Fusion of Multidimensional CNN and Handcrafted Features for Small-Sample Hyperspectral Image Classification
Abstract
:1. Introduction
- (1)
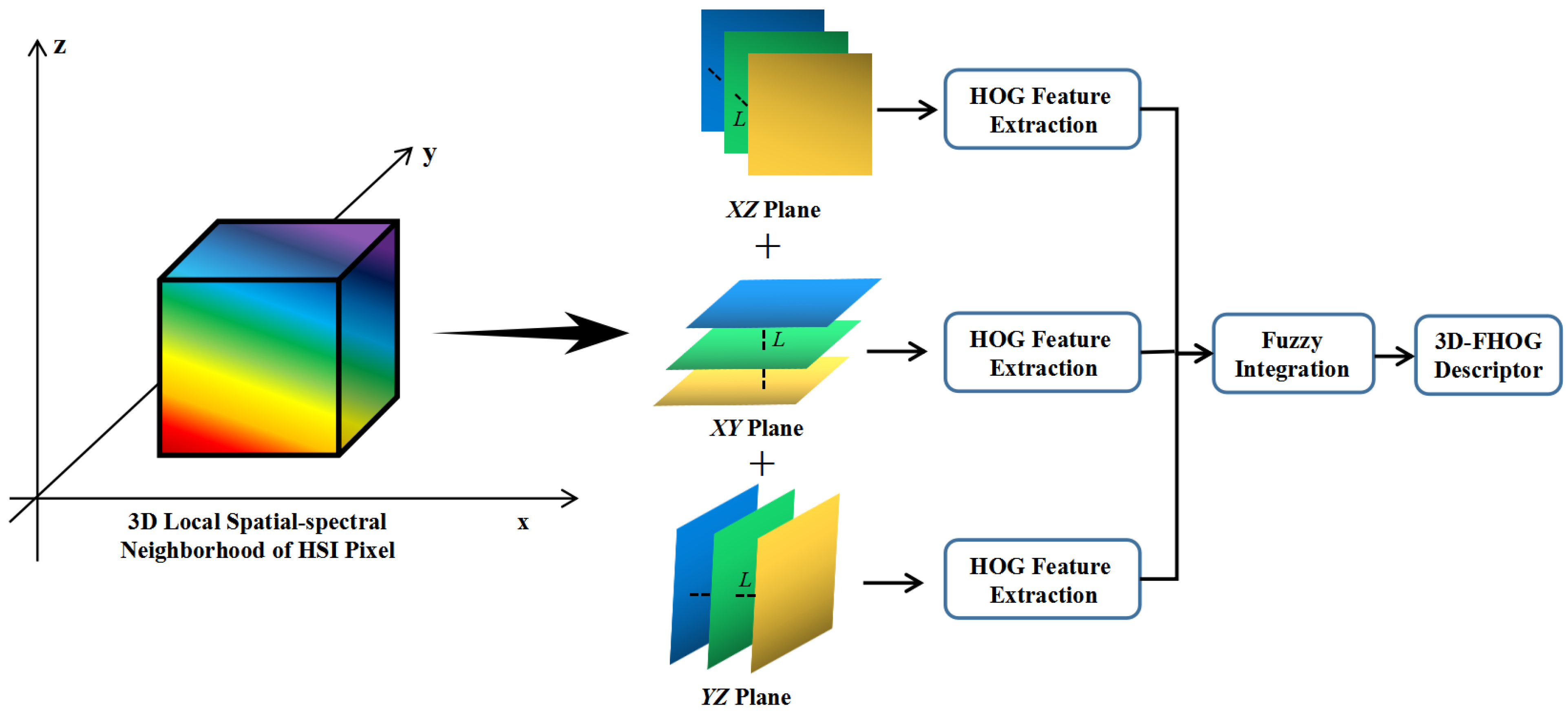
- A 3D-FHOG descriptor is proposed to fully extract the handcrafted spatial–spectral feature of HSI pixels. It calculates the HOG features from three orthogonal planes to generate the final 3D-FHOG descriptor based on fuzzy fusion operation, which is able to overcome the local spatial–spectral feature uncertainty;
- (2)
- An effective Siamese network, i.e., MDSN is designed for further exploiting the multidimensional CNN-based spatial–spectral feature in the scenery of small-scale labeled samples. It mainly utilizes the hybrid 3D-2D-1D CNN to learn the spatial–spectral feature from multiple dimensions and is updated by minimizing both contrastive loss and classification loss. Compared with the single-dimensional CNN-based networks, the performance of MDSN is significantly better in small-sample HSI classification;
- (3)
- It provides a novel extensible fusion framework for the combination of hand- crafted and multidimensional CNN-based spatial–spectral features. More importantly, experimental results indicate that our proposed MDSN combined with 3D-FHOG features can achieve better performance than the handcrafted features-based and CNN-based algorithms, which in turn verifies the superiority of the proposed fusion framework.
2. Related Works
2.1. Histogram of Oriented Gradients
2.2. Siamese Network
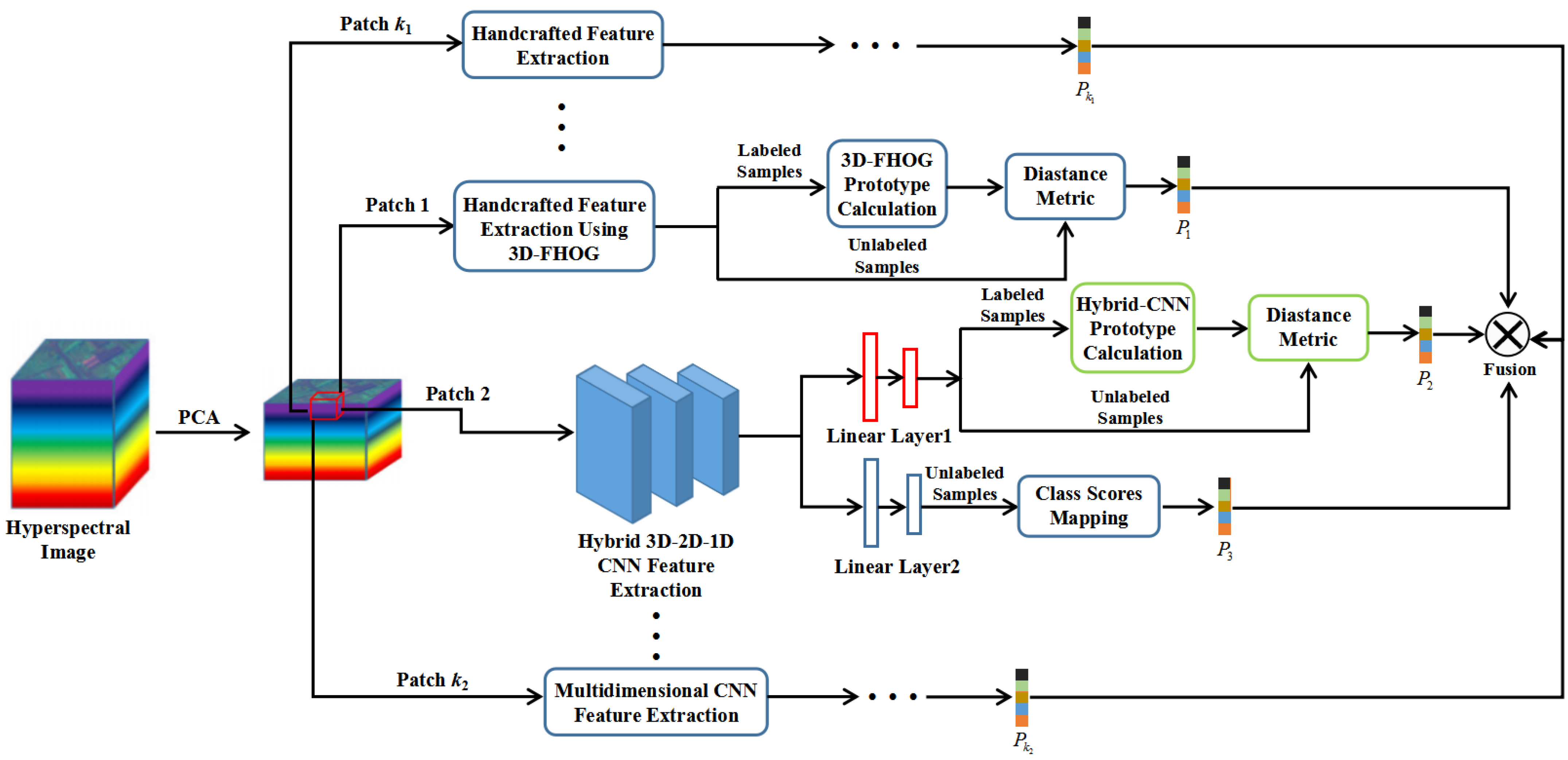
3. Methodology
3.1. The Proposed 3D-FHOG
3.2. The Proposed MDSN
3.3. MDSN Combined with 3D-FHOG for Small-Sample HSI Classification
| Algorithm 1. MDSN combined with 3D-FHOG for small-sample HSI classification |
| Input: HSI pixels x. Output: The fused class probability P. Step 1. Generating the 3D patches x1 ∈ ℝW1×W1×K1 and x2 ∈ ℝW2×W2×K2 from the local spatial–spectral neighborhood of x. Step 2. Performing the 3D-FHOG feature extraction and MDSN feature extraction on x1 and x2, respectively, by using F(·) and g1(·). Step 3. Computing the distance metric between F(x1) and ck to obtain the class probability P1. Step 4. Calculating the distance metric between g1(x2) and to obtain the class probability P2. Step 5. Getting the class probability P3 output from h(g2(x2)). Step 6. Fusing the class probability P1, P2 and P3 by using Equation (24). |
4. Experiments and Results
4.1. Data Sets
4.2. Experimental Setup
4.3. Experimental Result and Analysis
4.3.1. Influence of the Input Patch Size for 3D-FHOG
4.3.2. Compared with Handcrafted Feature-Based Methods
4.3.3. Compared with CNN-Based Methods
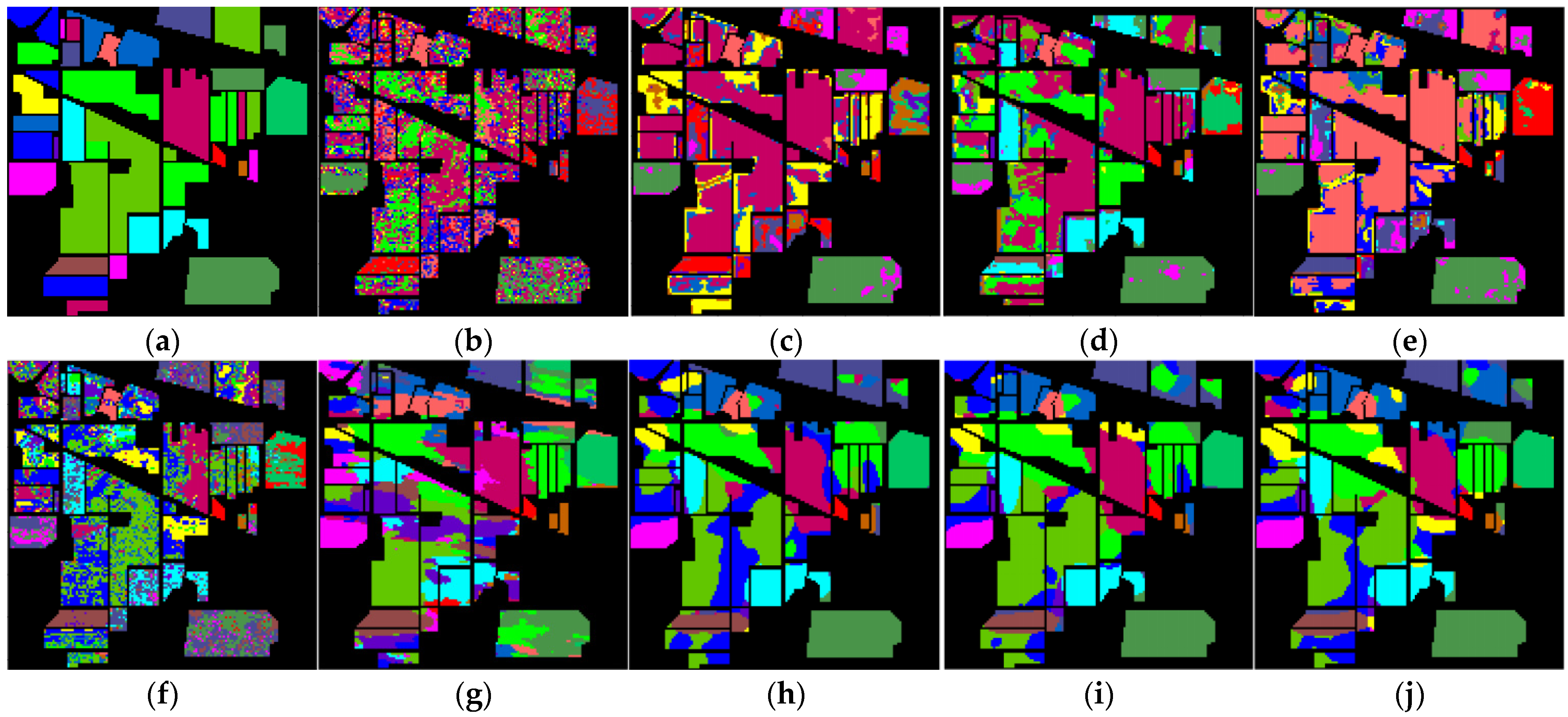
4.3.4. Classification Maps
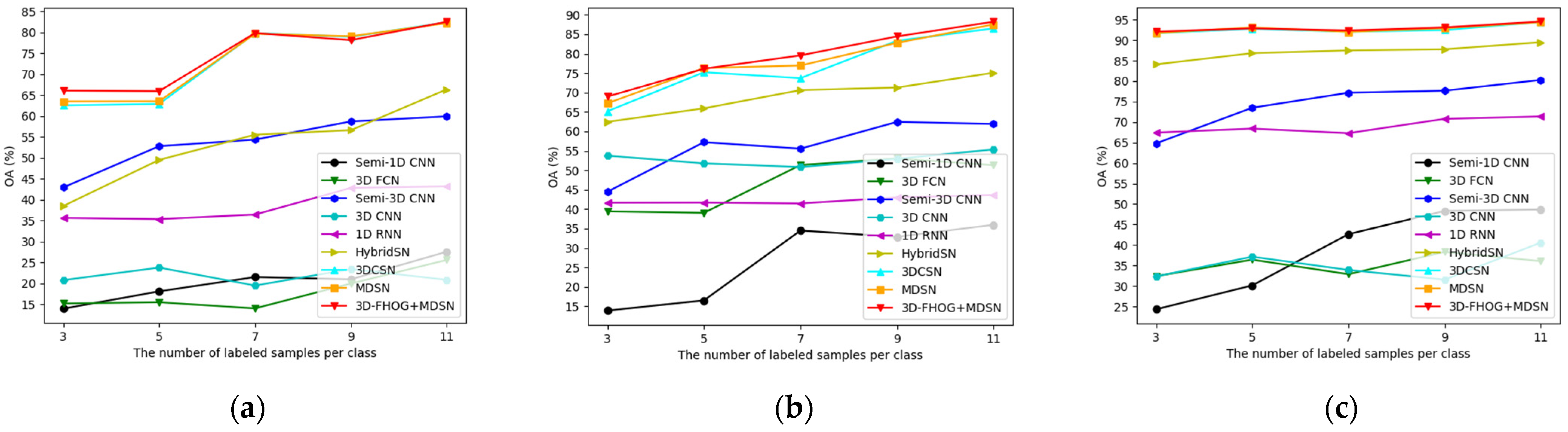
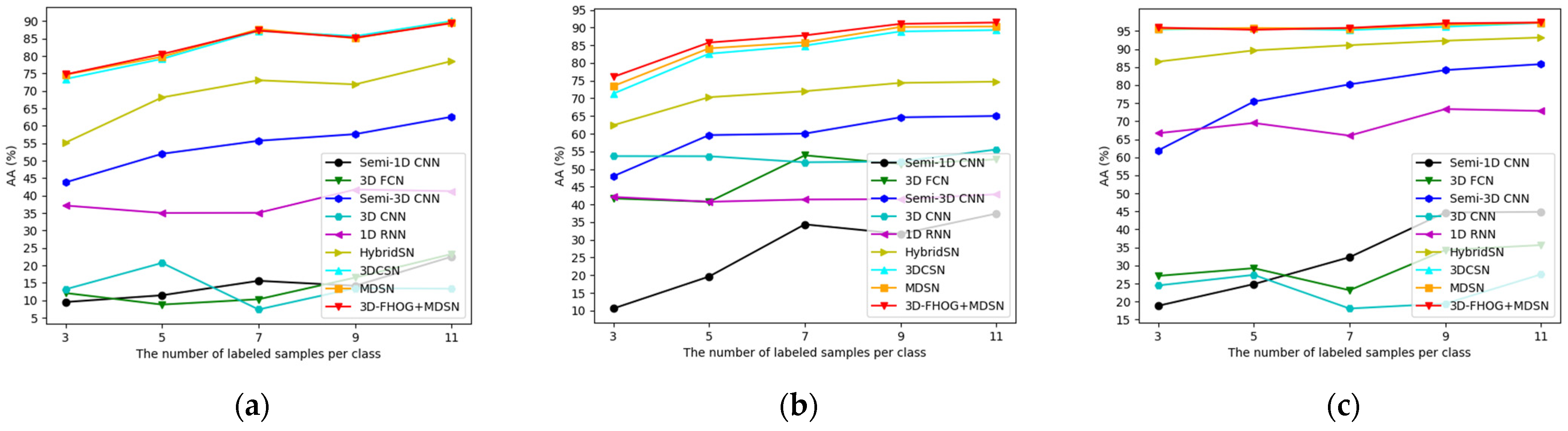
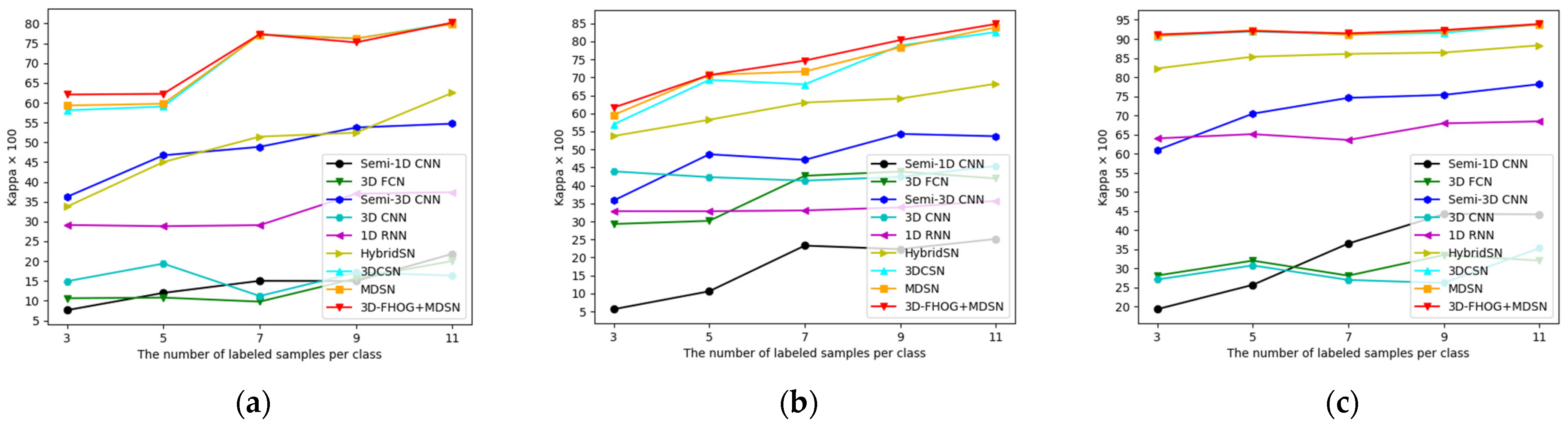
4.3.5. Influence of Training Sample Size
4.3.6. Time Consumption
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chakraborty, T.; Trehan, U. Spectralnet: Exploring spatial-spectral waveletcnn for hyperspectral image classification. arXiv 2021, arXiv:2104.00341. [Google Scholar]
- Zhang, Y.; Liu, K.; Dong, Y.; Wu, K.; Hu, X. Semisupervised classification based on SLIC segmentation for hyperspectral image. IEEE Geosci. Remote Sens. Lett. 2019, 17, 1440–1444. [Google Scholar] [CrossRef]
- Li, Y.; Tang, H.; Xie, W.; Luo, W. Multidimensional local binary pattern for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–13. [Google Scholar] [CrossRef]
- Luo, F.; Zou, Z.; Liu, J.; Lin, Z. Dimensionality reduction and classification of hyperspectral image via multistructure unified discriminative embedding. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–16. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, P.; Liu, N.; Yin, Q.; Zhang, F. Graph-Embedding Balanced Transfer Subspace Learning for Hyperspectral Cross-Scene Classification. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2022, 15, 2944–2955. [Google Scholar] [CrossRef]
