Cross-Comparison of Individual Tree Detection Methods Using Low and High Pulse Density Airborne Laser Scanning Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. ALS Data and Preprocessing
2.3. Field Validation Dataset
2.4. Individual Tree Detection Methods
2.4.1. Point-Cloud-Based Approach
2.4.2. Canopy-Height-Model-Based Approaches
2.4.3. Hybrid Approach
2.5. Matching ALS Detected and Reference Trees
2.6. Accuracy Assessment
3. Results
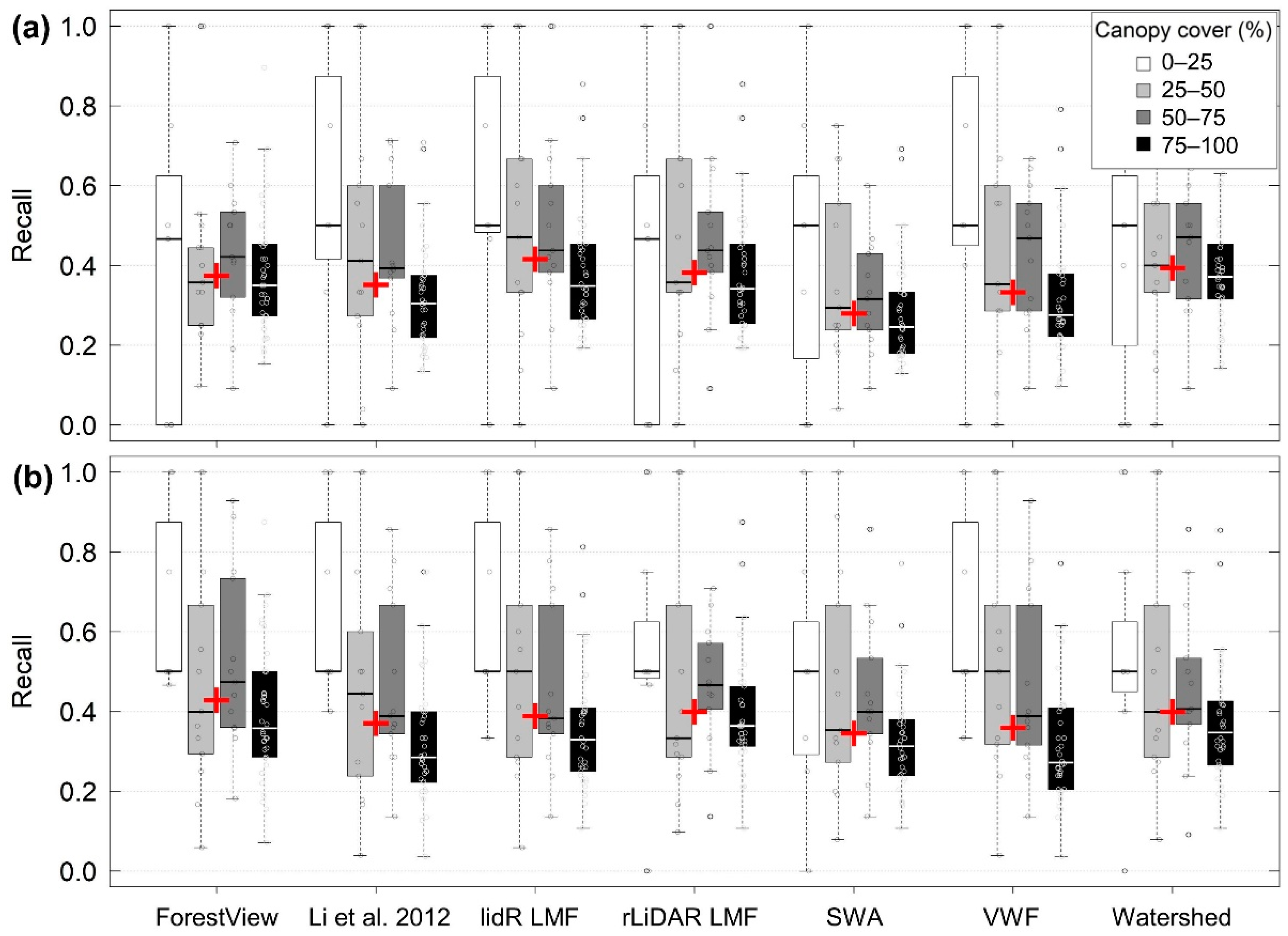
3.1. Individual Tree Detection Accuracy
3.2. Effects of Stand Density on ITD Method Accuracy
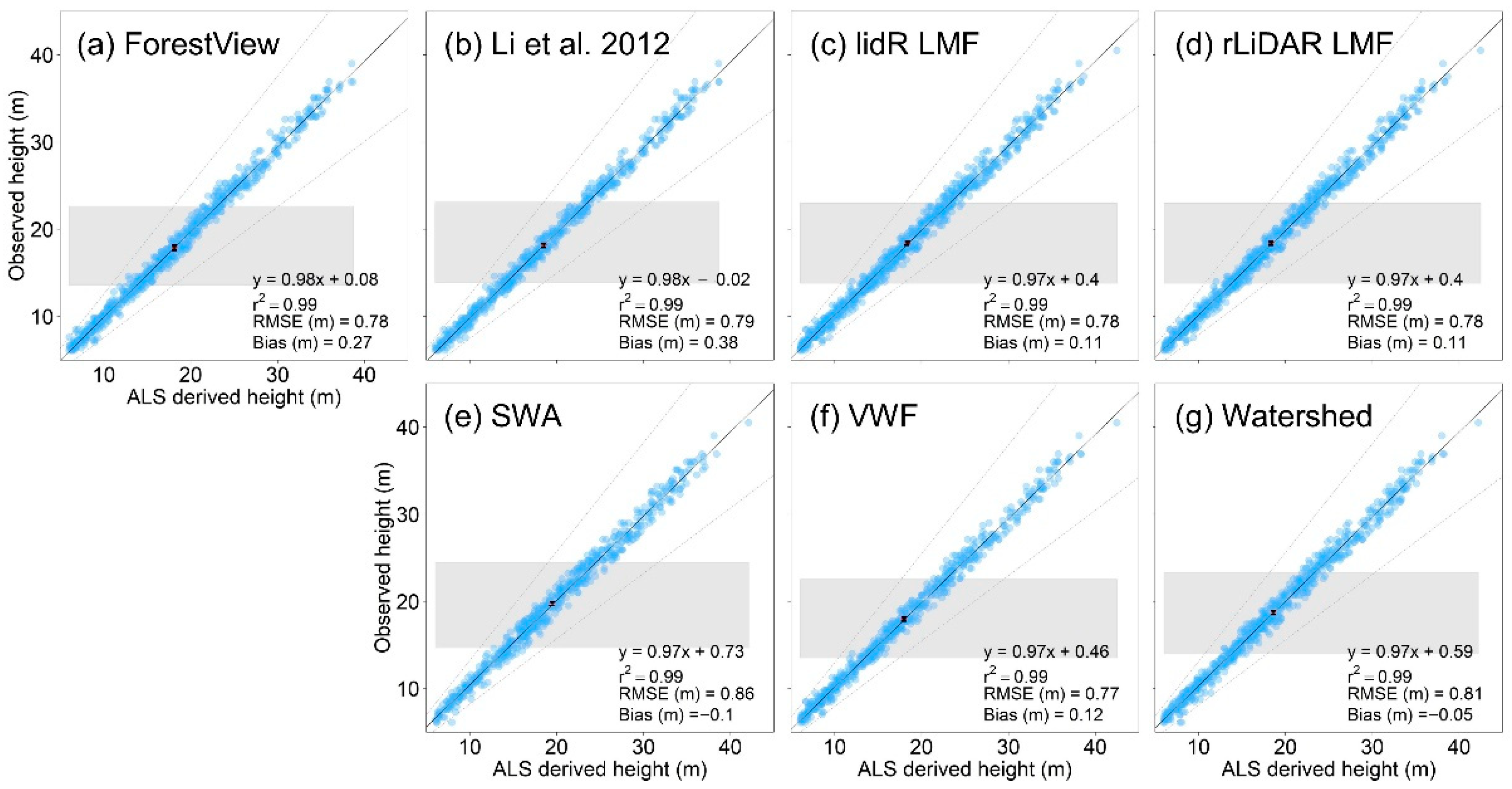
3.3. ITD Method Height Measurement Accuracy
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luoma, V.; Saarinen, N.; Wulder, M.A.; White, J.C.; Vastaranta, M.; Holopainen, M.; Hyyppä, J. Assessing precision in conventional field measurements of individual tree attributes. Forests 2017, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Durrieu, S.; Vega, C.; Bouvier, M.; Gosselin, F.; Renaud, J.P.; Saint-Andre, L. Optical remote sensing of tree and stand heights. In Land Resources Monitoring, Modeling, and Mapping with Remote Sensing; CRC Press: Boca Raton, FL, USA, 2015; pp. 449–485. [Google Scholar] [CrossRef]
- Persson, A.; Holmgren, J.; Söderman, U. Detecting and Measuring Individual Trees Using an Airborne Laser Scanner. Photogram. Eng. Remote Sens. 2002, 68, 925–932. [Google Scholar]
- Falkowski, M.J.; Smith, A.M.S.; Gessler, P.E.; Hudak, A.T.; Vierling, L.A.; Evans, J.S. The influence of conifer forest canopy cover on the accuracy of two individual tree measurement algorithms using lidar data. Can. J. Remote Sens. 2008, 34, S338–S350. [Google Scholar] [CrossRef]
- Dalla Corte, A.P.; Souza, D.V.; Rex, F.E.; Sanquetta, C.R.; Mohan, M.; Silva, C.A.; Zambrano, A.M.A.; Prata, G.; de Almeida, D.R.A.; Trautenmüller, J.W.; et al. Forest inventory with high-density UAV-Lidar: Machine learning approaches for predicting individual tree attributes. Comput. Electron. Agric. 2020, 179, 105815. [Google Scholar] [CrossRef]
- Tatum, J.; Wallin, D. Using Discrete-Point LiDAR to Classify Tree Species in the Riparian Pacific Northwest, USA. Remote Sens. 2021, 13, 2647. [Google Scholar] [CrossRef]
- Borgogno Mondino, E.; Fissore, V.; Falkowski, M.J.; Palik, B. How far can we trust forestry estimates from low-density LiDAR acquisitions? The Cutfoot Sioux experimental forest (MN, USA) case study. Int. J. Remote Sens. 2020, 41, 4551–4569. [Google Scholar] [CrossRef]
- Corrao, M.V.; Sparks, A.M.; Smith, A.M.S. A Conventional Cruise and Felled-Tree Validation of Individual Tree Diameter, Height and Volume Derived from Airborne Laser Scanning Data of a Loblolly Pine (P. taeda) Stand in Eastern Texas. Remote Sens. 2022, 14, 2567. [Google Scholar] [CrossRef]
