Comparing CNNs and Random Forests for Landsat Image Segmentation Trained on a Large Proxy Land Cover Dataset
Abstract
:1. Introduction
- U-Net CNNs can produce land cover maps of comparable or better accuracy than RFs; and
- An imperfect proxy variable can be used to train a well-performing machine-learning classifier.
2. Material and Methods
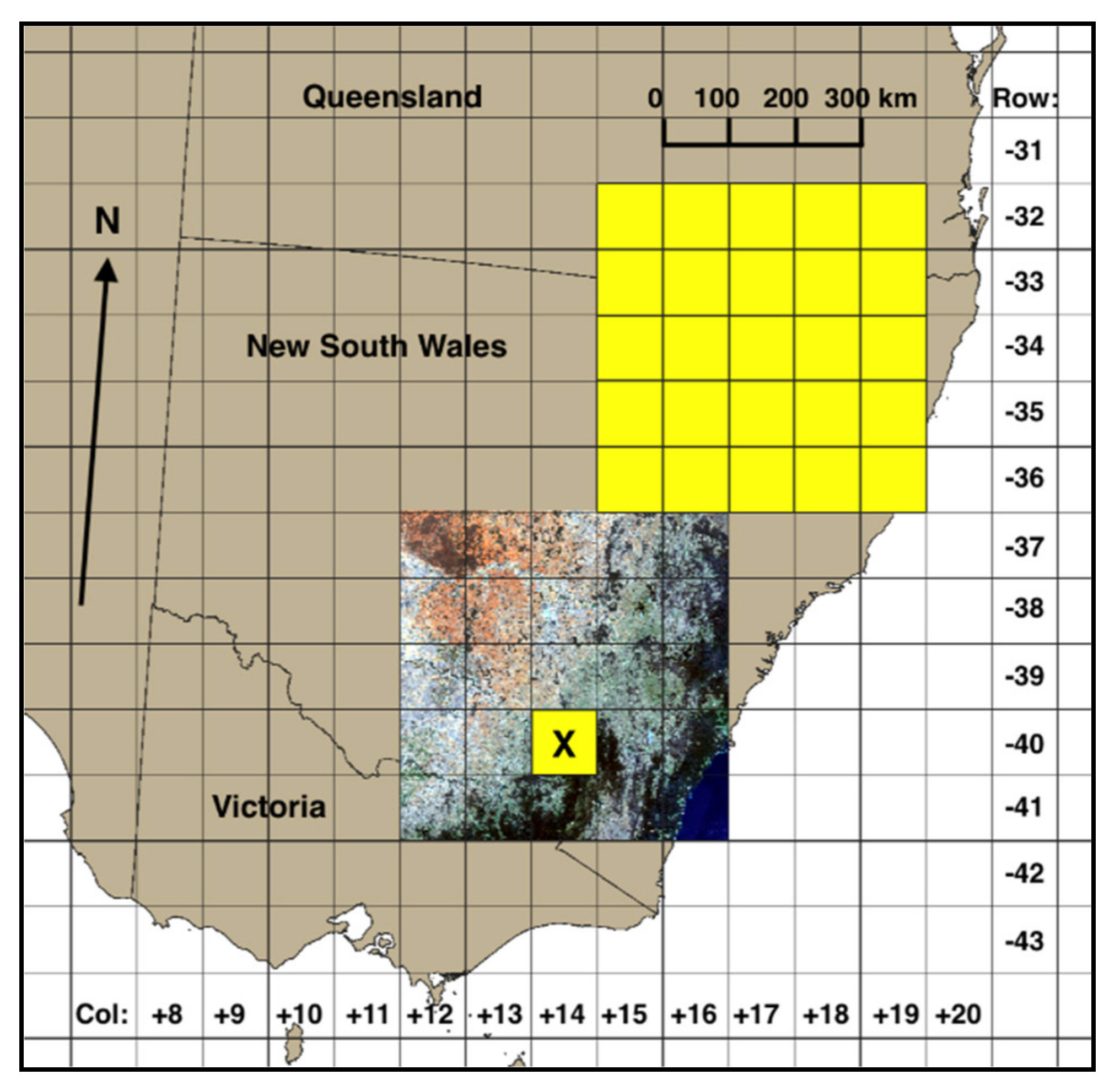
2.1. Input Data Preparation
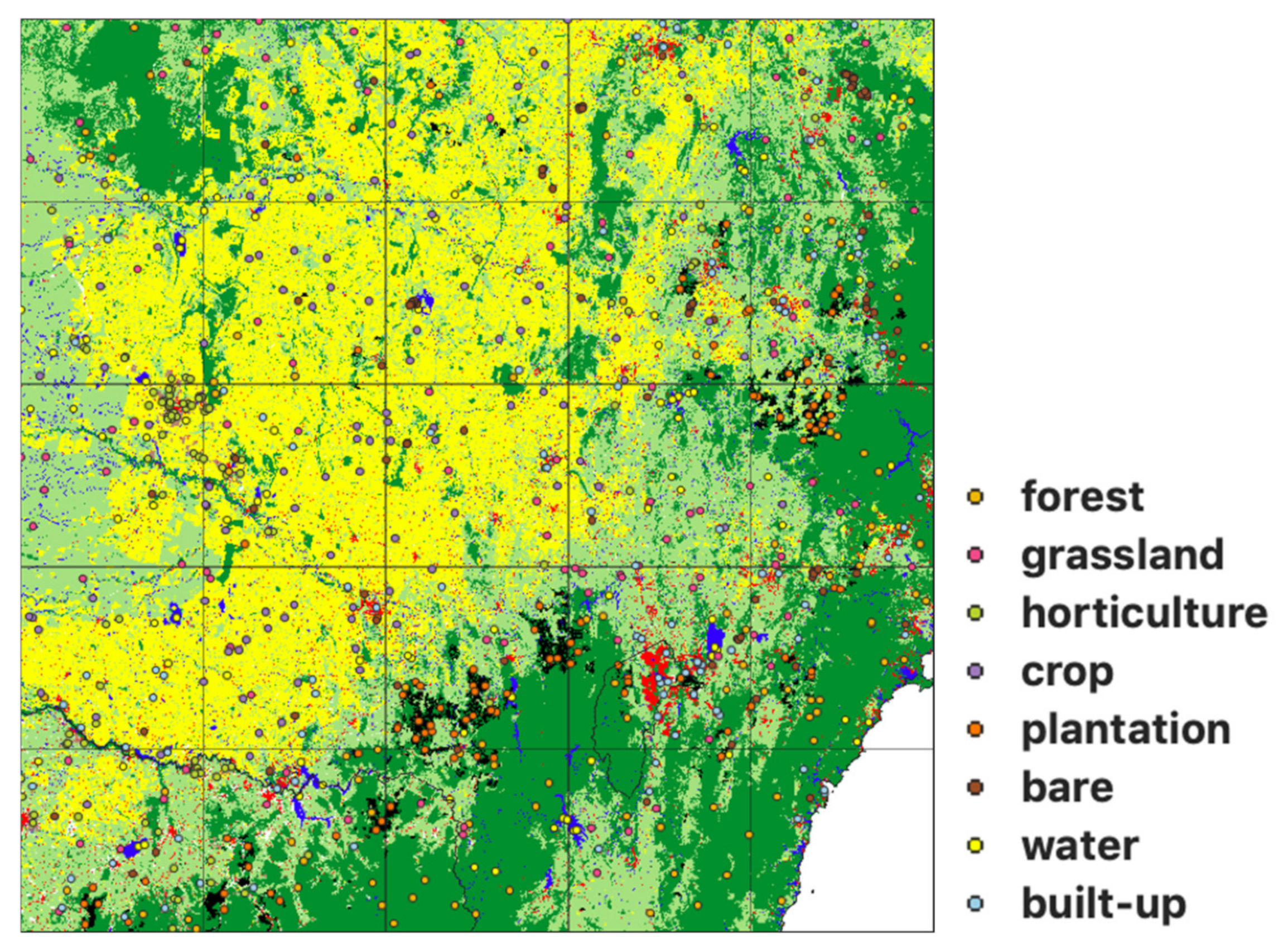
2.2. Label Data Preparation
2.3. Modelling Approach
- Autoencoder: a simple six-layer neural network that learns a representation of reduced dimensionality then decodes it to the original image dimensions. The last layer of the neural network predicts a class label of the pixel of interest (Figure S1);
- U-Net: a CNN encoder–decoder model where symmetric connections between the convolution and deconvolution layers are used to capture spatial context and build a final segmented image of the same resolution as the input. The depth of the base U-Net model was five, with dimension reductions of 128 × 128, 64 × 64, 32 × 32, 16 × 16, 8 × 8 (Figure 3). The CNNs were coded using the segmentation modelling software developed by [47]; and
- Random Forests (RF): an algorithm for classification and regression in which decision tree classifiers are fitted on various subsets of the dataset based on an attribute test. Predictions for each pixel were output by the RF classifier trees based on the class label that received majority support.
