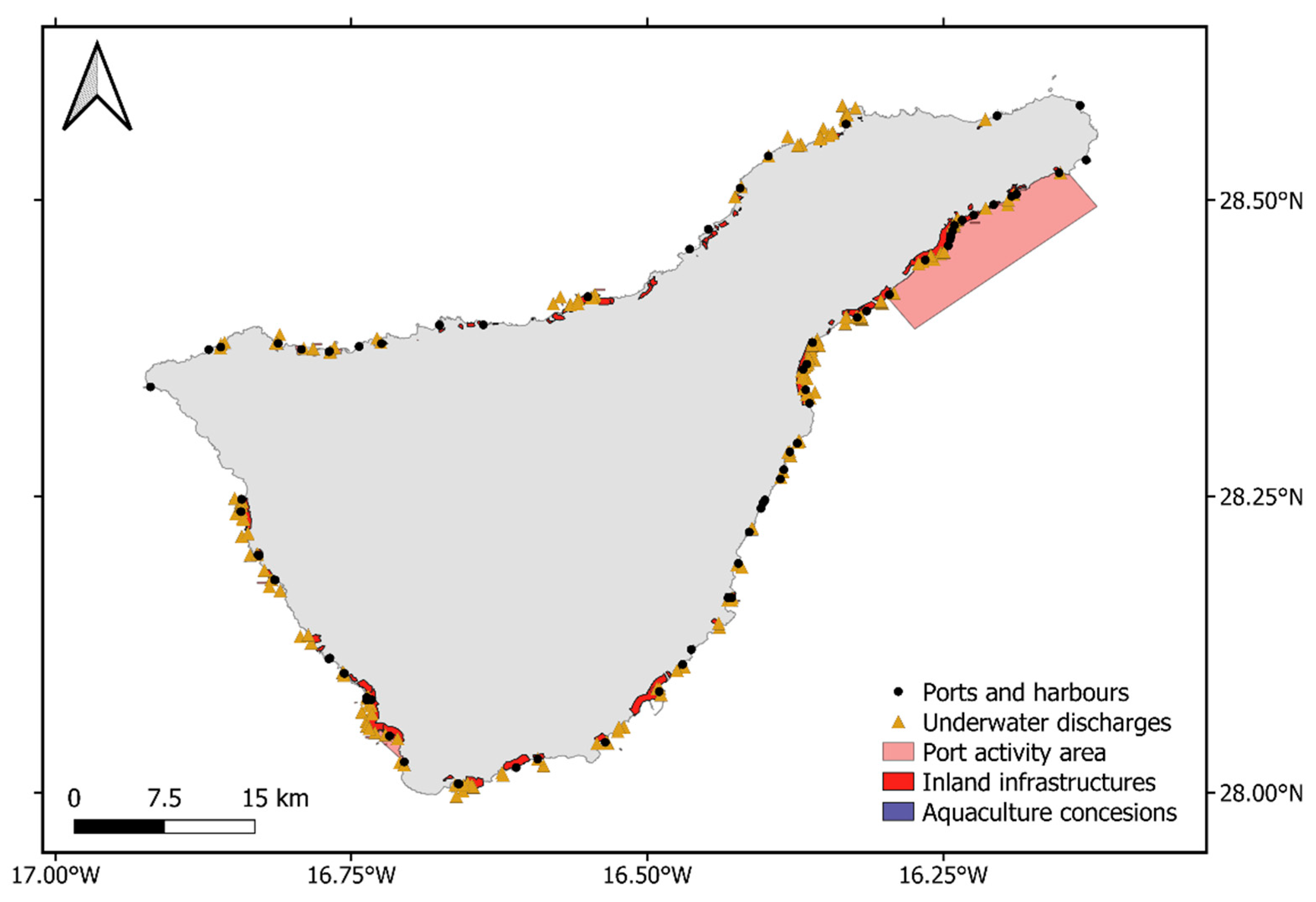
105]. This is an expected discrepancy as these 14,047 ha refer to potential areas for the species to establish, considering no human presence whatsoever. Coastal infrastructures such as ports are usually placed in naturally sheltered areas, also preferred for the species to establish, and habitat suitability in areas surrounding these infrastructures was especially jeopardized. After the degradation index was applied, the total suitable area decreased to 9131 ha, much closer to the values found in the Marine Phanerogams Atlas from Spain. It is likely that the coastal activities dataset lacked information about human impacts influencing the species distribution, mainly due to data availability, but it still allowed us to better comprehend the scope of the potential habitat loss due to these kinds of coastal impacts.