- Sparks, A.M.; Smith, A.M.S. Accuracy of a LiDAR-Based Individual Tree Detection and Attribute Measurement Algorithm Developed to Inform Forest Products Supply Chain and Resource Management. Forests 2022, 13, 3. [Google Scholar] [CrossRef]
- Tinkham, W.T.; Smith, A.M.S.; Affleck, D.L.; Saralecos, J.D.; Falkowski, M.J.; Hoffman, C.M.; Hudak, A.T.; Wulder, M.A. Development of height-volume relationships in second growth Abies grandis for use with aerial LiDAR. Can. J. Remote Sens. 2016, 42, 400–410. [Google Scholar] [CrossRef]
- Jeronimo, S.M.A.; Kane, V.R.; Churchill, D.J.; McGaughey, R.J.; Franklin, J.F. Applying LiDAR individual tree detection to management of structurally diverse forest landscapes. J. For. 2018, 116, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Tinkham, W.T.; Mahoney, P.R.; Smith, A.M.S.; Falkowski, M.J.; Woodall, C.; Donke, G.; Hudak, A.T. Applications of the United States Forest Service Forest Inventory and Analysis dataset: A review and future directions. Can. J. For. Res. 2018, 48, 1251–1268. [Google Scholar] [CrossRef]
- Duncanson, L.; Dubayah, R. Monitoring individual tree-based change with airborne lidar. Ecol. Evol. 2018, 8, 5079–5089. [Google Scholar] [CrossRef] [PubMed]
- Keefe, R.F.; Zimbelman, E.G.; Picchi, G. Use of Individual Tree and Product Level Data to Improve Operational Forestry. Curr. For. Rep. 2022, 8, 148–165. [Google Scholar] [CrossRef]
- Larsen, M.; Eriksson, M.; Descombes, X.; Perrin, G.; Brandtberg, T.; Gougeon, F.A. Comparison of six individual tree crown detection algorithms evaluated under varying forest conditions. Int. J. Remote Sens. 2011, 32, 5827–5852. [Google Scholar] [CrossRef]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An international comparison of individual tree detection and extraction using airborne laser scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- Eysn, L.; Hollaus, M.; Lindberg, E.; Berger, F.; Monnet, J.M.; Dalponte, M.; Kobal, M.; Pellegrini, M.; Lingua, E.; Mongus, D.; et al. A benchmark of lidar-based single tree detection methods using heterogeneous forest data from the alpine space. Forests 2015, 6, 1721–1747. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hyyppä, J.; Liang, X.; Kaartinen, H.; Yu, X.; Lindberg, E.; Holmgren, J.; Qin, Y.; Mallet, C.; Ferraz, A.; et al. International benchmarking of the individual tree detection methods for modeling 3-D canopy structure for silviculture and forest ecology using airborne laser scanning. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5011–5027. