Cuscuta spp. Segmentation Based on Unmanned Aerial Vehicles (UAVs) and Orthomasaics Using a U-Net Xception-Style Model
Abstract
1. Introduction
2. Materials and Methods
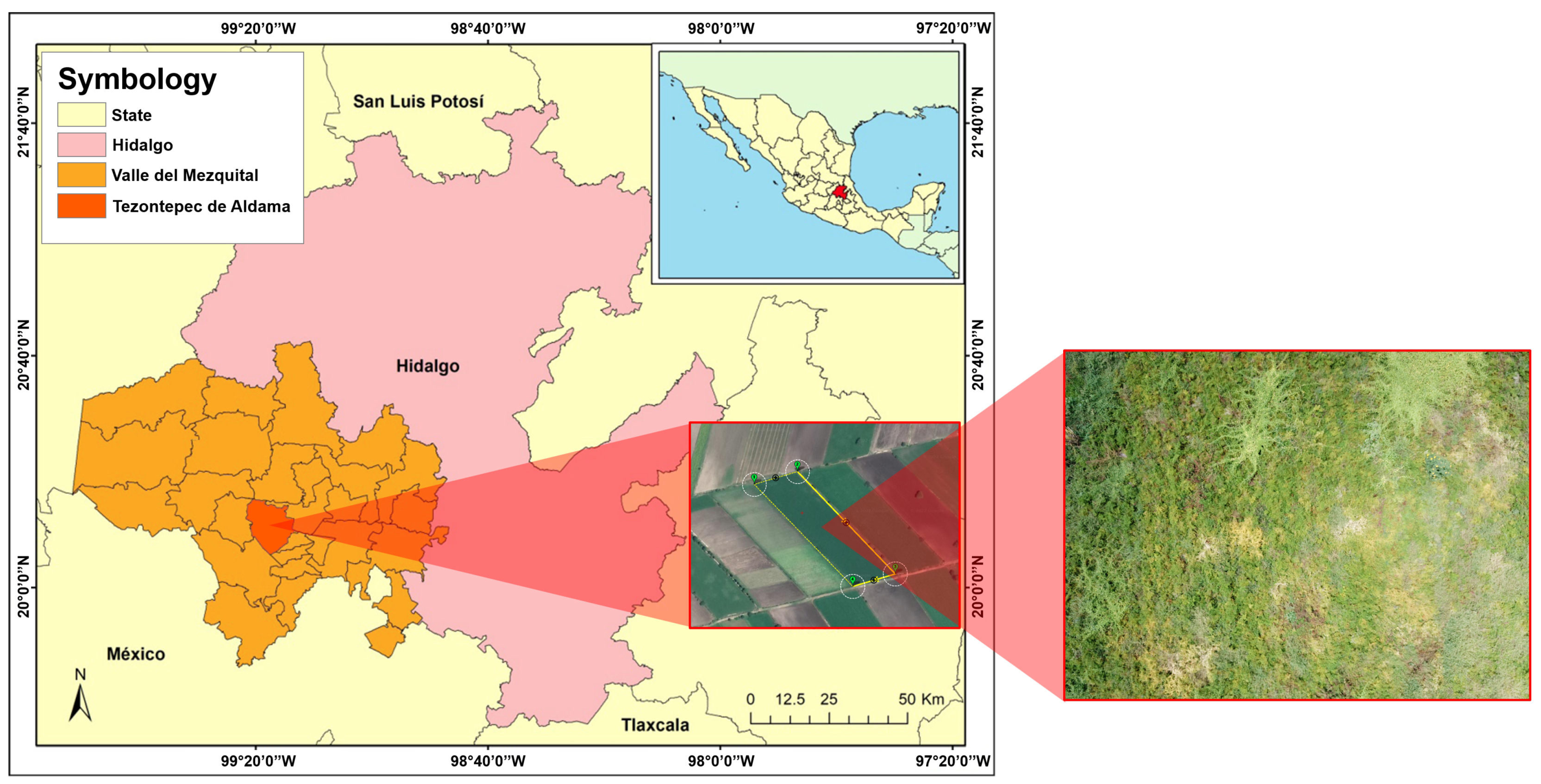
2.1. Study Area
2.2. Data Acquisition
2.3. Generation of Datasets
2.4. Orthomosaic
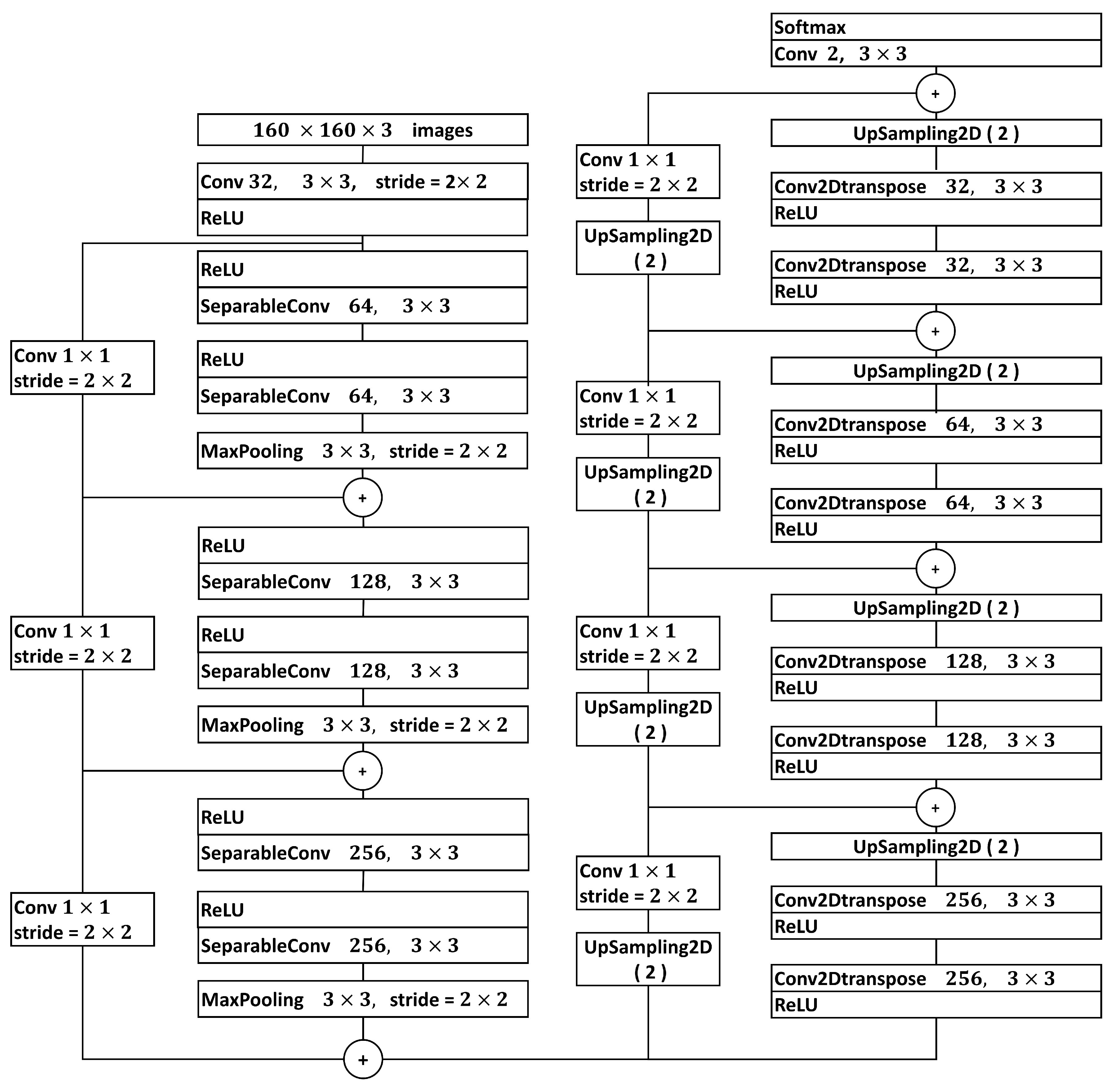