- Ye, M.; Qian, Y.; Zhou, J.; Tang, Y. Dictionary learning-based feature-level domain adaptation for cross-scene hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 1544–1562. [Google Scholar] [CrossRef] [Green Version]
- Mustafa, G.; Zheng, H.; Khan, I.H.; Tian, L.; Jia, H.; Li, G.; Cheng, T.; Tian, Y.; Cao, W.; Zhu, Y.; et al. Hyperspectral Reflectance Proxies to Diagnose In-Field Fusarium Head Blight in Wheat with Machine Learning. Remote Sens. 2022, 14, 2784. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A review of advanced technologies and development for hyperspectral-based plant disease detection in the past three decades. Remote Sens. 2020, 12, 3188. [Google Scholar] [CrossRef]
- Uzair, M.; Mahmood, A.; Mian, A. Hyperspectral Face Recognition with Spatiospectral Information Fusion and PLS Regression. IEEE Trans. Image Process. 2015, 24, 1127–1137. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, H. Hyperspectral-cube-based mobile face recognition: A comprehensive review. Inf. Fusion 2021, 74, 132–150. [Google Scholar] [CrossRef]
- Dobler, G.; Ghandehari, M.; Koonin, S.E.; Sharma, M.S. A hyperspectral survey of New York City lighting technology. Sensors 2016, 16, 2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baur, J.; Dobler, G.; Bianco, F.; Sharma, M.; Karpf, A. Persistent hyperspectral observations of the urban lightscape. In Proceedings of the 2018 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Anaheim, CA, USA, 26–29 November 2018; pp. 983–987. [Google Scholar]
- Lowe, D. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Zhao, G.; Pietikainen, M. Dynamic texture recognition using local binary patterns with an application to facial expressions. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 915–928. [Google Scholar] [CrossRef] [Green Version]
- Jia, S.; Hu, J.; Zhu, J.; Jia, X.; Li, Q. Three-dimensional local binary patterns for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2399–2413. [Google Scholar] [CrossRef]
- He, L.; Li, J.; Plaza, A.; Li, Y. Discriminative Low-Rank Gabor Filtering for Spectral-Spatial Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 1381–1395. [Google Scholar] [CrossRef]
- Cao, X.; Xu, L.; Meng, D.; Zhao, Q.; Xu, Z. Integration of 3-dimensional discrete wavelet transform and Markov random field for hyperspectral image classification. Neurocomputing 2017, 226, 90–100. [Google Scholar] [CrossRef]
- Sharma, V.; Diba, A.; Tuytelaars, T.; Gool, L. Hyperspectral CNN for Image Classification & Band Selection, with Application to Face Recognition; Tech. Rep. KUL/ESAT/PSI/1604; KU Leuven: Leuven, Belgium, 2016. [Google Scholar]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep convolutional neural networks for hyperspectral image classification. J. Sens. 2015, 2015, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Boulch, A.; Audebert, N.; Dubucq, D. Autoencodeurs pour la visualisation d’images hyperspectrales. In Proceedings of the 25th Colloque Gretsi, Juan-les-Pins, France, 5–8 September 2017; pp. 1–4. [Google Scholar]
- Mou, L.; Ghamisi, P.; Zhu, X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kwon, H. Going deeper with contextual CNN for hyperspectral image classification. IEEE Trans. Image Process. 2017, 26, 4843–4855. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Yu, X.; Zhang, P.; Tan, X.; Yu, A.; Xue, Z. A semi-supervised convolutional neural network for hyperspectral image classification. Remote Sens. Lett. 2017, 8, 839–848. [Google Scholar] [CrossRef]
- Luo, Y.; Zou, J.; Yao, C.; Zhao, X.; Li, T.; Bai, G. HSI-CNN: A novel convolution neural network for hyperspectral image. In Proceedings of the 2018 International Conference on Audio, Language and Image Processing (ICALIP), Shanghai, China, 16–17 July 2018; pp. 464–469. [Google Scholar]