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A new method for segmenting individual trees from the lidar point cloud. Photogram. Eng. Remote Sens. 2012, 10, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Ayrey, E.; Fraver, S.; Kershaw, J.A., Jr.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer stacking: A novel algorithm for individual forest tree segmentation from LiDAR point clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Hyyppä, J.; Schardt, M.; Haggrén, H.; Koch, B.; Lohr, U.; Scherrer, H.U.; Paananen, R.; Luukkonen, H.; Ziegler, M.; Hyyppä, H.; et al. HIGH-SCAN: The first European-wide attempt to derive single-tree information from laserscanner data. Photogramm. J. Finl. 2001, 17, 58–68. [Google Scholar]
- Popescu, S.C.; Wynne, R.H. Seeing the trees in the forest. Photogram. Eng. Remote Sens. 2004, 16, 589–604. [Google Scholar] [CrossRef] [Green Version]
- Falkowski, M.J.; Smith, A.M.S.; Hudak, A.T.; Gessler, P.E.; Vierling, L.A.; Crookston, N.L. Automated estimation of individual conifer tree height and crown diameter via two-dimensional spatial wavelet analysis of lidar data. Can. J. Remote Sens. 2006, 32, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.A.; Hudak, A.T.; Vierling, L.A.; Loudermilk, E.L.; O’Brien, J.J.; Hiers, J.K.; Jack, S.B.; Gonzalez-Benecke, C.; Lee, H.; Falkowski, M.J.; et al. Imputation of individual longleaf pine (Pinus palustris Mill.) tree attributes from field and LiDAR data. Can. J. Remote Sens. 2016, 42, 554–573. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in automatic individual tree crown detection and delineation—Evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef] [Green Version]
- Lindberg, E.; Holmgren, J. Individual Tree Crown Methods for 3D Data from Remote Sensing. Curr. For. Rep. 2017, 3, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Silva, C.A.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, A.T.; Dia, M. Individual tree detection from unmanned aerial vehicle (UAV) derived canopy height model in an open canopy mixed conifer forest. Forests 2017, 8, 340. [Google Scholar] [CrossRef] [Green Version]
- Leite, R.V.; Amaral, C.H.D.; Pires, R.D.P.; Silva, C.A.; Soares, C.P.B.; Macedo, R.P.; Silva, A.A.L.D.; Broadbent, E.N.; Mohan, M.; Leite, H.G. Estimating stem volume in eucalyptus plantations using airborne LiDAR: A comparison of area-and individual tree-based approaches. Remote Sens. 2020, 12, 1513. [Google Scholar] [CrossRef]