2.5. Image Segmentation with U-Net Xception-Style
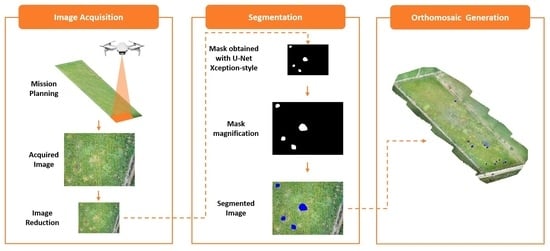
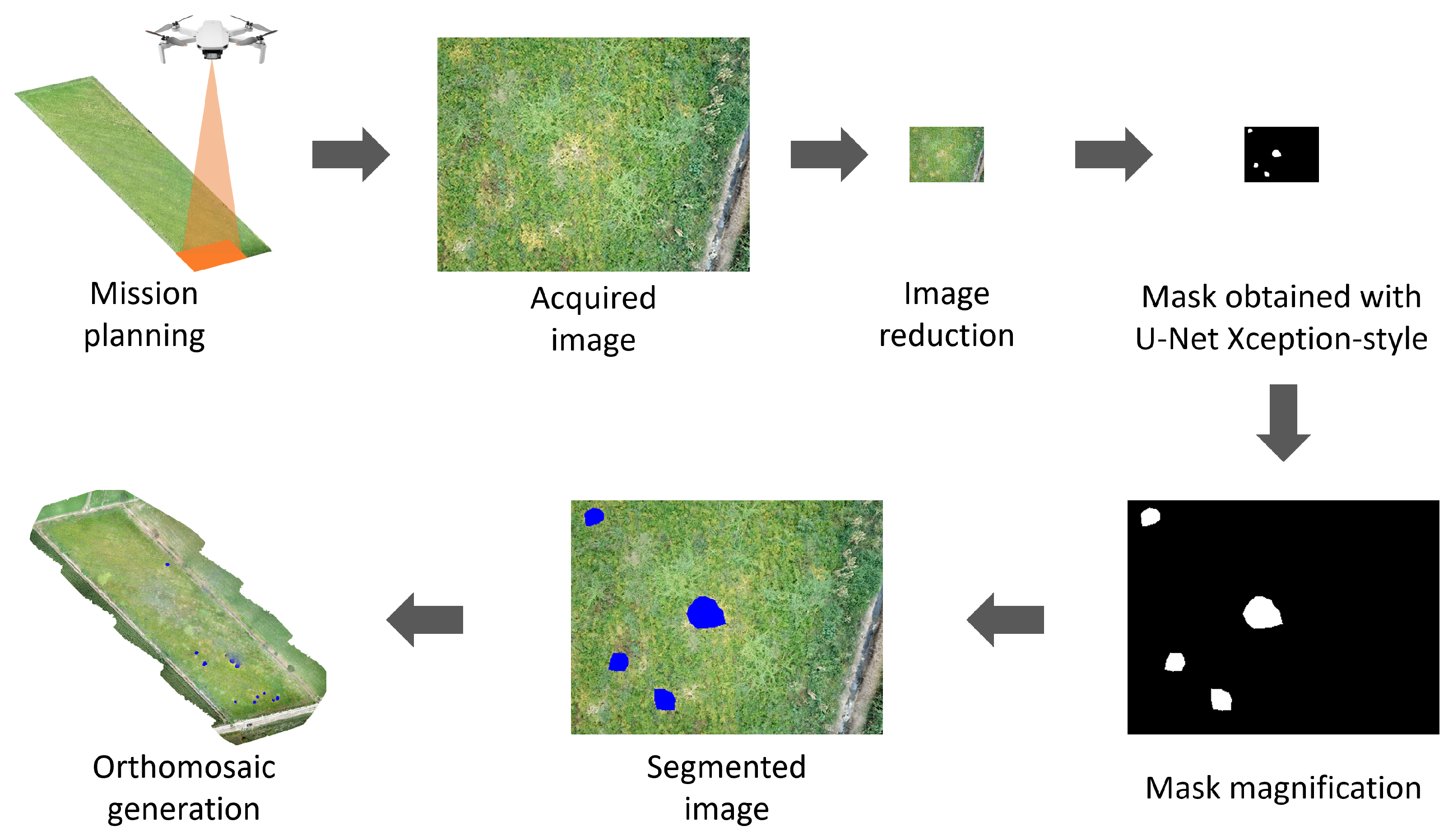
2.6. Methods
- Image acquisition by the UAV.