- Roy, S.; Krishna, G.; Dubey, S.; Bidyut, B. HybridSN: Exploring 3-D–2-D CNN feature hierarchy for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2019, 17, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Rostami, M.; Kolouri, S.; Eaton, E.; Kim, k. Deep transfer learning for few-shot SAR image classification. Remote Sens. 2019, 11, 1374. [Google Scholar] [CrossRef] [Green Version]
- Alajaji, D.; Alhichri, H.S.; Ammour, N.; Alajlan, N. Few-shot learning for remote sensing scene classification. In Proceedings of the 2020 Mediterranean and Middle-East Geoscience and Remote Sensing Symposium (M2GARSS), Tunis, Tunisia, 9–11 March 2020; pp. 81–84. [Google Scholar]
- Yuan, Z.; Huang, W. Multi-attention DeepEMD for few-shot learning in remote sensing. In Proceedings of the 2020 IEEE 9th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 11–13 December 2020; Volume 9, pp. 1097–1102. [Google Scholar]
- Kim, J.; Chi, M. SAFFNet: Self-attention-based feature fusion network for remote sensing few-shot scene classification. Remote Sens. 2021, 13, 2532. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 886–893. [Google Scholar]
- Surasak, T.; Takahiro, I.; Cheng, C.; Wang, C.; Sheng, P. Histogram of oriented gradients for human detection in video. In Proceedings of the 2018 5th International Conference on Business and Industrial Research (ICBIR), Bangkok, Thailand, 17–18 May 2018; pp. 172–176. [Google Scholar]
- Mao, L.; Xie, M.; Huang, Y.; Zhang, Y. Preceding vehicle detection using histograms of oriented gradients. In Proceedings of the 2010 International Conference on Communications, Circuits and Systems (ICCCAS), Chengdu, China, 28–30 July 2010; pp. 354–358. [Google Scholar]
- Qi, S.; Ma, J.; Lin, J.; Li, Y.; Tian, J. Unsupervised Ship Detection Based on Saliency and S-HOG Descriptor from Optical Satellite Images. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1451–1455. [Google Scholar]
- Chen, G.; Krzyzak, A.; Xie, W. Hyperspectral face recognition with histogram of oriented gradient features and collaborative representation-based classifier. Multimed. Tools. Appl. 2022, 81, 2299–2310. [Google Scholar] [CrossRef]
- Chopra, S.; Hadsell, R.; LeCun, Y. Learning a similarity metric discriminatively, with application to face verification. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 539–546. [Google Scholar]
- Melekhov, I.; Kannala, J.; Rahtu, E. Siamese network features for image matching. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 378–383. [Google Scholar]
- Tao, R.; Gavves, E.; Smeulders, A. Siamese instance search for tracking. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Cancun, Mexico, 4–8 December 2016; pp. 1420–1429. [Google Scholar]
- Bertinetto, L.; Valmadre, J.; Henriques, J.; Vedaldi, A.; Torr, P. Fully-convolutional siamese networks for object tracking. In Proceedings of the European Conference on Computer Vision, Amsterdam, the Netherlands, 8–16 October 2016; pp. 850–865. [Google Scholar]
- Koch, G.; Zemel, R.; Salakhutdinov, R. Siamese neural networks for one-shot image recognition. ICML Deep Learn. Workshop 2015, 2, 1–30. [Google Scholar]
- Zhao, S.; Li, W.; Du, Q.; Ran, Q. Hyperspectral classification based on siamese neural network using spectral-spatial feature. In Proceeding of 2018 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Valencia, Spain, 22–27 July 2018; pp. 2567–2570. [Google Scholar]
- Liu, B.; Yu, X.; Zhang, P.; Yu, A.; Fu, Q.; Wei, X. Supervised deep feature extraction for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 56, 1909–1921. [Google Scholar] [CrossRef]
- Cao, Z.; Li, X.; Jiang, J.; Zhao, L. 3D convolutional siamese network for few-shot hyperspectral classification. J. Appl. Remote Sens. 2020, 14, 048504. [Google Scholar] [CrossRef]
- Zeiler, M.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 5–12 September 2014; pp. 818–833. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Zadeh, L. Fuzzy set theory. Inf. Control. 1965, 8, 338–353. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Zhang, X. A Track-to-Track Association Algorithm Based on Fuzzy Synthetical Function and Its Application. Syst. Eng. Electron. 2003, 25, 1401–1403. [Google Scholar]