- Kolendo, Ł.; Kozniewski, M.; Ksepko, M.; Chmur, S.; Neroj, B. Parameterization of the Individual Tree Detection Method Using Large Dataset from Ground Sample Plots and Airborne Laser Scanning for Stands Inventory in Coniferous Forest. Remote Sens. 2021, 13, 2753. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkänen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative testing of single-tree detection algorithms under different types of forest. Forestry 2012, 85, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Pearse, G.D.; Watt, M.S.; Dash, J.P.; Stone, C.; Caccamo, G. Comparison of models describing forest inventory attributes using standard and voxel-based lidar predictors across a range of pulse densities. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 341–351. [Google Scholar] [CrossRef]
- Eitel, J.U.; Höfle, B.; Vierling, L.A.; Abellán, A.; Asner, G.P.; Deems, J.S.; Glennie, C.L.; Joerg, P.C.; LeWinter, A.L.; Magney, T.S.; et al. Beyond 3-D: The new spectrum of lidar applications for earth and ecological sciences. Remote Sens. Environ. 2016, 186, 372–392. [Google Scholar] [CrossRef] [Green Version]
- Roussel, J.R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.; Meador, A.S.; Bourdon, J.F.; de Boissieu, F.; Achim, A. lidR: An R package for analysis of Airborne Laser Scanning (ALS) data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- McGaughey, R.J. FUSION/LDV: Software for LIDAR Data Analysis and Visualization. USDA Forest Service, Pacific Northwest Research Station. 2022. Available online: http://forsys.cfr.washington.edu/fusion/fusion_overview.html (accessed on 11 April 2022).
- Smith, A.M.S.; Falkowski, M.J.; Hudak, A.T.; Evans, J.S.; Robinson, A.P.; Steele, C.M. A cross-comparison of field, spectral, and lidar estimates of forest canopy cover. Can. J. Remote Sens. 2009, 35, 447–459. [Google Scholar] [CrossRef]
- Hudak, A.T.; Strand, E.K.; Vierling, L.A.; Byrne, J.C.; Eitel, J.U.H.; Martinuzzi, S.; Falkiwski, M.J. Quantifying aboveground forest carbon pools and fluxes from repeat LiDAR surveys. Remote Sens. Environ. 2012, 123, 25–40. [Google Scholar] [CrossRef] [Green Version]
- United States Geological Survey. Lidar Base Specification Version 2.1. 2019. Available online: https://www.usgs.gov/3DEP/lidarspec (accessed on 6 June 2022).
- Kraft, G. Beiträge zur Lehre von den Durchforstungen, Schlagstellungen und Lichtungshieben; Klindsworth’s Verlag: Hannover, Germany, 1884. [Google Scholar]
- Silva, C.A.; Crookston, N.L.; Hudak, A.T.; Vierling, L.A. rLiDAR: An R Package for Reading, Processing and Visualizing LiDAR (Light Detection and Ranging) Data, Version 0.1.5. 2021. Available online: https://cran.r-project.org/web/packages/rLiDAR/index.html (accessed on 11 April 2022).
- Plowright, A.; Roussel, J. Forest Tools. R Package Version 0.2.5. 2019. Available online: https://github.com/andrew-plowright/ForestTools (accessed on 20 January 2022).