- Only one of the spectral bands, i.e., red (R), green (G), blue (B), near-infrared (N), or one of the two shortwave infrared bands (S1 and S2);
- Only one of the TMADs bands: Euclidean distance (edev), Spectral distance (sdev) and Bray–Curtis dissimilarity (bcdev);
- Selected combinations of three bands: RGB, NS1S2 and TMADs;
- The three visible bands plus near-infrared (RGBN);
- All six bands; and
- All six bands and three TMADs.
3. Results
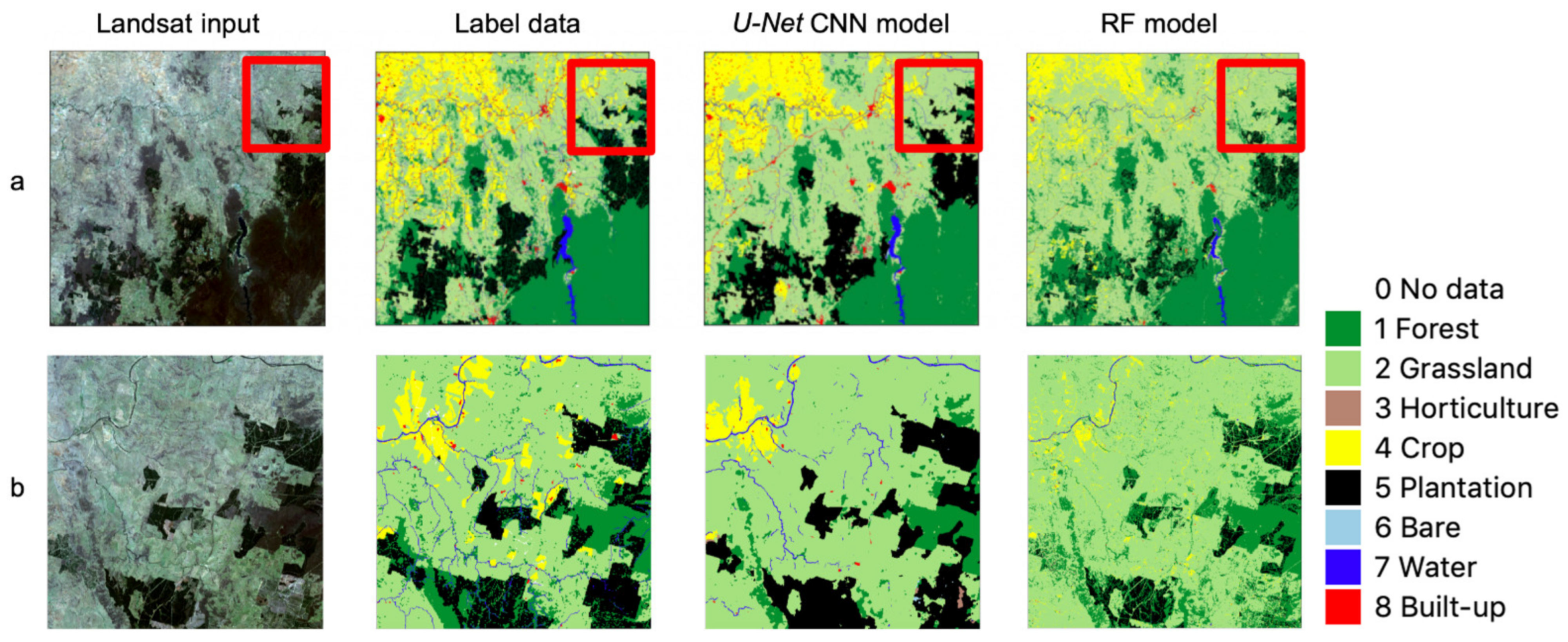
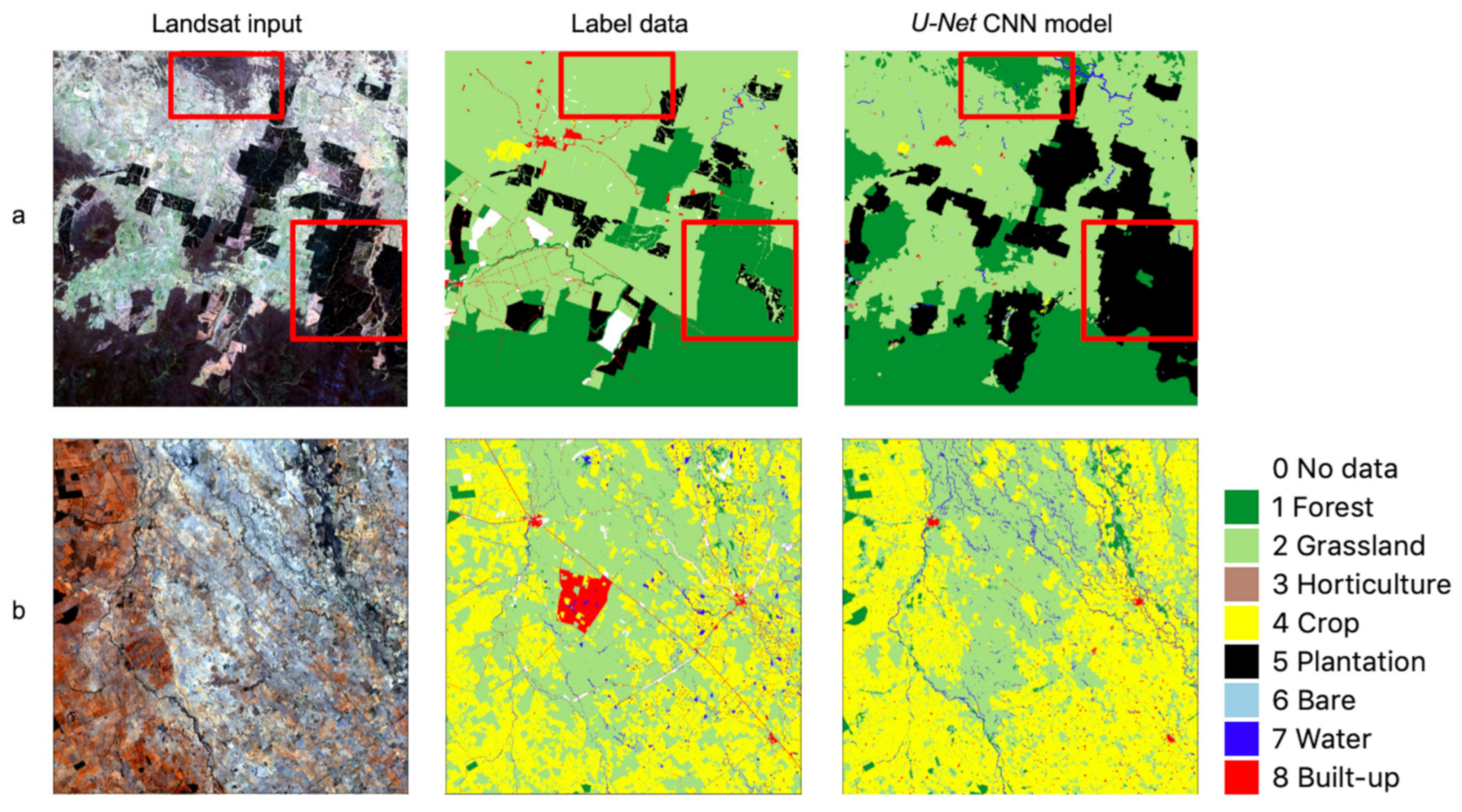
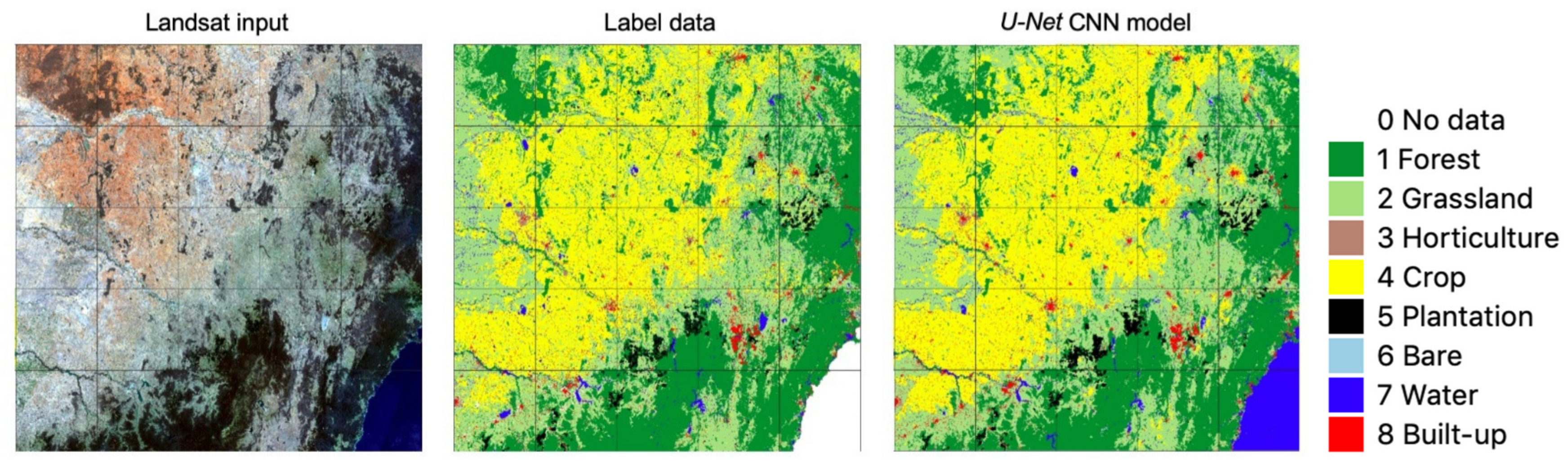
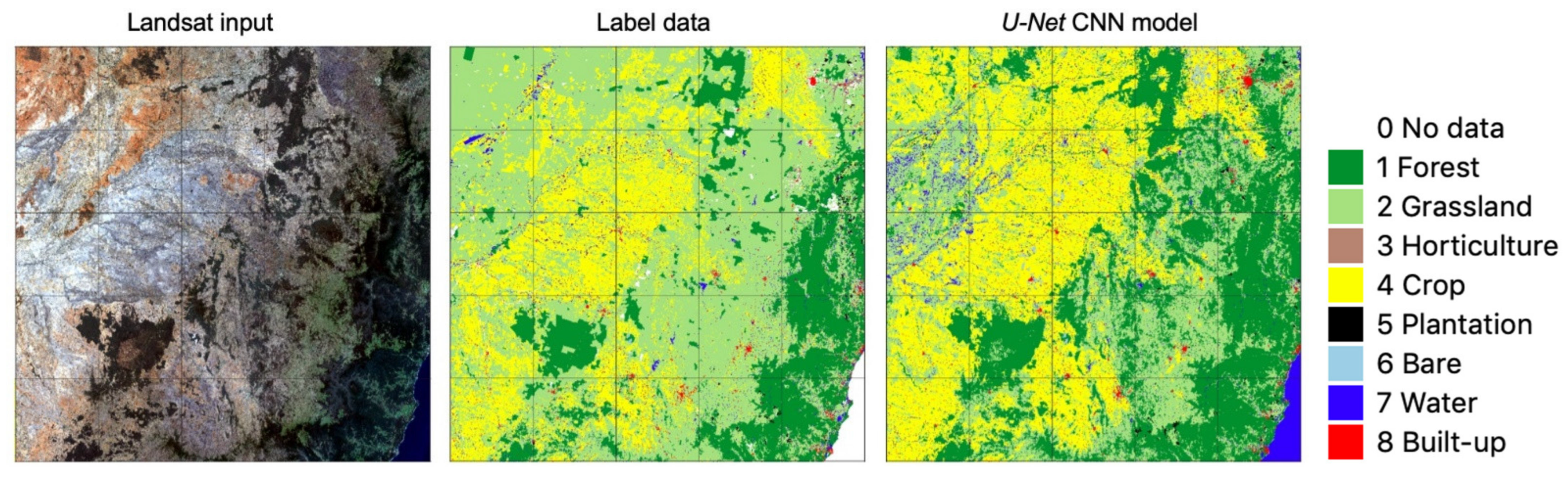
3.1. Accuracy of Label Data and CNN Model Output
4. Discussion
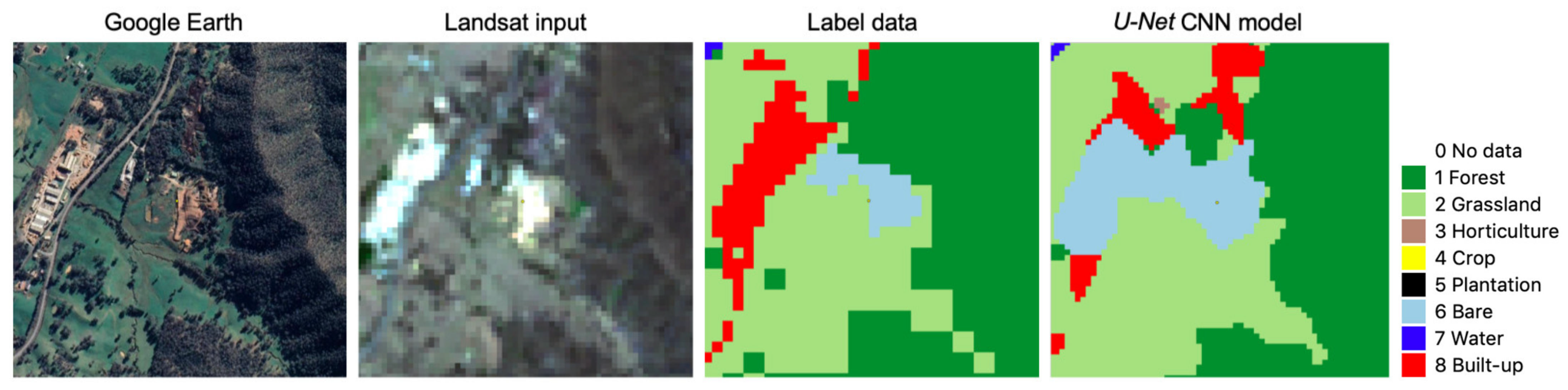
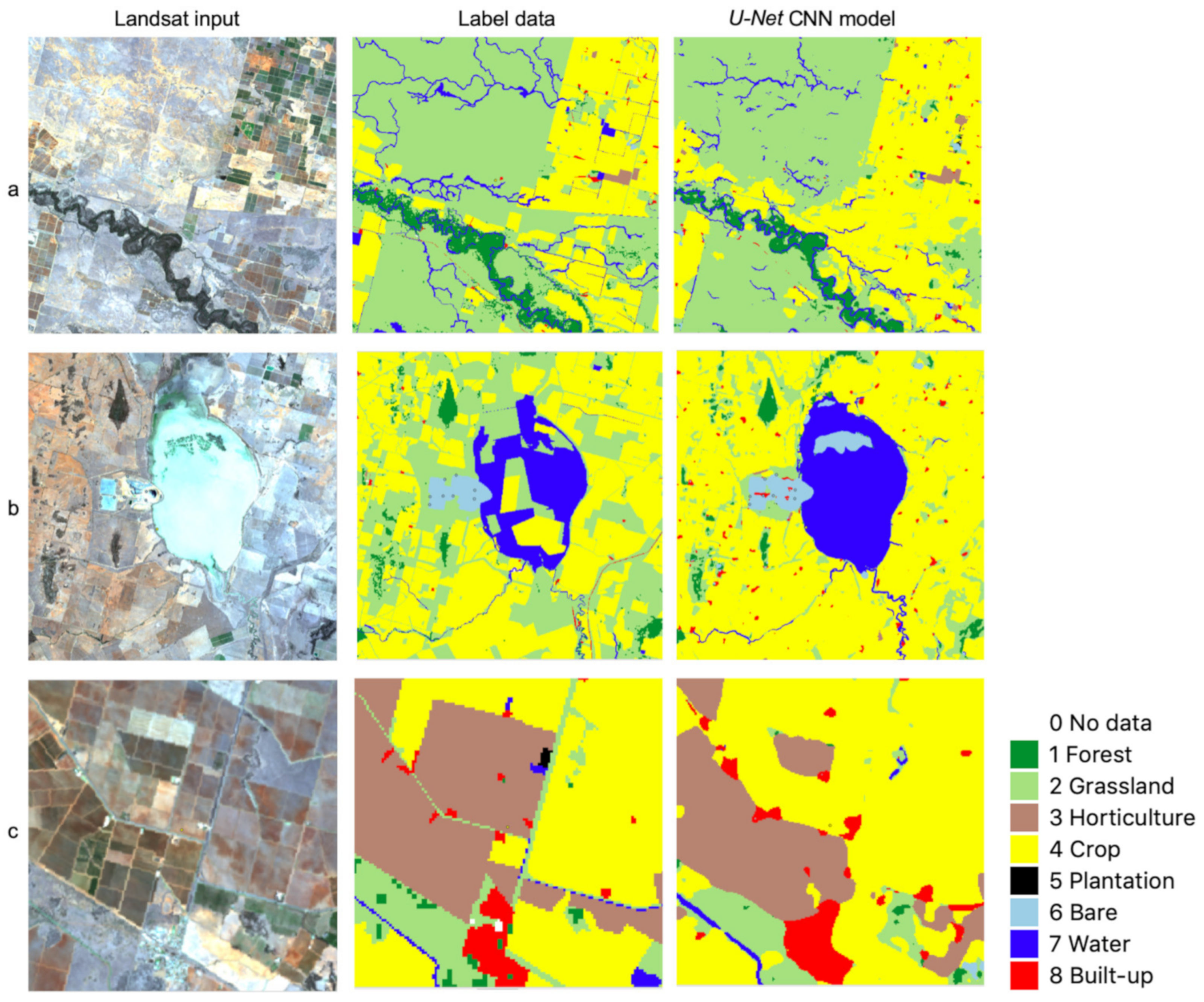
4.1. Some Observations on U-Net CNN Model Output
4.2. Spatial Transferability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ellis, E.C. Anthropogenic transformation of the terrestrial biosphere. Proc. R. Soc. A 2011, 369, 1010–1035. [Google Scholar] [CrossRef]
- Di Gregorio, A.; Jansen, L.J.M. Land cover classification system: Classification concepts and user manual: LCCS, Software version 2. In 8 Environment and Natural Resources Series; Food and Agriculture Organization of the United Nations: Rome, Italy, 2005. [Google Scholar]
- Bojinski, S.; Verstraete, M.; Peterson, T.C.; Richter, C.; Simmons, A.; Zemp, M. The Concept of Essential Climate Variables in Support of Climate Research, Applications, and Policy. Bull. Am. Meteorol. Soc. 2014, 95, 1431–1443. [Google Scholar] [CrossRef]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential Biodiversity Variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Lesslie, R.; Mewett, J. Reprint: Land Use and Management—The Australian Context. In Land Use in Australia: Past, Present and Future; Thackway, R., Ed.; ANU Press: Canberra, Australia, 2018; Available online: http://press-files.anu.edu.au/downloads/press/n4113/pdf/ch03.pdf (accessed on 17 May 2022).