- Image size reduction.
- Obtaining a mask with the U-Net Xception-style model.
- Increasing the size of the mask.
- Segmenting the infected areas in blue.
- Generation of an orthomosaic with the set of images.
3. Results
3.1. U-Net Xception-Style Training
3.2. Model Evaluation
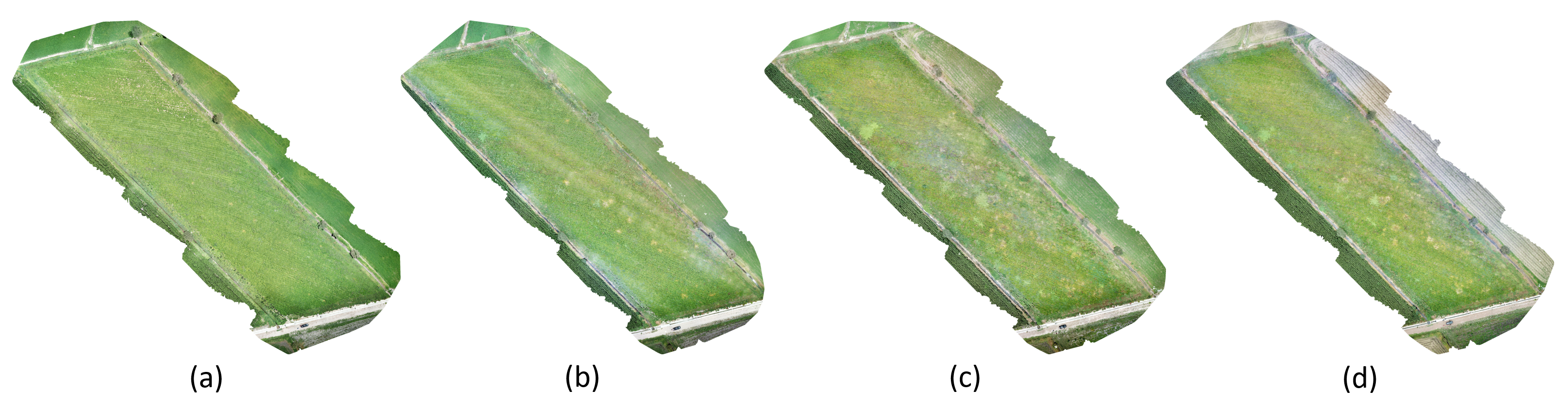
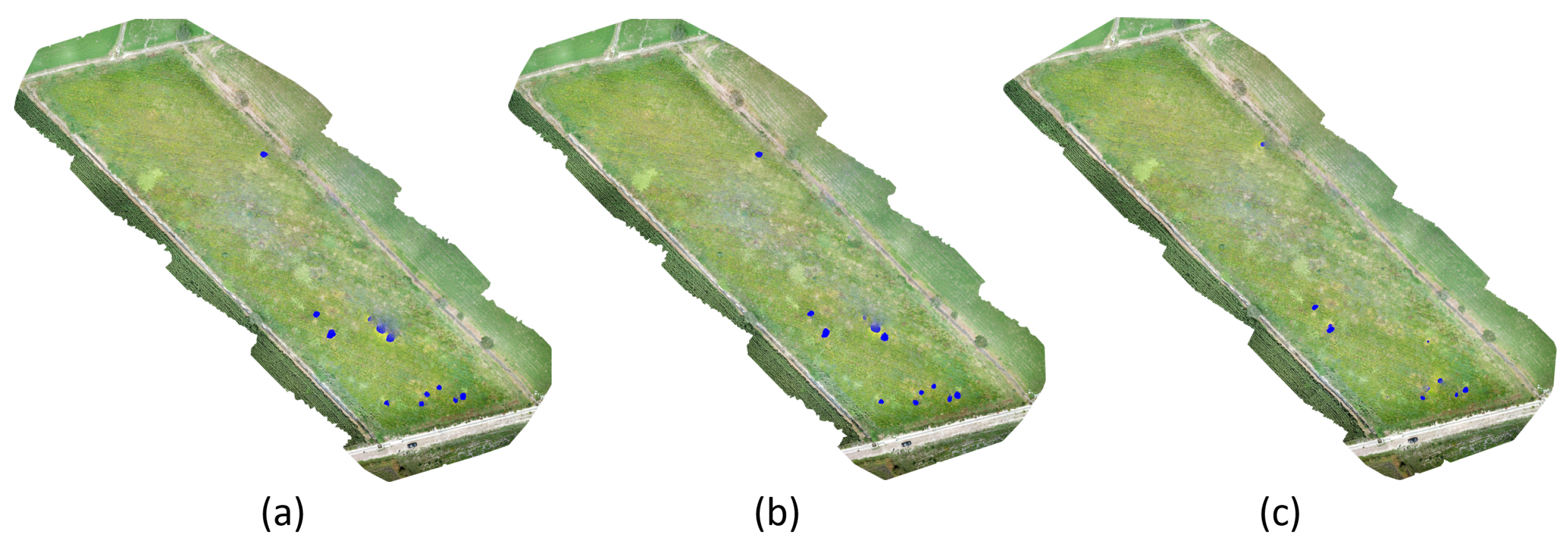
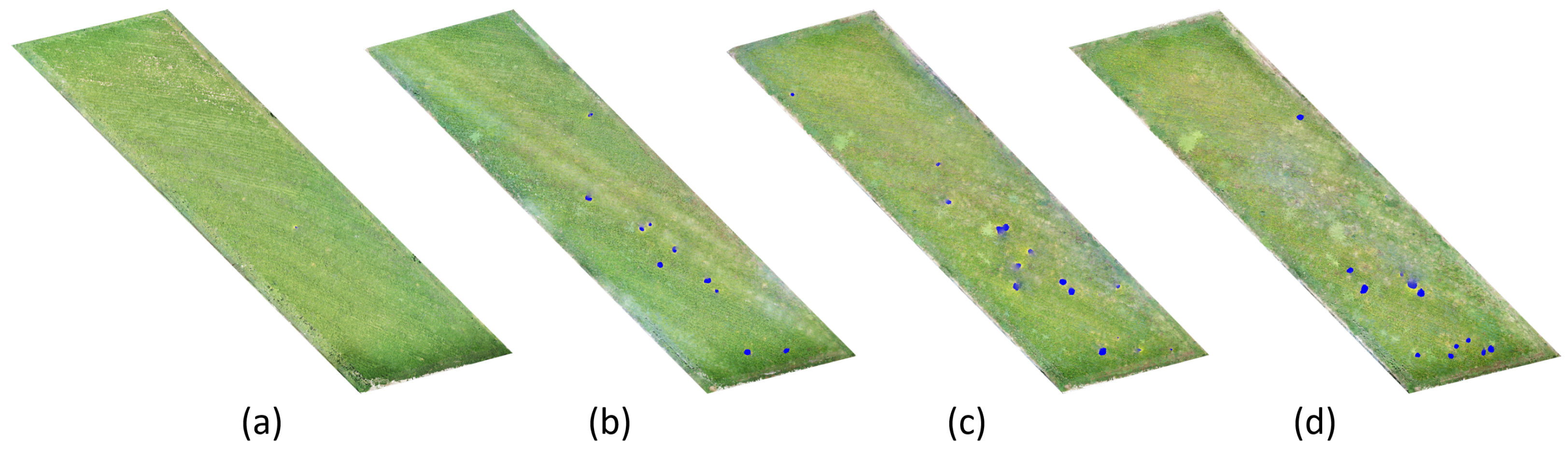
3.3. Cuscuta spp. Segmentation in the Cultivation of Arbol Peppers
4. Discussion
5. Conclusions
- The analysis considered a single arbol pepper crop, which caused the overfitting of the trained model.
- There is still no exact quantification of Cuscuta spp. that allows an objective comparison.
- There was no substantial change in the prediction times by decreasing the size of the input images; this only significantly reduced the training times.