- Garrity, S.R.; Vierling, L.A.; Smith, A.M.S.; Hann, D.B.; Falkowski, M.J. Automatic detection of shrub location, crown area, and cover using spatial wavelet analysis and aerial photography. Can. J. Remote Sens. 2008, 34, S376–S384. [Google Scholar] [CrossRef]
- Poznanovic, A.J.; Falkowski, M.J.; MacLean, A.L.; Evans, J.S.; Smith, A.M.S. An accuracy assessment of tree detection algorithms in juniper woodlands. Photogram. Eng. Remote Sens. 2014, 80, 45–55. [Google Scholar] [CrossRef]
- Smith, A.M.S.; Strand, E.K.; Steele, C.M.; Hann, D.B.; Garrity, S.R.; Falkowski, M.J.; Evans, J.S. Production of vegetation spatial-structure maps by per-object analysis of juniper encroachment in multi-temporal aerial photographs. Can. J. Remote Sens. 2008, 34, S268–S285. [Google Scholar] [CrossRef] [Green Version]
- Strand, E.K.; Smith, A.M.S.; Bunting, S.C.; Vierling, L.A.; Hann, D.B.; Gessler, P.E. Wavelet estimation of plant spatial patterns in multi-temporal aerial photography. Int. J. Remote Sens. 2006, 27, 2049–2054. [Google Scholar] [CrossRef]
- Strand, E.K.; Vierling, L.A.; Smith, A.M.S.; Bunting, S.C. Net Changes in Above Ground Woody Carbon Stock in Western Juniper Woodlands, 1946–1998. J. Geophys. Res. 2008, 113, G01013. [Google Scholar] [CrossRef] [Green Version]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Patt. Analys. Mach. Intel. 1991, 13, 583–598. [Google Scholar] [CrossRef] [Green Version]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.; Hussin, Y.A. Generating pit-free canopy height models from airborne lidar. Photogram. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Yang, Q.; Su, Y.; Jin, S.; Kelly, M.; Hu, T.; Ma, Q.; Li, Y.; Song, S.; Zhang, J.; Xu, G.; et al. The influence of vegetation characteristics on individual tree segmentation methods with airborne LiDAR data. Remote Sens. 2019, 11, 2880. [Google Scholar] [CrossRef] [Green Version]
- Vastaranta, M.; Saarinen, N.; Kankare, V.; Holopainen, M.; Kaartinen, H.; Hyyppä, J.; Hyyppä, H. Multisource single-tree inventory in the prediction of tree quality variables and logging recoveries. Remote Sens. 2014, 6, 3475–3491. [Google Scholar] [CrossRef] [Green Version]
- Pentz, M.; Shott, M. Handling Experimental Data; Open University Press: Maidenhead, UK, 1988; ISBN 12-978-0335158973. [Google Scholar]
- Burkhart, H.E.; Tomé, M. Modeling Forest Trees and Stands; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Robinson, A.P.; Duursma, R.A.; Marshall, J.D. A regression-based equivalence test for model validation: Shifting the burden of proof. Tree Phys. 2005, 25, 903–913. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org (accessed on 6 June 2022).
- Robinson, A. Equivalence: Provides Tests and Graphics for Assessing Tests of Equivalence, Version 0.7.2. 2016. Available online: https://cran.r-project.org/web/packages/equivalence (accessed on 24 January 2022).
- Hudak, A.T.; Evans, J.S.; Smith, A.M.S. Review: LiDAR Utility for Natural Resource Managers. Remote Sens. 2009, 1, 934–951. [Google Scholar] [CrossRef] [Green Version]
- Martinuzzi, S.; Vierling, L.A.; Gould, W.A.; Falkowski, M.J.; Evans, J.S.; Hudak, A.T.; Vierling, K.T. Mapping snags and understory shrubs for a LiDAR-based assessment of wildlife habitat suitability. Remote Sens. Environ. 2009, 113, 2533–2546. [Google Scholar] [CrossRef] [Green Version]