- Liu, J.; Li, R.; Liu, Y.; Zhang, Z. Multi-sensor data fusion based on correlation function and fuzzy integration function. Syst. Eng. Electron. 2006, 28, 1006–1009. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R. Prototypical networks for few-shot learning. Adv. Neural Inf. Processing Syst. 2017, 30, 1–11. [Google Scholar]
- Tang, H.; Li, Y.; Han, X.; Huang, Q.; Xie, W. A spatial–spectral prototypical network for hyperspectral remote sensing image. IEEE Geosci. Remote Sens. Lett. 2019, 17, 167–171. [Google Scholar] [CrossRef]
- Mura, M.; Benediktsson, J.; Waske, B.; Bruzzone, L. Extended profiles with morphological attribute filters for the analysis of hyperspectral data. Int. J. Remote Sens. 2010, 31, 5975–5991. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]


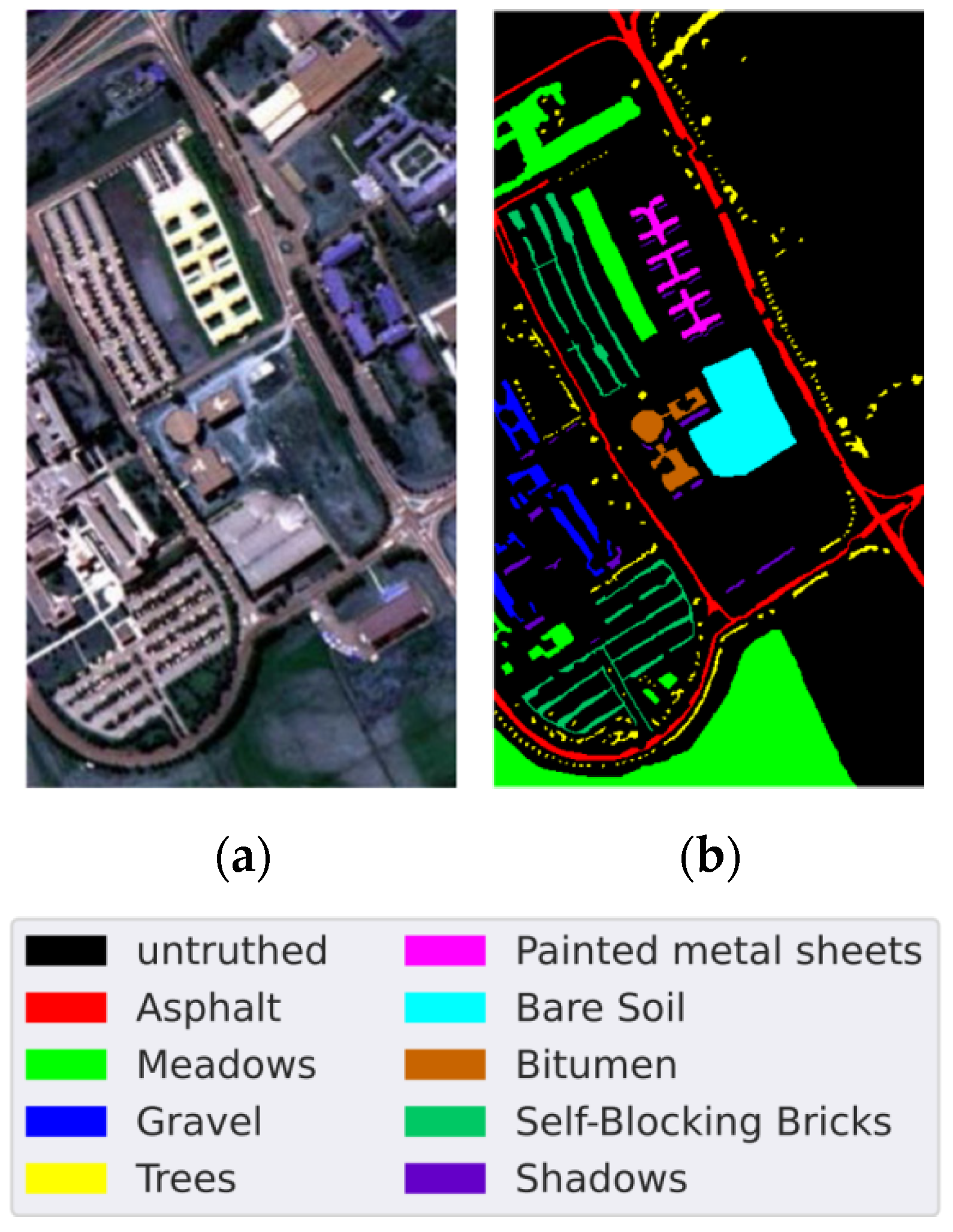

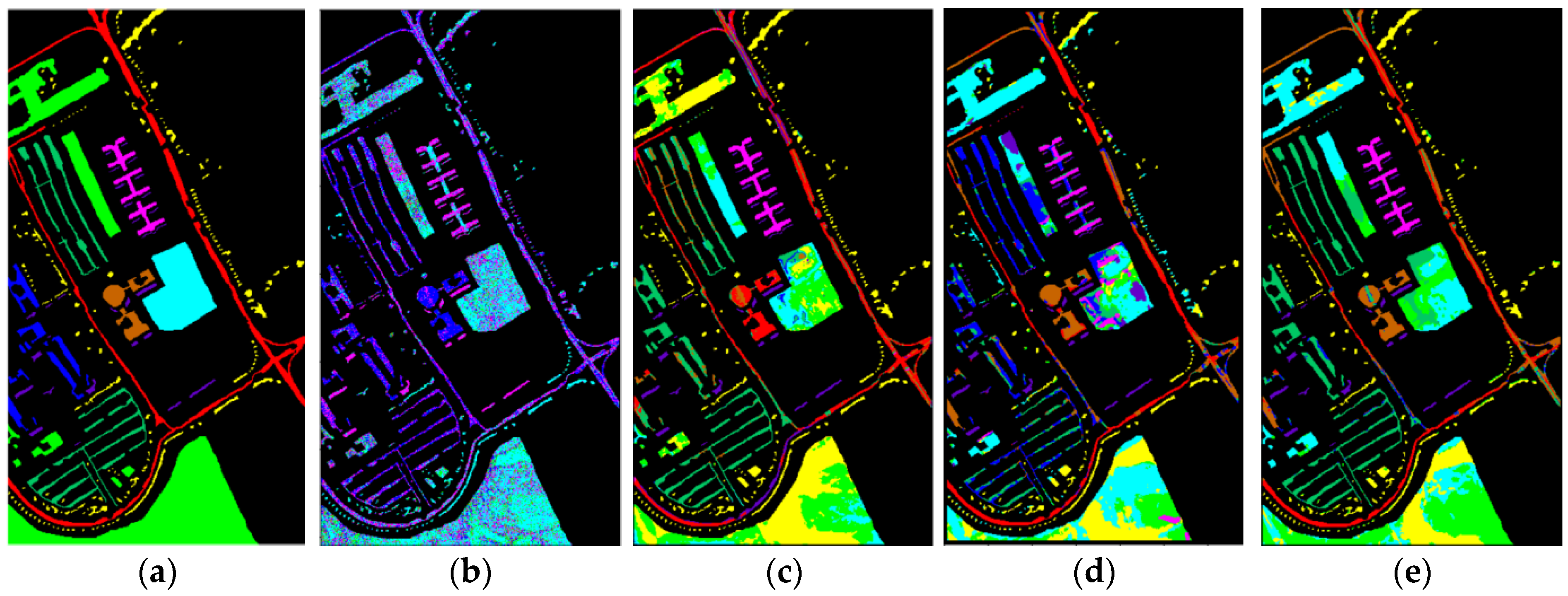

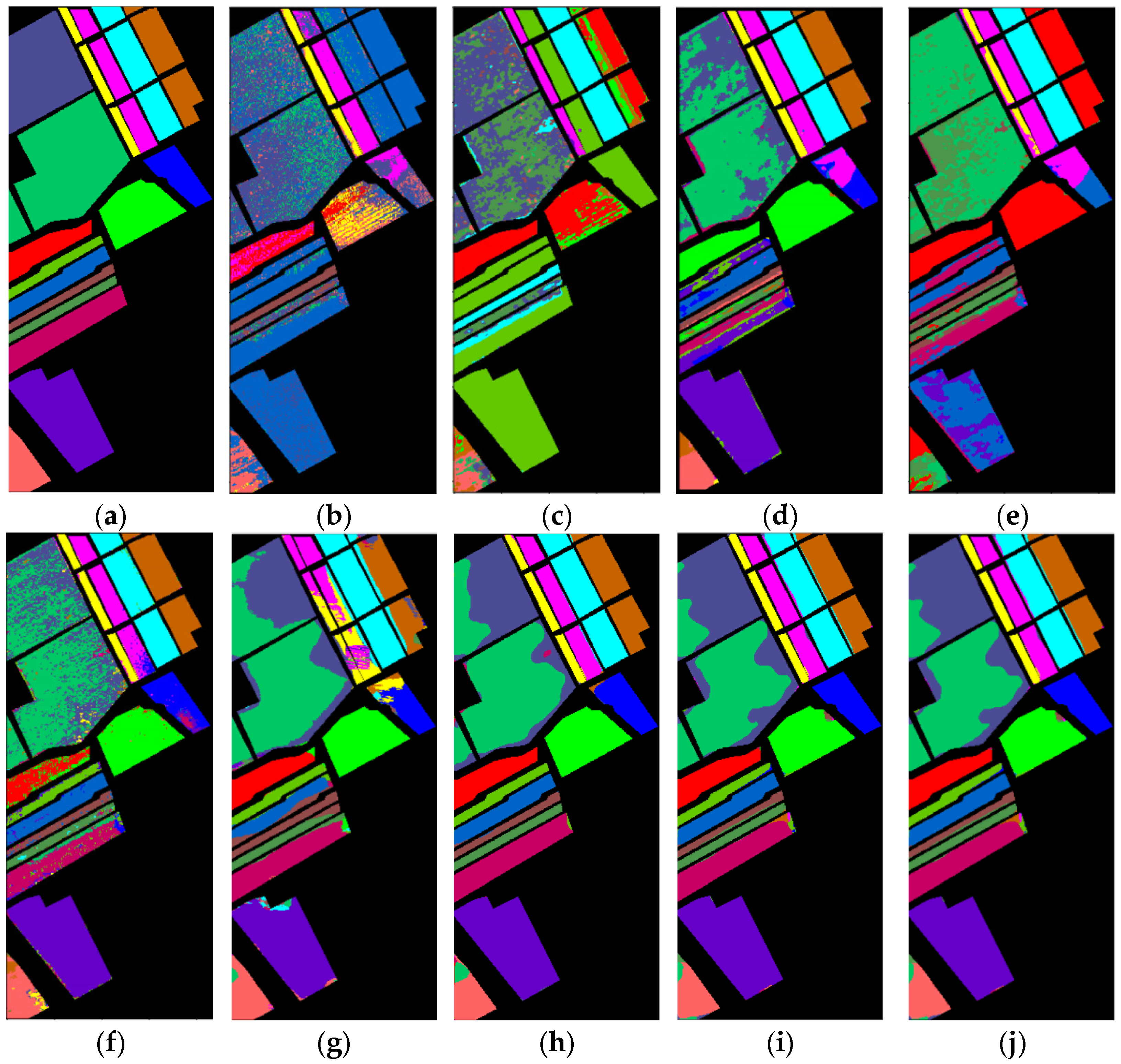
| Class | Name | Samples | Training Samples | Testing Samples |
|---|---|---|---|---|
| 1 | Alfalfa | 46 | 3 | 43 |
| 2 | Corn-notill | 1428 | 3 | 1425 |
| 3 | Corn-mintill | 830 | 3 | 827 |
| 4 | Corn | 237 | 3 | 234 |
| 5 | Grass-pasture | 483 | 3 | 480 |
| 6 | Grass-trees | 730 | 3 | 727 |
| 7 | Grass-pasture-mowed | 28 | 3 | 25 |
| 8 | Hay-windrowed | 478 | 3 | 475 |
| 9 | Oats | 20 | 3 | 17 |
| 10 | Soybean-notill | 972 | 3 | 969 |
| 11 | Soybean-mintill | 2455 | 3 | 2452 |
| 12 | Soybean-clean | 593 | 3 | 590 |
| 13 | Wheat | 205 | 3 | 202 |
| 14 | Woods | 1265 | 3 | 1262 |
| 15 | Buildings-Grass-Trees-Drives | 386 | 3 | 383 |
| 16 | Stone-Steel-Towers | 93 | 3 | 90 |
| Total | 10,249 | 48 | 10,201 |
| Class | Name | Samples | Training Samples | Testing Samples |
|---|---|---|---|---|
| 1 | Asphalt | 6631 | 3 | 6628 |
| 2 | Meadows | 18,649 | 3 | 18,646 |
| 3 | Gravel | 2099 | 3 | 2096 |
| 4 | Trees | 3064 | 3 | 3061 |
| 5 | Sheets | 1345 | 3 | 1342 |
| 6 | Bare soil | 5029 | 3 | 5026 |
| 7 | Bitumen | 1330 | 3 | 1327 |
| 8 | Bricks | 3682 | 3 | 3679 |
| 9 | Shadow | 947 | 3 | 944 |
| Total | 42,776 | 27 | 42,749 |
| Class | Name | Samples | Training Samples | Testing Samples |
|---|---|---|---|---|
| 1 | Brocoli_green_weeds_1 | 2009 | 3 | 2006 |
| 2 | Brocoli_green_weeds_2 | 3726 | 3 | 3723 |
| 3 | Fallow | 1976 | 3 | 1973 |
| 4 | Fallow_rough_plow | 1394 | 3 | 1391 |
| 5 | Fallow_smooth | 2678 | 3 | 2675 |
| 6 | Stubble | 3959 | 3 | 3956 |
| 7 | Celery | 3579 | 3 | 3576 |
| 8 | Grapes_untrained | 11,271 | 3 | 11,268 |
| 9 | Soil_vinyard_develop | 6203 | 3 | 6200 |
| 10 | Corn_senesced_green_weeds | 3278 | 3 | 3275 |
| 11 | Lettuce_romaine_4wk | 1068 | 3 | 1065 |
| 12 | Lettuce_romaine_5wk | 1927 | 3 | 1924 |
| 13 | Lettuce_romaine_6wk | 916 | 3 | 913 |
| 14 | Lettuce_romaine_7wk | 1070 | 3 | 1067 |
| 15 | Vinyard_untrained | 7268 | 3 | 7265 |
| 16 | Vinyard_vertical_trellis | 1807 | 3 | 1804 |
| Total | 54,129 | 48 | 54,081 |
| Input Patch Size | 7 × 7 × 7 | 9 × 9 × 9 | 11 × 11 × 11 | 13 × 13 × 13 | 15 × 15 × 15 | 17 × 17 × 17 | |
|---|---|---|---|---|---|---|---|
| Evaluation Metric | |||||||
| OA | 36.30 ± 0.15 | 39.70 ± 0.04 | 42.66 ± 0.03 | 48.71 ± 0.02 | 51.89 ± 0.01 | 50.97 ± 0.01 | |
| AA | 42.27 ± 0.07 | 46.56 ± 0.05 | 49.23 ± 0.09 | 54.23 ± 0.09 | 57.61 ± 0.05 | 57.12 ± 0.06 | |
| κ | 28.49 ± 0.17 | 32.39 ± 0.05 | 35.70 ± 0.04 | 42.02 ± 0.03 | 45.71 ± 0.01 | 44.81 ± 0.01 | |
| Class | Spectral | EMAP | HOG | SIFT | 3D-LBP | 3D-Gabor | 3D-DWT | 3D-FHOG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | |
| 1 | 78.60 | 2.04 | 73.02 | 4.29 | 67.44 | 4.82 | 99.53 | 0.01 | 100.00 | 0.00 | 36.28 | 2.91 | 59.53 | 6.70 | 73.95 | 3.19 |
| 2 | 24.60 | 0.59 | 11.84 | 2.15 | 20.39 | 0.31 | 31.90 | 2.00 | 27.37 | 2.07 | 39.26 | 3.59 | 23.24 | 2.24 | 28.69 | 0.40 |
| 3 | 21.69 | 2.59 | 34.32 | 3.42 | 17.34 | 0.68 | 25.92 | 1.22 | 23.70 | 0.85 | 24.93 | 1.54 | 23.43 | 3.30 | 23.53 | 1.15 |
| 4 | 20.85 | 2.70 | 33.85 | 0.91 | 23.42 | 0.16 | 55.47 | 1.57 | 44.87 | 8.25 | 37.26 | 5.11 | 25.30 | 0.38 | 31.54 | 0.95 |