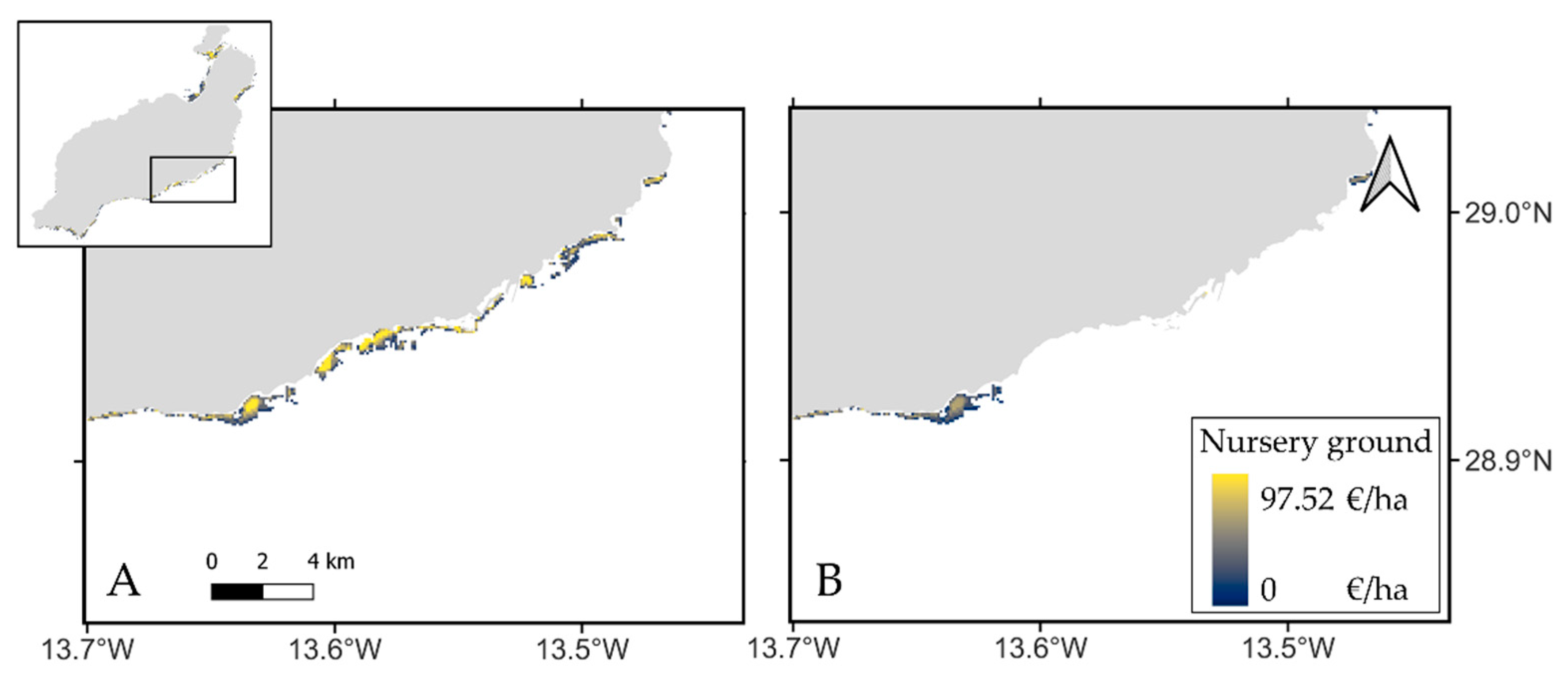
This study represents the first attempt to estimate the monetary value of both Blue carbon and Nursery grounds at an archipelago scale in the Canary Islands. For the species studied, we estimated the monetary value for
C. nodosa’s role as nursery grounds at 97.52 €*ha
−1, a slightly higher value compared to previous studies (89.13 €*ha
−1) [
4]. This value was then estimated at 1,369,863 € for the whole archipelago considering a pristine scenario, and 890,455 € after the degradation index was applied. The economic estimation of
C. nodosa’s role as nursery grounds is extremely difficult to compare with the known real economic valuation of coastal fisheries (
https://www.gobiernodecanarias.org/agp/sgt/galerias/doc/estadisticas/pesca/2007_2021-especie_meses-valor.ods, accessed on 4 July 2022). These published values relate to the economic market profit of the extracted fish, which represent a small fraction of the whole fish population. The economic estimation presented in this study relates to the total potential economic value of the whole fish stocks that use
C. nodosa as nursery ground, and both assessments refer to different Ecosystem Services. Nevertheless, the value of this work lies especially in highlighting the economic loss of ES for
Cymodocea meadows in the Canary Islands as a whole, which is a result to be taken into account in future decision-making for the management of these communities.