- Casas, A.; Garcia, M.; Siegel, R.B.; Koltunov, A.; Ramirez, C.; Ustin, S. Burned forest characterization at single-tree level with airborne laser scanning for assessing wildlife habitat. Remote Sens. Environ. 2016, 175, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Kramer, H.A.; Collins, B.M.; Kelly, M.; Stephens, S.L. Quantifying Ladder Fuels: A New Approach Using LiDAR. Forests 2014, 5, 1432–1453. [Google Scholar] [CrossRef]
- Olszewski, J.H.; Bailey, J.D. LiDAR as a Tool for Assessing Change in Vertical Fuel Continuity Following Restoration. Forests 2022, 13, 503. [Google Scholar] [CrossRef]
- Stenzel, J.E.; Bartowitz, K.J.; Hartman, M.D.; Lutz, J.A.; Smith, A.M.S.; Kolden, C.A.; Law, B.E.; Swanson, M.E.; Larson, A.J.; Parton, W.J.; et al. Fixing a snag in estimating carbon emissions from wildfires. Glob. Change Biol. 2019, 25, 3985–3994. [Google Scholar] [CrossRef] [PubMed]
- Jarron, L.R.; Coops, N.C.; MacKenzie, W.H.; Dykstra, P. Detection and Quantification of Coarse Woody Debris in Natural Forest Stands Using Airborne LiDAR. For. Sci. 2021, 67, 550–563. [Google Scholar] [CrossRef]
- Hanan, E.J.; Kennedy, M.C.; Ren, J.; Johnson, M.C.; Smith, A.M.S. Missing climate feedbacks in fire models: Limitations and uncertainties in fuel loadings and the role of decomposition in fine fuel succession. J. Adv. Model. Earth Sys. 2022, 14, e2021MS002818. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Hudak, A.T.; Crookston, N.; Gessler, P.E.; Ubeler, E.H.; Smith, A.M.S. Landscape-scale parameterization of a tree-level forest growth model: A k-NN imputation approach incorporating LiDAR data. Can. J. For. Res. 2010, 40, 184–199. [Google Scholar] [CrossRef]
- Hudak, A.T.; Fekety, P.A.; Kane, V.R.; Kennedy, R.E.; Filippelli, S.K.; Falkowski, M.J.; Tinkham, W.T.; Smith, A.M.S.; Crookston, N.L.; Domke, G.M.; et al. A carbon monitoring system for mapping regional, annual aboveground biomass across the northwestern USA. Environ. Res. Lett. 2020, 15, 095003. [Google Scholar] [CrossRef]
- McCarley, T.R.; Kolden, C.A.; Vaillant, N.M.; Hudak, A.T.; Smith, A.M.S.; Wing, B.M.; Kellogg, B.S.; Kreitler, J. Multi-temporal LiDAR and Landsat quantification of fire-induced changes to forest structure. Remote Sens. Environ. 2017, 191, 419–432. [Google Scholar] [CrossRef] [Green Version]
- McCarley, T.R.; Kolden, C.A.; Vaillant, N.M.; Hudak, A.T.; Smith, A.M.S.; Kreitler, J. Landscape-scale quantification of fire-induced change in canopy cover following mountain pine beetle outbreak and timber harvest. For. Ecol. Manag. 2017, 391, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Ma, Q.; Su, Y.; Battles, J.J.; Collins, B.M.; Stephens, S.L.; Kelly, M.; Gui, Q. A simple and integrated approach for fire severity assessment using bi-temporal airborne LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 25–38. [Google Scholar] [CrossRef]
- Hillman, S.; Hally, B.; Wallace, L.; Turner, D.; Lucieer, A.; Reinke, K.; Jones, S. High-Resolution Estimates of Fire Severity—An Evaluation of UAS Image and LiDAR Mapping Approaches on a Sedgeland Forest Boundary in Tasmania, Australia. Fire 2021, 4, 14. [Google Scholar] [CrossRef]
- Smith, A.M.S.; Kolden, C.A.; Tinkham, W.T.; Talhelm, A.F.; Marshall, J.D.; Hudak, A.T.; Boschetti, L.; Falkowski, M.J.; Greenberg, J.A.; Anderson, J.W.; et al. Remote sensing the vulnerability of vegetation in natural terrestrial ecosystems. Remote Sens. Environ. 2014, 154, 322–337. [Google Scholar] [CrossRef]
- Li, X.; Chen, W.Y.; Sanesi, G.; Lafortezza, R. Remote sensing in urban forestry: Recent applications and future directions. Remote Sens. 2019, 11, 1144. [Google Scholar] [CrossRef] [Green Version]
- Jarron, L.R.; Coops, N.C.; MacKenzie, W.H.; Tompalski, P.; Dykstra, P. Detection of sub-canopy forest structure using airborne LiDAR. Remote Sens. Environ. 2020, 244, 111770. [Google Scholar] [CrossRef]