- Due to its characteristic yellow color, the proposed model was exclusively adapted for Cuscuta spp.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| UAV | Unmanned Aerial Vehicle |
| MIoU | Mean Intersection-over-Union |
| CNN | Convolutional Neural Networks |
| FOV | Field-of-View |
| GPS | Global Positioning System |
| GCS | Ground Control System |
| DEM | Digital Elevation Model |
| IoU | Intersection-over-Union |
References
- Costea, M.; García-Ruiz, I.; Dockstader, K.; Stefanović, S. More problems despite bigger flowers: Systematics of Cuscuta tinctoria clade (subgenus Grammica, Convolvulaceae) with description of six new species. Syst. Bot. 2013, 4, 1160–1187. [Google Scholar] [CrossRef]
- CABI. Datasheets Cuscuta. 2022. Available online: https://www.cabi.org/isc/search/index?q=cuscuta (accessed on 19 July 2022).
- Costea, M.; García, M.A.; Stefanović, S. A phylogenetically based infrageneric classification of the parasitic plant genus Cuscuta (Dodder, Convolvulaceae). Syst. Bot. 2015, 1, 269–285. [Google Scholar] [CrossRef]
- Ahmadi, K.; Omidi, H.; Dehaghi, M.A. A Review on the Botanical, Phytochemical and Pharmacological Characteristics of Cuscuta spp. In Parasitic Plants; IntechOpen: London, UK, 2022. [Google Scholar]
- Le, Q.V.; Tennakoon, K.U.; Metali, F.; Lim, L.B.; Bolin, J.F. Impact of Cuscuta australis infection on the photosynthesis of the invasive host, Mikania micrantha, under drought condition. Weed Biol. Manag. 2015, 15, 138–146. [Google Scholar]
- Stefanović, S.; Kuzmina, M.; Costea, M. Delimitation of major lineages within Cuscuta subgenus Grammica (convolvulaceae) using plastid and nuclear DNA sequences. Am. J. Bot. 2007, 4, 568–589. [Google Scholar]
- Iqbal, M.; Hussain, M.; Abid, A.; Ali, M.; Nawaz, R.; Qaqar, M.; Asghar, M.; Iqbal, Z. A review: Cuscuta (Cuscuta planifora) major weed threat in Punjab–Pakistan. Int. J. Adv. Res. Biol. Sci. 2014, 4, 42–46. [Google Scholar]
- Kogan, M.; Lanini, W. Biology and management of Cuscuta in crops. Cienc. E Investig. Agrar. Rev. Latinoam. Cienc. Agric. 2005, 32, 165–180. [Google Scholar]
- Dawson, J.H.; Musselman, L.; Wolswinkel, P.; Dörr, I. Biology and control of Cuscuta. Rev. Weed Sci. 1994, 6, 265–317. [Google Scholar]
- Carranza, E. Flora del Bajío y de Regiones Adyacentes; Instituto de Ecología: Mexico City, Mexico, 2008; Volume 155. [Google Scholar]
- Ríos, V.; Luis, J.; García, E. Catálogo de Malezas de México; Fondo de Cultura Económico: Mexico City, Mexico, 1998. [Google Scholar]
- Aly, R.; Dubey, N.K. Weed management for parasitic weeds. In Recent Advances in Weed Management; Springer: Berlin/Heidelberg, Germany, 2014; pp. 315–345. [Google Scholar]
- Kannan, C.; Kumar, B.; Aditi, P.; Gharde, Y. Effect of native Trichoderma viride and Pseudomonas fluorescens on the development of Cuscuta campestris on chickpea, Cicer arietinum. J. Appl. Nat. Sci. 2014, 2, 844–851. [Google Scholar] [CrossRef][Green Version]
- Mishra, J. Biology and management of Cuscuta species. Indian J. Weed Sci. 2009, 41, 1–11. [Google Scholar]
- Hazaymeh, K.; Sahwan, W.; Al Shogoor, S.; Schütt, B. A Remote Sensing-Based Analysis of the Impact of Syrian Crisis on Agricultural Land Abandonment in Yarmouk River Basin. Sensors 2022, 22, 3931. [Google Scholar]
- Chen, M.; Tang, Y.; Zou, X.; Huang, K.; Li, L.; He, Y. High-accuracy multi-camera reconstruction enhanced by adaptive point cloud correction algorithm. Opt. Lasers Eng. 2019, 122, 170–183. [Google Scholar] [CrossRef]
- Wu, F.; Duan, J.; Chen, S.; Ye, Y.; Ai, P.; Yang, Z. Multi-target recognition of bananas and automatic positioning for the inflorescence axis cutting point. Front. Plant Sci. 2021, 12, 705021. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lin, Y.; Xu, X.; Chen, Z.; Wu, Z.; Tang, Y. A Study on Long-Close Distance Coordination Control Strategy for Litchi Picking. Agronomy 2022, 12, 1520. [Google Scholar] [CrossRef]