| 5 | 40.92 | 4.54 | 34.08 | 4.10 | 36.54 | 0.67 | 36.75 | 1.72 | 15.12 | 0.50 | 19.46 | 5.71 | 12.79 | 8.18 | 58.42 | 0.30 |
| 6 | 27.54 | 1.41 | 69.52 | 1.36 | 36.73 | 1.79 | 54.22 | 4.07 | 49.57 | 0.92 | 29.32 | 2.91 | 49.90 | 2.52 | 67.98 | 0.94 |
| 7 | 93.60 | 0.05 | 89.60 | 0.37 | 66.40 | 3.11 | 100.00 | 0.00 | 100.00 | 0.00 | 100.00 | 0.00 | 76.00 | 0.88 | 72.00 | 4.35 |
| 8 | 49.39 | 2.33 | 38.19 | 1.13 | 28.21 | 1.05 | 64.21 | 2.87 | 71.62 | 2.74 | 91.96 | 0.13 | 66.90 | 4.38 | 63.49 | 3.59 |
| 9 | 71.76 | 0.76 | 64.70 | 0.69 | 87.38 | 1.34 | 100.00 | 0.00 | 100.00 | 0.00 | 75.29 | 1.80 | 100.00 | 0.00 | 94.44 | 1.23 |
| 10 | 37.21 | 1.12 | 36.62 | 4.73 | 21.63 | 0.21 | 32.57 | 4.61 | 34.08 | 3.93 | 17.87 | 7.04 | 50.18 | 2.16 | 46.81 | 0.92 |
| 11 | 32.28 | 1.46 | 37.75 | 4.18 | 14.69 | 0.21 | 17.35 | 0.94 | 26.66 | 5.48 | 26.32 | 10.73 | 18.67 | 3.28 | 67.11 | 0.16 |
| 12 | 12.27 | 2.31 | 11.97 | 0.38 | 19.56 | 0.24 | 24.54 | 1.96 | 31.93 | 1.12 | 14.81 | 1.46 | 22.85 | 2.53 | 28.95 | 1.45 |
| 13 | 94.36 | 0.18 | 92.97 | 0.30 | 67.03 | 1.38 | 73.66 | 3.38 | 96.44 | 0.09 | 75.74 | 0.51 | 53.17 | 11.96 | 74.26 | 1.71 |
| 14 | 59.30 | 5.30 | 59.49 | 9.23 | 18.49 | 0.29 | 45.91 | 8.80 | 62.98 | 1.61 | 67.84 | 8.33 | 88.73 | 2.74 | 63.72 | 2.45 |
| 15 | 12.64 | 0.63 | 28.41 | 1.36 | 28.82 | 0.75 | 57.18 | 2.32 | 36.55 | 1.91 | 27.36 | 0.61 | 12.74 | 0.49 | 46.42 | 1.74 |
| 16 | 82.00 | 1.16 | 91.11 | 0.07 | 50.67 | 3.11 | 86.67 | 2.03 | 96.44 | 0.39 | 93.33 | 0.44 | 81.33 | 0.49 | 80.44 | 2.62 |
| OA | 34.95 | 0.08 | 38.50 | 0.51 | 22.90 | 0.02 | 35.98 | 0.15 | 38.60 | 0.21 | 36.80 | 0.32 | 37.41 | 0.16 | 51.89 | 0.01 |
| AA | 47.44 | 0.05 | 50.46 | 0.07 | 37.80 | 0.05 | 56.62 | 0.14 | 57.33 | 0.05 | 48.57 | 0.05 | 47.80 | 0.09 | 57.61 | 0.05 |
| κ | 27.48 | 0.07 | 31.79 | 0.46 | 15.82 | 0.01 | 29.28 | 0.17 | 33.77 | 0.20 | 29.89 | 0.24 | 30.68 | 0.18 | 45.71 | 0.01 |
| Class | Spectral | EMAP | HOG | SIFT | 3D-LBP | 3D-Gabor | 3D-DWT | 3D-FHOG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | |
| 1 | 38.97 | 10.24 | 49.72 | 0.90 | 24.34 | 2.03 | 56.90 | 1.91 | 35.26 | 13.03 | 17.06 | 1.93 | 34.06 | 3.05 | 58.95 | 0.83 |
| 2 | 30.82 | 10.07 | 44.82 | 4.46 | 21.17 | 0.18 | 24.07 | 4.58 | 44.38 | 11.18 | 29.50 | 2.82 | 46.16 | 3.51 | 44.84 | 3.00 |
| 3 | 31.14 | 0.64 | 73.39 | 0.76 | 38.27 | 1.60 | 29.37 | 0.95 | 50.13 | 6.87 | 25.53 | 5.45 | 76.81 | 1.25 | 71.87 | 1.47 |
| 4 | 75.48 | 2.30 | 76.67 | 0.74 | 54.13 | 0.66 | 28.35 | 3.07 | 49.63 | 7.55 | 56.37 | 4.45 | 96.49 | 0.01 | 75.51 | 0.76 |
| 5 | 71.47 | 0.08 | 99.15 | 0.00 | 55.75 | 2.11 | 64.59 | 2.94 | 93.02 | 0.34 | 99.40 | 0.01 | 100.00 | 0.00 | 77.03 | 1.48 |
| 6 | 40.17 | 11.70 | 57.73 | 4.51 | 26.43 | 0.79 | 33.97 | 1.48 | 54.91 | 2.95 | 77.59 | 2.68 | 44.16 | 9.67 | 49.71 | 2.76 |
| 7 | 88.45 | 0.81 | 69.15 | 3.63 | 30.25 | 1.02 | 25.64 | 0.62 | 82.14 | 0.56 | 80.47 | 1.02 | 77.27 | 10.03 | 79.74 | 3.67 |
| 8 | 73.11 | 0.39 | 32.06 | 2.06 | 57.73 | 0.47 | 31.22 | 2.64 | 85.67 | 1.12 | 50.43 | 3.94 | 22.23 | 0.69 | 79.36 | 1.28 |
| 9 | 99.89 | 0.00 | 95.05 | 0.05 | 85.93 | 0.48 | 43.28 | 2.27 | 30.95 | 3.66 | 33.11 | 6.88 | 65.42 | 0.93 | 76.52 | 0.94 |
| OA | 44.63 | 0.63 | 53.25 | 0.80 | 31.42 | 0.02 | 33.25 | 0.43 | 50.82 | 3.49 | 40.61 | 0.64 | 50.18 | 0.42 | 56.89 | 0.57 |
| AA | 61.06 | 0.18 | 66.41 | 0.14 | 43.78 | 0.03 | 37.49 | 0.08 | 58.46 | 0.42 | 52.16 | 0.11 | 62.51 | 0.09 | 68.17 | 0.20 |
| κ | 35.29 | 0.42 | 44.5 | 0.73 | 21.64 | 0.02 | 21.20 | 0.11 | 42.52 | 3.12 | 31.16 | 0.51 | 40.26 | 0.33 | 48.13 | 0.56 |
| Class | Spectral | EMAP | HOG | SIFT | 3D-LBP | 3D-Gabor | 3D-DWT | 3D-FHOG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | |
| 1 | 98.28 | 0.02 | 97.90 | 0.05 | 49.29 | 0.84 | 41.40 | 0.78 | 57.43 | 10.06 | 68.32 | 11.18 | 95.65 | 0.05 | 96.96 | 0.08 |
| 2 | 70.20 | 4.32 | 96.10 | 0.14 | 36.37 | 0.76 | 37.82 | 4.48 | 63.12 | 0.64 | 76.93 | 7.91 | 85.01 | 0.12 | 92.46 | 0.34 |