- Popescu, S.C. Estimating biomass of individual pine trees using airborne lidar. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]




| Metric | Minimum | Maximum | Median | Mean | SD |
|---|---|---|---|---|---|
| Tree Density (trees/ha) | 18.8 | 1846.7 | 508.8 | 542.7 | 386.7 |
| Basal Area (m2/ha) | 0.4 | 111.6 | 31.3 | 31.9 | 23.0 |
| DBH (cm) | 5.1 | 137.2 | 19.1 | 23.2 | 14.6 |
| Height (m) | 6.1 | 40.9 | 14 | 15.9 | 7.6 |
| Method | ALS Pulse Density (ppm2) | Input Parameters | Total # of Method Approaches |
|---|---|---|---|
| ForestView® | 8 | Default CHM and point cloud metrics | 1 |
| 22 | Default CHM and point cloud metrics | 1 | |
| [19] | 8 | 3 spacing thresholds | 3 |
| 22 | 3 spacing thresholds | 3 | |
| lidR LMF | 8 | 2 CHM resolutions, 3 SW sizes, 2 SW shapes | 12 |
| 22 | 3 CHM resolutions, 3 SW sizes, 2 SW shapes | 18 | |
| rLiDAR LMF | 8 | 2 CHM resolutions, 3 SW sizes, 1 SW shape | 6 |
| 22 | 3 CHM resolutions, 3 SW sizes, 1 SW shape | 9 | |
| SWA | 8 | 2 CHM resolutions | 2 |
| 22 | 3 CHM resolutions | 3 | |
| VWF | 8 | 2 CHM resolutions, 2 SW size algorithms | 4 |
| 22 | 3 CHM resolutions, 2 SW size algorithms | 6 | |
| Watershed | 8 | 2 CHM resolutions | 2 |
| 22 | 3 CHM resolutions | 3 |
| Metric | Description | Equation |
|---|---|---|
| CC | ALS-derived canopy cover | |
| TPH | Trees per hectare | |
| BA | Basal area (m2) per hectare | |
| SDI | Stand density index |
| Method and Input Parameters | 8 ppm2 ALS Data | 22 ppm2 ALS Data | ||||
|---|---|---|---|---|---|---|
| Recall | Precision | F-Score | Recall | Precision | F-Score | |
| ForestView® | ||||||
| Default CHM and point cloud metrics | 0.39 | 0.58 | 0.45 | 0.47 | 0.62 | 0.50 |
| [19] | ||||||
| Spacing thresh: 0.75 m | 0.46 | 0.55 | 0.48 | 0.46 | 0.56 | 0.47 |
| Spacing thresh: 1.25 m | 0.40 | 0.67 | 0.48 | 0.41 | 0.64 | 0.47 |
| Spacing thresh: 1.75 m | 0.35 | 0.73 | 0.46 | 0.37 | 0.67 | 0.45 |
| lidR LMF | ||||||
| CHM: 10 cm, SW: circular 1.5 m | 0.55 | 0.44 | 0.45 | |||
| CHM: 10 cm, SW: circular 2.5 m | 0.46 | 0.66 | 0.51 | |||
| CHM: 10 cm, SW: circular 3.5 m | 0.40 | 0.71 | 0.48 | |||
| CHM: 10 cm, SW: square 1.5 m | 0.52 | 0.51 | 0.48 | |||
| CHM: 10 cm, SW: square 2.5 m | 0.43 | 0.69 | 0.50 | |||
| CHM: 10 cm, SW: square 3.5 m | 0.37 | 0.72 | 0.46 | |||
| CHM: 25 cm, SW: circular 1.5 m | 0.48 | 0.51 | 0.47 | 0.53 | 0.46 | 0.46 |
| CHM: 25 cm, SW: circular 2.5 m | 0.41 | 0.71 | 0.51 | 0.45 | 0.66 | 0.51 |
| CHM: 25 cm, SW: circular 3.5 m | 0.36 | 0.74 | 0.47 | 0.40 | 0.71 | 0.48 |
| CHM: 25 cm, SW: square 1.5 m | 0.45 | 0.61 | 0.50 | 0.48 | 0.58 | 0.49 |
| CHM: 25 cm, SW: square 2.5 m | 0.38 | 0.73 | 0.49 | 0.42 | 0.70 | 0.49 |
| CHM: 25 cm, SW: square 3.5 m | 0.33 | 0.76 | 0.44 | 0.36 | 0.71 | 0.44 |
| CHM: 50 cm, SW: circular 1.5 m | 0.47 | 0.54 | 0.48 | 0.50 | 0.60 | 0.52 |
| CHM: 50 cm, SW: circular 2.5 m | 0.41 | 0.69 | 0.50 | 0.44 | 0.71 | 0.52 |
| CHM: 50 cm, SW: circular 3.5 m | 0.36 | 0.73 | 0.47 | 0.40 | 0.73 | 0.49 |
| CHM: 50 cm, SW: square 1.5 m | 0.47 | 0.54 | 0.48 | 0.50 | 0.60 | 0.52 |
| CHM: 50 cm, SW: square 2.5 m | 0.39 | 0.71 | 0.50 | 0.43 | 0.71 | 0.51 |
| CHM: 50 cm, SW: square 3.5 m | 0.34 | 0.74 | 0.45 | 0.38 | 0.73 | 0.47 |
| rLiDAR LMF | ||||||
| CHM: 10 cm, SW: square 1.5 m | 0.44 | 0.58 | 0.49 | |||
| CHM: 10 cm, SW: square 2.5 m | 0.22 | 0.69 | 0.36 | |||
| CHM: 10 cm, SW: square 3.5 m | 0.13 | 0.71 | 0.24 | |||
| CHM: 25 cm, SW: square 1.5 m | 0.41 | 0.63 | 0.49 | 0.44 | 0.63 | 0.50 |