- Yu, J.; Sharpe, S.M.; Schumann, A.W.; Boyd, N.S. Deep learning for image-based weed detection in turfgrass. Eur. J. Agron. 2019, 104, 78–84. [Google Scholar] [CrossRef]
- You, J.; Liu, W.; Lee, J. A DNN-based semantic segmentation for detecting weed and crop. Comput. Electron. Agric. 2020, 178, 105750. [Google Scholar] [CrossRef]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A Review on UAV-Based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Selvi, C.T.; Subramanian, R.S.; Ramachandran, R. Weed Detection in Agricultural fields using Deep Learning Process. In Proceedings of the 2021 7th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 19–20 March 2021; Volume 1, pp. 1470–1473. [Google Scholar]
- Abouzahir, S.; Sadik, M.; Sabir, E. Lightweight Computer Vision System for Automated Weed Mapping. In Proceedings of the 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), Virtual, 26–29 January 2022. [Google Scholar]
- Neupane, K.; Baysal-Gurel, F. Automatic Identification and Monitoring of Plant Diseases Using Unmanned Aerial Vehicles: A Review. Remote Sens. 2021, 13, 3841. [Google Scholar] [CrossRef]
- Ahmadi, P.; Mansor, S.; Farjad, B.; Ghaderpour, E. Unmanned Aerial Vehicle (UAV)-Based Remote Sensing for Early-Stage Detection of Ganoderma. Remote Sens. 2022, 14, 1239. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.; Gloor, E.; Phillips, O.L.; Aragao, L.E. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef]
- Reder, S.; Mund, J.P.; Albert, N.; Waßermann, L.; Miranda, L. Detection of Windthrown Tree Stems on UAV-Orthomosaics Using U-Net Convolutional Networks. Remote Sens. 2021, 14, 75. [Google Scholar] [CrossRef]
- Yao, X.; Yang, H.; Wu, Y.; Wu, P.; Wang, B.; Zhou, X.; Wang, S. Land use classification of the deep convolutional neural network method reducing the loss of spatial features. Sensors 2019, 19, 2792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ke, Y.; Zhang, Z.; Wang, M.; Li, P.; Zhang, S. Urban land use and land cover classification using novel deep learning models based on high spatial resolution satellite imagery. Sensors 2018, 18, 3717. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, W.; Yang, L.; Sun, S.; Hu, W.; Zhang, F.; Li, W. DeepUNet: A deep fully convolutional network for pixel-level sea-land segmentation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3954–3962. [Google Scholar] [CrossRef]
- Giang, T.L.; Dang, K.B.; Le, Q.T.; Nguyen, V.G.; Tong, S.S.; Pham, V.M. U-Net convolutional networks for mining land cover classification based on high-resolution UAV imagery. IEEE Access 2020, 8, 186257–186273. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Wada, K. Labelme: Image Polygonal Annotation with Python. 2016. Available online: https://github.com/wkentaro/labelme (accessed on 19 July 2022).
- Cheng, Y.; Xue, D.; Li, Y. A fast mosaic approach for remote sensing images. In Proceedings of the 2007 International Conference on Mechatronics and Automation, Harbin, China, 5–8 August 2007; pp. 2009–2013. [Google Scholar]
- Lam, O.H.Y.; Dogotari, M.; Prüm, M.; Vithlani, H.N.; Roers, C.; Melville, B.; Zimmer, F.; Becker, R. An open source workflow for weed mapping in native grassland using unmanned aerial vehicle: Using Rumex obtusifolius as a case study. Eur. J. Remote Sens. 2021, 54, 71–88. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Chollet, F. Image Segmentation with a U-Net-Like Architecture. 2019. Available online: https://keras.io/examples/vision/oxford_pets_image_segmentation (accessed on 19 July 2022).