| 3 | 49.98 | 0.40 | 76.34 | 2.26 | 31.42 | 1.21 | 26.34 | 2.72 | 49.49 | 4.18 | 21.28 | 1.00 | 50.85 | 7.75 | 93.36 | 0.68 |
| 4 | 98.13 | 0.02 | 92.38 | 0.34 | 69.89 | 1.89 | 85.74 | 0.73 | 95.87 | 0.01 | 77.94 | 6.14 | 97.44 | 0.01 | 84.02 | 2.35 |
| 5 | 97.70 | 0.00 | 81.14 | 4.44 | 50.31 | 2.77 | 31.50 | 2.25 | 93.81 | 0.05 | 60.26 | 5.94 | 79.58 | 10.32 | 78.65 | 8.17 |
| 6 | 96.67 | 0.02 | 99.59 | 0.00 | 50.69 | 1.76 | 51.69 | 2.30 | 56.15 | 10.42 | 90.36 | 0.02 | 77.50 | 17.58 | 97.56 | 0.04 |
| 7 | 97.75 | 0.05 | 99.61 | 0.00 | 40.75 | 1.07 | 18.79 | 0.23 | 85.70 | 0.05 | 32.38 | 6.32 | 64.80 | 1.68 | 95.22 | 0.05 |
| 8 | 45.40 | 1.86 | 44.12 | 4.26 | 25.96 | 1.05 | 28.69 | 10.42 | 39.31 | 10.00 | 32.26 | 11.49 | 46.18 | 13.01 | 68.29 | 0.41 |
| 9 | 75.46 | 7.87 | 90.75 | 1.64 | 35.71 | 1.04 | 14.99 | 0.71 | 75.41 | 3.07 | 97.64 | 0.01 | 75.34 | 17.42 | 100.00 | 0.01 |
| 10 | 29.34 | 6.40 | 63.81 | 3.48 | 36.71 | 4.02 | 18.04 | 1.09 | 64.05 | 1.06 | 22.00 | 4.32 | 25.95 | 6.86 | 64.18 | 5.06 |
| 11 | 77.35 | 0.18 | 80.02 | 2.14 | 63.83 | 3.27 | 89.56 | 0.06 | 92.60 | 0.00 | 61.67 | 7.09 | 78.33 | 4.84 | 76.73 | 2.54 |
| 12 | 73.79 | 0.40 | 75.55 | 1.79 | 62.85 | 3.87 | 60.57 | 9.16 | 61.23 | 13.09 | 54.07 | 4.06 | 65.97 | 1.20 | 81.05 | 0.43 |
| 13 | 95.64 | 0.36 | 76.12 | 3.00 | 64.82 | 2.13 | 39.01 | 6.72 | 36.17 | 16.42 | 95.44 | 0.06 | 89.18 | 0.02 | 89.35 | 0.90 |
| 14 | 76.94 | 3.91 | 75.35 | 6.23 | 75.00 | 2.57 | 77.56 | 0.76 | 74.13 | 1.31 | 62.38 | 10.54 | 66.64 | 1.66 | 82.19 | 2.28 |
| 15 | 66.34 | 2.49 | 73.11 | 3.68 | 18.79 | 0.43 | 45.66 | 10.26 | 77.79 | 1.12 | 63.14 | 8.10 | 40.54 | 8.41 | 38.67 | 1.68 |
| 16 | 20.45 | 0.95 | 72.66 | 1.57 | 26.53 | 1.46 | 24.70 | 0.98 | 41.35 | 4.88 | 52.93 | 0.37 | 21.44 | 4.00 | 57.35 | 0.66 |
| OA | 67.96 | 0.19 | 76.03 | 0.14 | 37.37 | 0.02 | 35.74 | 0.17 | 63.78 | 0.18 | 57.82 | 0.10 | 60.35 | 0.71 | 76.95 | 0.03 |
| AA | 73.09 | 0.05 | 80.91 | 0.16 | 46.18 | 0.09 | 43.25 | 0.06 | 66.48 | 0.16 | 60.56 | 0.12 | 66.27 | 0.54 | 80.94 | 0.08 |
| κ | 64.59 | 0.23 | 73.51 | 0.16 | 31.49 | 0.04 | 29.70 | 0.14 | 60.14 | 0.19 | 53.64 | 0.10 | 56.35 | 0.84 | 74.46 | 0.04 |
| Class | Semi-1D CNN | 3D FCN | Semi-3D CNN | 3D CNN | 1D RNN | HybridSN | 3DCSN | MDSN | 3D-FHOG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +MDSN | ||||||||||||||||||
| Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | |
| 1 | 3.22 | 0.09 | 4.74 | 0.22 | 42.14 | 2.70 | 7.80 | 0.57 | 24.69 | 1.00 | 73.49 | 1.35 | 100.00 | 0.00 | 100.00 | 0.00 | 99.53 | 0.01 |
| 2 | 6.36 | 0.68 | 4.96 | 0.55 | 28.22 | 1.21 | 10.85 | 1.98 | 24.71 | 0.22 | 19.87 | 1.14 | 46.29 | 0.03 | 53.38 | 0.22 | 51.21 | 0.10 |
| 3 | 8.33 | 0.95 | 10.88 | 1.58 | 20.13 | 0.48 | 5.09 | 0.40 | 23.73 | 0.52 | 27.64 | 0.65 | 53.01 | 1.49 | 59.54 | 0.19 | 63.41 | 0.22 |
| 4 | 3.81 | 0.11 | 12.17 | 0.79 | 22.93 | 0.86 | 12.16 | 1.00 | 18.54 | 0.70 | 17.61 | 0.80 | 50.60 | 0.54 | 57.52 | 0.38 | 55.13 | 0.52 |
| 5 | 11.33 | 2.96 | 6.91 | 1.20 | 42.58 | 0.33 | 15.32 | 1.15 | 46.47 | 0.15 | 58.46 | 1.55 | 55.50 | 0.04 | 54.58 | 0.02 | 57.54 | 0.02 |
| 6 | 7.14 | 2.04 | 3.03 | 0.29 | 69.21 | 0.94 | 15.00 | 4.01 | 55.14 | 2.00 | 70.84 | 2.34 | 88.09 | 0.21 | 90.76 | 0.02 | 91.77 | 0.02 |
| 7 | 7.99 | 0.49 | 4.00 | 0.05 | 40.29 | 2.39 | 0.75 | 0.02 | 29.69 | 1.08 | 100.00 | 0.00 | 100.00 | 0.00 | 100.00 | 0.00 | 100.00 | 0.00 |
| 8 | 8.80 | 0.69 | 7.49 | 2.24 | 78.91 | 2.95 | 15.47 | 9.57 | 63.07 | 3.37 | 89.39 | 0.19 | 99.12 | 0.02 | 98.36 | 0.01 | 99.49 | 0.00 |
| 9 | 2.14 | 0.03 | 4.66 | 0.06 | 25.82 | 0.29 | 4.67 | 0.34 | 23.85 | 3.75 | 95.29 | 0.33 | 100.00 | 0.00 | 100.00 | 0.00 | 100.00 | 0.00 |
| 10 | 5.87 | 0.52 | 11.50 | 1.42 | 32.52 | 0.13 | 8.41 | 2.34 | 29.13 | 0.44 | 24.99 | 3.59 | 58.39 | 0.13 | 58.02 | 0.10 | 59.86 | 0.09 |
| 11 | 25.22 | 2.83 | 10.57 | 4.47 | 38.19 | 3.38 | 20.45 | 3.52 | 26.45 | 1.59 | 17.19 | 1.99 | 49.35 | 0.63 | 45.53 | 0.11 | 53.60 | 0.08 |