| CHM: 25 cm, SW: square 2.5 m | 0.28 | 0.72 | 0.42 | 0.28 | 0.70 | 0.41 |
| CHM: 25 cm, SW: square 3.5 m | 0.21 | 0.75 | 0.35 | 0.17 | 0.68 | 0.30 |
| CHM: 50 cm, SW: square 1.5 m | 0.45 | 0.55 | 0.48 | 0.50 | 0.61 | 0.52 |
| CHM: 50 cm, SW: square 2.5 m | 0.35 | 0.71 | 0.47 | 0.38 | 0.72 | 0.49 |
| CHM: 50 cm, SW: square 3.5 m | 0.25 | 0.74 | 0.39 | 0.25 | 0.70 | 0.39 |
| SWA | ||||||
| CHM: 10 cm | 0.39 | 0.71 | 0.49 | |||
| CHM: 25 cm | 0.33 | 0.68 | 0.42 | 0.38 | 0.70 | 0.47 |
| CHM: 50 cm | 0.27 | 0.68 | 0.38 | 0.34 | 0.71 | 0.43 |
| VWF | ||||||
| CHM: 10 cm, SW: default function | 0.44 | 0.67 | 0.49 | |||
| CHM: 25 cm, SW: default function | 0.39 | 0.72 | 0.49 | 0.44 | 0.67 | 0.49 |
| CHM: 50 cm, SW: default function | 0.39 | 0.71 | 0.49 | 0.43 | 0.71 | 0.50 |
| CHM: 10 cm, SW: Palouse Range function | 0.33 | 0.71 | 0.42 | |||
| CHM: 25 cm, SW: Palouse Range function | 0.31 | 0.77 | 0.42 | 0.34 | 0.70 | 0.42 |
| CHM: 50 cm, SW: Palouse Range function | 0.31 | 0.76 | 0.42 | 0.34 | 0.73 | 0.42 |
| Watershed | ||||||
| CHM: 10 cm | 0.62 | 0.30 | 0.37 | |||
| CHM: 25 cm | 0.51 | 0.40 | 0.42 | 0.51 | 0.52 | 0.48 |
| CHM: 50 cm | 0.41 | 0.51 | 0.45 | 0.44 | 0.68 | 0.52 |
| Accuracy Metric | Canopy Cover | Trees per Hectare | Basal Area per Hectare | Stand Density Index | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 | Slope | p | r2 | Slope | p | r2 | Slope | p | r2 | Slope | p | |
| 8 ppm2 ALS dataset | ||||||||||||
| Recall | 0.05 | −0.15 | 0.07 | 0.19 | −0.0002 | <0.01 | 0.16 | −0.003 | <0.01 | 0.20 | −0.0002 | <0.01 |
| Precision | 0.01 | 0.06 | 0.43 | 0.07 | 0.0001 | <0.05 | 0.01 | −0.0002 | 0.82 | 0.01 | −0.0001 | 0.87 |
| F−score | 0.08 | −0.16 | <0.05 | 0.14 | −0.0001 | <0.01 | 0.19 | −0.003 | <0.01 | 0.21 | −0.0001 | <0.01 |
| RMSEhgt | 0.01 | −0.05 | 0.66 | 0.03 | −0.0001 | 0.17 | 0.02 | −0.001 | 0.28 | 0.02 | −0.0001 | 0.22 |
| Biashgt | 0.02 | 0.33 | 0.22 | 0.01 | 0.0001 | 0.49 | 0.20 | 0.01 | <0.01 | 0.16 | 0.0005 | <0.01 |
| 22 ppm2 ALS dataset | ||||||||||||
| Recall | 0.16 | −0.29 | <0.01 | 0.27 | −0.0002 | <0.01 | 0.38 | −0.006 | <0.01 | 0.40 | −0.0003 | <0.01 |
| Precision | 0.01 | −0.002 | 0.98 | 0.02 | −0.0001 | 0.19 | 0.02 | −0.001 | 0.17 | 0.01 | −0.0005 | 0.36 |
| F−score | 0.08 | −0.21 | <0.05 | 0.13 | −0.0001 | <0.01 | 0.30 | −0.004 | <0.01 | 0.30 | −0.0002 | <0.01 |
| RMSEhgt | 0.07 | 0.26 | <0.05 | 0.03 | 0.0001 | 0.16 | 0.23 | 0.005 | <0.01 | 0.20 | 0.0002 | <0.01 |
| Biashgt | 0.07 | 0.42 | <0.05 | 0.03 | 0.0002 | 0.13 | 0.20 | 0.009 | <0.01 | 0.17 | 0.0004 | <0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sparks, A.M.; Corrao, M.V.; Smith, A.M.S. Cross-Comparison of Individual Tree Detection Methods Using Low and High Pulse Density Airborne Laser Scanning Data. Remote Sens. 2022, 14, 3480. https://doi.org/10.3390/rs14143480
Sparks AM, Corrao MV, Smith AMS. Cross-Comparison of Individual Tree Detection Methods Using Low and High Pulse Density Airborne Laser Scanning Data. Remote Sensing. 2022; 14(14):3480. https://doi.org/10.3390/rs14143480
Chicago/Turabian StyleSparks, Aaron M., Mark V. Corrao, and Alistair M. S. Smith. 2022. "Cross-Comparison of Individual Tree Detection Methods Using Low and High Pulse Density Airborne Laser Scanning Data" Remote Sensing 14, no. 14: 3480. https://doi.org/10.3390/rs14143480
APA StyleSparks, A. M., Corrao, M. V., & Smith, A. M. S. (2022). Cross-Comparison of Individual Tree Detection Methods Using Low and High Pulse Density Airborne Laser Scanning Data. Remote Sensing, 14(14), 3480. https://doi.org/10.3390/rs14143480