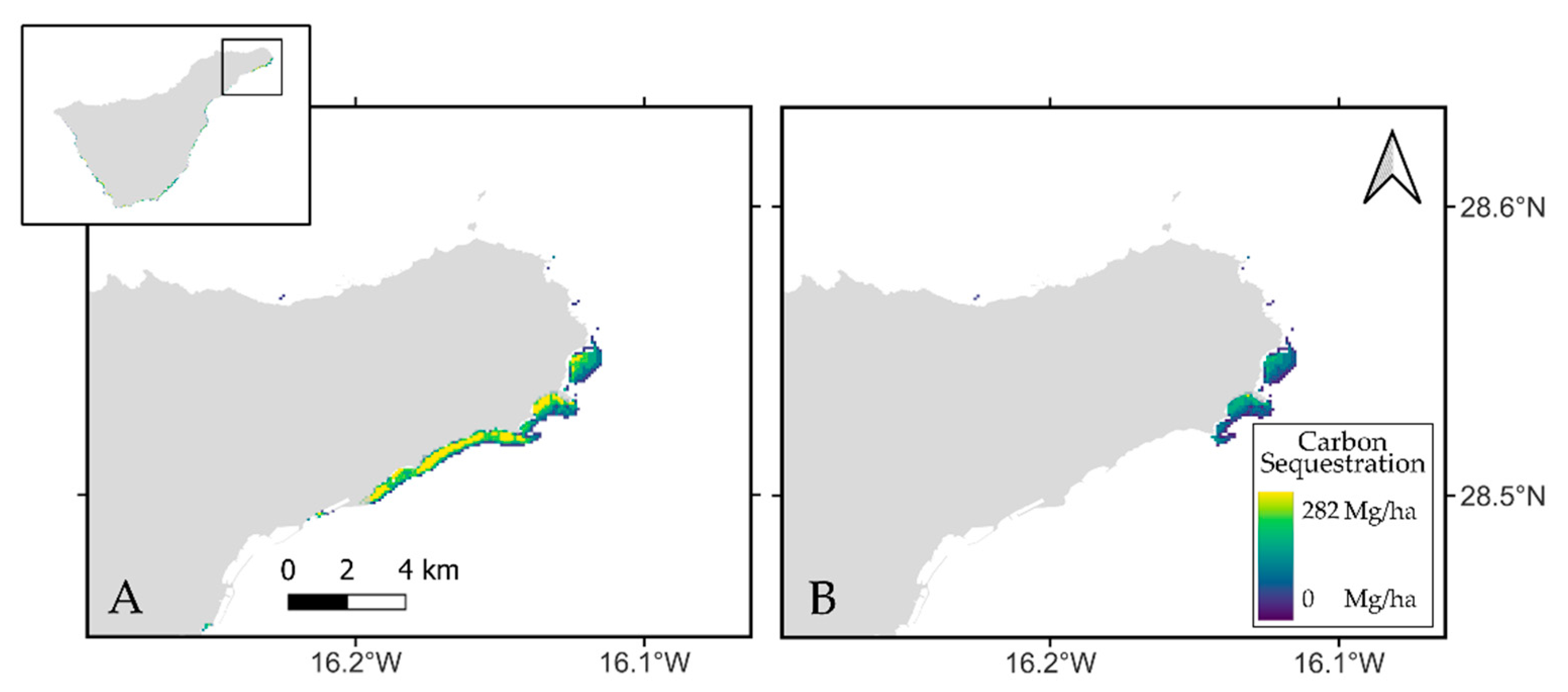
The Blue Carbon estimations presented in this study are very consistent with previous studies using similar methodologies [
40], where they assessed the carbon stock related to this species in 3,173,469 Mg. Our estimations resulted in 3,961,254 Mg CO
2 and this stock was then valued at 192,715,007€. When considering a degraded scenario, these values dropped down to 2,574,942 Mg CO
2, resulting in an economic assessment of 125,270,928€. Overall, studies assessing the monetary social value of carbon stocks in different regions and other seagrass species, present certain discrepancies. ES economic values carried out for the Spanish territory [
40] are lower than for the European region [
106], which is probably related to the use of different carbon prices, induced by carbon price instability over time and the regional particularities of the species studied.
5. Conclusions
The analysis of the input parameters of the modeling, as well as the application of different algorithms, allowed us to obtain a reliable and representative distribution model of the potential habitat of Cymodocea nodosa. This model provides us with valuable information not only about its distribution in pristine conditions but also about the relationship of the species with the environmental variables analyzed.
At the same time, the distribution model allowed us to quantify the impact of human activities on this species and its ES provision. Our results, although based only on two ES, demonstrate its great socio-economic importance, setting precedents and highlighting the need for further studies of this kind.
The degraded habitat produced in this study, although approaching a realistic scenario for the distribution of the species in the archipelago, needs to be further explored. Many variables describing other types of human activities jeopardizing the distribution of C. nodosa have not been taken into account and should be taken into consideration for future studies. However, the proposed methodology helps us to understand in greater depth the capabilities that this type of analysis offers.
This methodology allowed us to observe the decrease in habitat suitability in a spatially explicit way, being a very useful tool for policy decision support, which has been overlooked, especially in European ORs. One of the reasons for the lack of studies that consider the spatially explicit dimension of ES assessments, as well as the geographical distribution of the different human impacts on the coast, is the availability of data. In this sense, the use of RS products and imagery is presented as a valuable tool to overcome this limitation.