| 12 | 9.83 | 0.71 | 5.14 | 0.31 | 20.00 | 0.07 | 7.79 | 1.45 | 17.44 | 0.18 | 30.61 | 1.00 | 60.10 | 0.31 | 55.05 | 0.27 | 62.14 | 0.02 |
| 13 | 24.44 | 2.92 | 18.83 | 1.93 | 65.11 | 0.26 | 25.52 | 7.23 | 65.46 | 2.49 | 81.88 | 7.62 | 81.78 | 0.87 | 87.23 | 0.11 | 83.56 | 0.16 |
| 14 | 7.16 | 1.59 | 34.84 | 13.17 | 74.91 | 1.32 | 46.28 | 14.50 | 66.19 | 0.34 | 65.55 | 0.38 | 83.00 | 0.35 | 85.48 | 0.22 | 85.86 | 0.25 |
| 15 | 9.03 | 0.46 | 7.72 | 0.65 | 24.92 | 0.23 | 8.04 | 0.68 | 27.61 | 0.45 | 50.39 | 2.92 | 76.60 | 0.00 | 76.66 | 0.00 | 76.76 | 0.00 |
| 16 | 11.64 | 4.40 | 46.29 | 6.65 | 74.71 | 3.11 | 7.09 | 0.23 | 52.40 | 2.57 | 59.56 | 0.92 | 72.67 | 0.05 | 72.00 | 0.16 | 55.56 | 0.05 |
| OA | 13.98 | 0.25 | 15.22 | 1.68 | 42.95 | 0.12 | 20.80 | 1.02 | 35.67 | 0.10 | 38.52 | 0.50 | 62.55 | 0.06 | 63.51 | 0.01 | 66.08 | 0.00 |
| AA | 9.52 | 0.06 | 12.11 | 0.58 | 43.79 | 0.04 | 13.17 | 0.69 | 37.16 | 0.15 | 55.17 | 0.25 | 73.40 | 0.02 | 74.63 | 0.00 | 74.72 | 0.01 |
| κ | 7.62 | 0.12 | 10.62 | 1.07 | 36.23 | 0.08 | 14.92 | 0.84 | 29.13 | 0.08 | 33.76 | 0.45 | 58.09 | 0.07 | 59.32 | 0.01 | 62.07 | 0.00 |
| Class | Semi-1D CNN | 3D FCN | Semi-3D CNN | 3D CNN | 1D RNN | HybridSN | 3DCSN | MDSN | 3D-FHOG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +MDSN | ||||||||||||||||||
| Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | |
| 1 | 1.00 | 0.02 | 33.05 | 4.85 | 61.66 | 1.80 | 75.39 | 0.31 | 42.33 | 9.42 | 53.59 | 1.17 | 52.58 | 0.50 | 63.63 | 0.35 | 67.44 | 0.24 |
| 2 | 11.90 | 0.86 | 39.22 | 5.49 | 41.14 | 4.97 | 55.24 | 1.37 | 44.65 | 1.35 | 67.38 | 2.70 | 65.48 | 0.27 | 63.51 | 0.33 | 64.01 | 0.23 |
| 3 | 11.07 | 1.05 | 18.40 | 1.35 | 33.01 | 0.27 | 17.02 | 1.86 | 19.47 | 5.48 | 76.12 | 1.22 | 87.97 | 0.78 | 87.36 | 0.27 | 89.83 | 0.11 |
| 4 | 11.93 | 1.04 | 49.64 | 1.66 | 41.05 | 0.87 | 48.31 | 5.91 | 58.79 | 0.54 | 18.63 | 0.11 | 34.45 | 1.93 | 37.84 | 0.56 | 41.80 | 0.58 |
| 5 | 23.68 | 8.97 | 80.47 | 6.07 | 73.99 | 2.68 | 99.39 | 0.00 | 73.57 | 2.84 | 100.00 | 0.00 | 99.73 | 0.00 | 99.97 | 0.00 | 99.79 | 0.00 |
| 6 | 10.74 | 1.74 | 34.57 | 0.06 | 34.79 | 0.17 | 36.20 | 0.09 | 25.79 | 0.03 | 77.96 | 0.92 | 74.46 | 2.07 | 78.12 | 0.86 | 79.32 | 0.81 |
| 7 | 13.05 | 2.12 | 26.46 | 1.90 | 33.64 | 0.78 | 22.89 | 3.03 | 21.71 | 0.85 | 74.35 | 1.33 | 94.06 | 0.52 | 91.94 | 0.54 | 97.66 | 0.08 |
| 8 | 10.35 | 3.32 | 32.58 | 0.56 | 50.49 | 0.74 | 30.97 | 8.60 | 38.20 | 5.41 | 47.12 | 1.78 | 61.66 | 0.20 | 69.22 | 0.34 | 70.55 | 0.20 |
| 9 | 1.55 | 0.04 | 61.04 | 11.43 | 62.02 | 1.84 | 97.49 | 0.08 | 54.68 | 1.19 | 46.46 | 1.26 | 71.48 | 0.18 | 69.56 | 0.06 | 74.02 | 0.11 |
| OA | 13.88 | 0.32 | 39.43 | 0.60 | 44.47 | 0.42 | 53.76 | 0.54 | 41.66 | 0.72 | 62.46 | 0.59 | 65.18 | 0.09 | 67.23 | 0.11 | 68.97 | 0.09 |
| AA | 10.58 | 0.04 | 41.71 | 0.30 | 47.98 | 0.13 | 53.66 | 0.39 | 42.13 | 0.37 | 62.40 | 0.10 | 71.32 | 0.08 | 73.46 | 0.03 | 76.05 | 0.04 |
| κ | 5.63 | 0.06 | 29.30 | 0.46 | 35.81 | 0.24 | 43.91 | 0.57 | 32.86 | 0.62 | 53.73 | 0.61 | 56.97 | 0.12 | 59.58 | 0.13 | 61.62 | 0.11 |
| Class | Semi-1D CNN | 3D FCN | Semi-3D CNN | 3D CNN | 1D RNN | HybridSN | 3DCSN | MDSN | 3D-FHOG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +MDSN | ||||||||||||||||||
| Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | Mean | Var | |
| 1 | 0.00 | 0.00 | 27.60 | 5.49 | 29.38 | 5.77 | 19.72 | 2.59 | 44.33 | 5.10 | 98.99 | 0.01 | 99.97 | 0.00 | 99.79 | 0.00 | 99.78 | 0.00 |
| 2 | 21.46 | 3.46 | 34.09 | 4.30 | 32.76 | 12.28 | 21.44 | 6.90 | 38.63 | 13.56 | 98.30 | 0.08 | 99.40 | 0.00 | 99.30 | 0.01 | 99.54 | 0.00 |
| 3 | 13.73 | 1.40 | 2.15 | 0.11 | 51.79 | 1.21 | 8.15 | 0.69 | 46.95 | 6.78 | 81.88 | 1.01 | 99.83 | 0.00 | 99.91 | 0.01 | 99.94 | 0.00 |
| 4 | 30.45 | 10.00 | 60.99 | 12.46 | 92.24 | 0.34 | 32.08 | 10.37 | 91.74 | 0.12 | 66.77 | 7.51 | 99.15 | 0.00 | 99.08 | 0.01 | 99.61 | 0.00 |
| 5 | 14.05 | 7.90 | 16.52 | 3.68 | 68.87 | 3.20 | 28.87 | 11.02 | 58.39 | 15.68 | 57.70 | 4.99 | 94.88 | 0.34 | 94.49 | 0.15 | 95.24 | 0.16 |