- Winston, R.; Schwarzländer, M.; Hinz, H.L.; Day, M.D.; Cock, M.J.; Julien, M.; Julien, M.H. Biological Control of Weeds: A World Catalogue of Agents and Their Target Weeds; USDA Forest Service, Forest Health Technology Enterprise Team: Morgantown, WV, USA, 2014.
| Original Image | Flip | Rotate | Flip |
|---|---|---|---|
 |  |  |  |
 |  |  |  |
 |  |  |  |
| Size | Training Time | Loss | Accuracy |
|---|---|---|---|
| 2 s 43 ms/step | 0.0375 | 98.87% | |
| 3 s 58 ms/step | 0.0298 | 98.99% | |
| 4 s 83 ms/step | 0.026 | 99.02% | |
| 5 s 122 ms/step | 0.024 | 99.10% | |
| 7 s 158 ms/step | 0.0242 | 99.10% | |
| 9 s 204 ms/step | 0.0249 | 99.10% | |
| 11 s 260 ms/step | 0.026 | 99.13% | |
| 15 s 340 ms/step | 0.0225 | 99.18% | |
| 19 s 433 ms/step | 0.0264 | 99.17% | |
| 23 s 533 ms/step | 0.0257 | 99.13% |
| Size | Time for U-Net Xception-Style | Time for DeepLabV3+ | MIoU for U-Net Xception-Style | MIoU for DeepLabV3+ |
|---|---|---|---|---|
| 0.03861 s | 0.04080 s | 47.67% | 42.92% | |
| 0.03910 s | 0.04145 s | 61.27% | 35.43% | |
| 0.03959 s | 0.04249 s | 56.11% | 47.84% | |
| 0.04038 s | 0.04453 s | 63.59% | 39.65% | |
| 0.04000 s | 0.04505 s | 60.22% | 43.83% | |
| 0.04075 s | 0.04667 s | 57.48% | 35.71% | |
| 0.04114 s | 0.04667 s | 52.34% | 42.01% | |
| 0.04261 s | 0.04961 s | 59.56% | 56.23% | |
| 0.04371 s | 0.05149 s | 71.20% | 52.07% | |
| 0.04505 s | 0.05343 s | 60.28% | 44.77% |
| Date | Pixels | Cuscuta spp. (m) | Cuscuta spp. (%) |
|---|---|---|---|
| 14 July 2021 | 182 | 0.46 m | 0.003% |
| 8 August 2021 | 17,741 | 44.35 m | 0.303% |
| 29 August 2021 | 28,376 | 70.94 m | 0.485% |
| 11 September 2021 | 33,218 | 83.05 m | 0.568% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez-Lazcano, L.; Camacho-Bello, C.J.; Cornejo-Velazquez, E.; Arroyo-Núñez, J.H.; Clavel-Maqueda, M. Cuscuta spp. Segmentation Based on Unmanned Aerial Vehicles (UAVs) and Orthomasaics Using a U-Net Xception-Style Model. Remote Sens. 2022, 14, 4315. https://doi.org/10.3390/rs14174315
Gutiérrez-Lazcano L, Camacho-Bello CJ, Cornejo-Velazquez E, Arroyo-Núñez JH, Clavel-Maqueda M. Cuscuta spp. Segmentation Based on Unmanned Aerial Vehicles (UAVs) and Orthomasaics Using a U-Net Xception-Style Model. Remote Sensing. 2022; 14(17):4315. https://doi.org/10.3390/rs14174315
Chicago/Turabian StyleGutiérrez-Lazcano, Lucia, César J. Camacho-Bello, Eduardo Cornejo-Velazquez, José Humberto Arroyo-Núñez, and Mireya Clavel-Maqueda. 2022. "Cuscuta spp. Segmentation Based on Unmanned Aerial Vehicles (UAVs) and Orthomasaics Using a U-Net Xception-Style Model" Remote Sensing 14, no. 17: 4315. https://doi.org/10.3390/rs14174315
APA StyleGutiérrez-Lazcano, L., Camacho-Bello, C. J., Cornejo-Velazquez, E., Arroyo-Núñez, J. H., & Clavel-Maqueda, M. (2022). Cuscuta spp. Segmentation Based on Unmanned Aerial Vehicles (UAVs) and Orthomasaics Using a U-Net Xception-Style Model. Remote Sensing, 14(17), 4315. https://doi.org/10.3390/rs14174315