| 6 | 30.00 | 8.63 | 65.66 | 9.94 | 98.60 | 0.00 | 76.07 | 3.42 | 98.96 | 0.00 | 99.42 | 0.00 | 98.59 | 0.02 | 99.14 | 0.01 | 99.67 | 0.00 |
| 7 | 26.15 | 7.92 | 8.12 | 2.64 | 93.31 | 0.03 | 0.03 | 0.00 | 93.07 | 0.22 | 96.26 | 0.10 | 98.90 | 0.01 | 99.08 | 0.01 | 99.50 | 0.00 |
| 8 | 12.54 | 5.76 | 11.91 | 2.25 | 60.37 | 3.67 | 30.07 | 7.50 | 59.36 | 3.60 | 76.19 | 0.85 | 85.97 | 0.01 | 84.86 | 0.03 | 84.47 | 0.02 |
| 9 | 33.27 | 6.81 | 29.99 | 7.45 | 81.87 | 0.34 | 41.37 | 13.06 | 72.43 | 13.37 | 95.95 | 0.09 | 96.61 | 0.21 | 97.12 | 0.26 | 99.30 | 0.01 |
| 10 | 10.73 | 0.90 | 8.20 | 0.75 | 38.28 | 0.39 | 15.78 | 4.98 | 58.19 | 5.03 | 87.66 | 0.13 | 95.47 | 0.02 | 94.54 | 0.02 | 94.52 | 0.01 |
| 11 | 2.13 | 0.18 | 27.21 | 5.42 | 37.03 | 0.68 | 10.97 | 2.49 | 36.12 | 6.11 | 97.39 | 0.01 | 99.66 | 0.00 | 99.59 | 0.00 | 99.91 | 0.00 |
| 12 | 30.11 | 6.40 | 31.57 | 2.50 | 60.37 | 11.17 | 23.35 | 5.02 | 72.69 | 7.13 | 80.83 | 0.40 | 98.34 | 0.02 | 98.40 | 0.02 | 98.11 | 0.01 |
| 13 | 15.41 | 1.88 | 17.41 | 4.81 | 80.12 | 1.26 | 42.65 | 8.33 | 89.82 | 0.54 | 95.60 | 0.67 | 99.87 | 0.00 | 99.96 | 0.00 | 99.96 | 0.00 |
| 14 | 41.57 | 13.93 | 13.52 | 0.13 | 70.73 | 4.34 | 26.72 | 10.96 | 86.54 | 0.38 | 87.52 | 1.69 | 98.67 | 0.00 | 98.74 | 0.01 | 99.02 | 0.00 |
| 15 | 6.36 | 0.96 | 34.87 | 3.76 | 44.37 | 2.95 | 2.28 | 0.20 | 39.59 | 3.99 | 65.97 | 2.80 | 71.67 | 0.29 | 72.73 | 0.52 | 72.82 | 0.45 |
| 16 | 13.11 | 1.00 | 43.96 | 0.21 | 49.94 | 6.54 | 11.81 | 4.48 | 79.97 | 0.15 | 97.55 | 0.10 | 90.30 | 0.42 | 93.10 | 0.22 | 93.72 | 0.11 |
| OA | 24.31 | 0.40 | 32.38 | 0.45 | 64.79 | 0.08 | 32.34 | 2.52 | 67.42 | 0.34 | 84.07 | 0.01 | 91.69 | 0.00 | 91.72 | 0.01 | 92.06 | 0.01 |
| AA | 18.82 | 0.36 | 27.11 | 0.08 | 61.88 | 0.23 | 24.46 | 2.03 | 66.67 | 0.48 | 86.50 | 0.02 | 95.45 | 0.01 | 95.61 | 0.00 | 95.94 | 0.01 |
| κ | 19.30 | 0.32 | 28.11 | 0.34 | 60.91 | 0.10 | 27.07 | 2.48 | 63.99 | 0.38 | 82.29 | 0.01 | 90.74 | 0.00 | 90.78 | 0.01 | 91.16 | 0.01 |
| Model | Semi-1D CNN | 3D FCN | Semi-3D CNN | 3D CNN | 1D RNN | HybridSN | 3D CSN | MDSN | 3D-FHOG + MDSN | |
|---|---|---|---|---|---|---|---|---|---|---|
| IP | Training Time (s) | 449.72 | 197.29 | 500.10 | 21.56 | 9.76 | 4.09 | 295.81 | 314.16 | 314.17 |
| Testing time (s) | 0.44 | 15.04 | 3.91 | 4.81 | 1.46 | 10.36 | 10.58 | 12.58 | 15.50 | |
| PU | Training Time (s) | 3659.20 | 735.33 | 3710.15 | 37.50 | 23.03 | 1.77 | 34.64 | 38.66 | 38.58 |
| Testing time (s) | 3.53 | 140.01 | 23.22 | 19.21 | 7.88 | 19.98 | 20.46 | 26.58 | 41.78 | |
| SA | Training Time (s) | 2443.70 | 990.52 | 2893.45 | 95.27 | 39.32 | 2.18 | 102.17 | 114.46 | 115.28 |
| Testing time (s) | 2.37 | 83.36 | 29.18 | 27.51 | 7.78 | 24.91 | 25.54 | 34.93 | 64.84 |
| Model | 3D-FHOG + MDSN | |
|---|---|---|
| IP | HFE time (s) | 722.57 |
| HFE time for each pixel (s) | 0.07 | |
| PU | HFE time (s) | 3040.99 |
| HFE time for each pixel (s) | 0.07 | |
| SA | HFE time (s) | 3815.43 |
| HFE time for each pixel (s) | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, H.; Li, Y.; Huang, Z.; Zhang, L.; Xie, W. Fusion of Multidimensional CNN and Handcrafted Features for Small-Sample Hyperspectral Image Classification. Remote Sens. 2022, 14, 3796. https://doi.org/10.3390/rs14153796
Tang H, Li Y, Huang Z, Zhang L, Xie W. Fusion of Multidimensional CNN and Handcrafted Features for Small-Sample Hyperspectral Image Classification. Remote Sensing. 2022; 14(15):3796. https://doi.org/10.3390/rs14153796
Chicago/Turabian StyleTang, Haojin, Yanshan Li, Zhiquan Huang, Li Zhang, and Weixin Xie. 2022. "Fusion of Multidimensional CNN and Handcrafted Features for Small-Sample Hyperspectral Image Classification" Remote Sensing 14, no. 15: 3796. https://doi.org/10.3390/rs14153796
APA StyleTang, H., Li, Y., Huang, Z., Zhang, L., & Xie, W. (2022). Fusion of Multidimensional CNN and Handcrafted Features for Small-Sample Hyperspectral Image Classification. Remote Sensing, 14(15), 3796. https://doi.org/10.3390/rs14153796