- Lowell, K.; Woodgate, P.; Jones, S.; Richards, G.P. National Carbon Accounting System: Continuous Improvement of the National Carbon Accounting System Land Cover Change Mapping; Technical Report No. 39; Australian Greenhouse Office: Canberra, Australia, 2003. Available online: http://pandora.nla.gov.au/pan/23322/20050218-0000/www.greenhouse.gov.au/ncas/reports/pubs/tr39final.pdf (accessed on 17 May 2022).
- Furby, S.L.; Caccetta, P.A.; Wallace, J.F.; Lehmann, E.A.; Zdunic, K. Recent development in vegetation monitoring products from Australia’s National Carbon Accounting System. In Proceedings of the 2009 IEEE International Geoscience and Remote Sensing Symposium, Cape Town, South Africa, 12–17 July 2009; pp. IV-276–IV-279. [Google Scholar] [CrossRef]
- ABARES. Forests of Australia. 2018. Available online: https://www.agriculture.gov.au/abares/forestsaustralia/forest-data-maps-and-tools/spatial-data/forest-cover (accessed on 17 May 2022).
- Thackway, R.; Lee, A.; Donohue, R.; Keenan, R.J.; Wood, M. Vegetation information for improved natural resource management in Australia. Landsc. Urban Plan. 2007, 79, 127–136. [Google Scholar] [CrossRef]
- NVIS Technical Working Group. Australian Vegetation Attribute Manual: National Vegetation Information System, Version 7.0.; Bolton, M.P., deLacey, C., Bossard, K.B., Eds.; Department of the Environment and Energy: Canberra, Australia, 2017. Available online: https://www.environment.gov.au/land/publications/australian-vegetation-attribute-manual-version-7 (accessed on 17 May 2022).
- Mutendeudzi, M.; Read, S.; Howell, C.; Davey, S.; Clancy, T. Improving Australia’s Forest Area Estimate Using a ‘Multiple Lines of Evidence’ Approach; Technical Report 13.07; Australian Bureau of Agricultural and Resource Economics and Sciences: Canberra, Australia, 2013. Available online: https://www.agriculture.gov.au/sites/default/files/documents/ImpAustForCovStats_20131119_v1.0.0.pdf (accessed on 17 May 2022).
- Van Dijk, A.; Summers, D. Australia’s Environment in 2018. 2018. Available online: http://wald.anu.edu.au/australias-environment/ (accessed on 17 May 2022).
- Thackway, R. (Ed.) Land Use in Australia: Past, Present and Future; ANU Press: Canberra, Australia, 2018; Available online: https://press.anu.edu.au/publications/land-use-australia (accessed on 17 May 2022).
- Macintosh, A. The National Greenhouse Accounts and Land Clearing: Do the Numbers Stack Up? Australia Institute Research Paper No. 38; Australia Institute: Canberra, Australia, 2007; Available online: https://australiainstitute.org.au/wp-content/uploads/2020/12/WP93_8.pdf (accessed on 17 May 2022).
- SLATS. Land Cover Change in Queensland. 2018. Available online: https://www.qld.gov.au/__data/assets/pdf_file/0031/91876/landcover-change-in-queensland-2016-17-and-2017-18.pdf (accessed on 17 May 2022).
- Atkinson, P.M.; Tatnall, A.R.L. Introduction Neural networks in remote sensing. Int. J. Remote Sens. 1997, 18, 699–709. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pelletier, C.; Valero, S.; Inglada, J.; Champion, N.; Dedieu, G. Assessing the robustness of Random Forests to map land cover with high resolution satellite image time series over large areas. Remote Sens. Environ. 2016, 187, 156–168. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random Forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A review of supervised object-based land-cover image classification. ISPRS J. Photogramm. Remote Sens. 2017, 130, 277–2933. [Google Scholar] [CrossRef]
- LeCun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional networks and applications in vision. In Proceedings of 2010 IEEE International Symposium on Circuits and Systems, Paris, France, 30 May–2 June 2010; pp. 253–256. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J.; Carvalhais, N.; Prabhat. Deep learning and process understanding for data-driven Earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef]
- Phiri, D.; Morgenroth, J. Developments in Landsat Land Cover Classification Methods: A Review. Remote Sens. 2017, 9, 967. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015. MICCAI 2015; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Rezaee, M.; Mahdianpari, M.; Zhang, Y.; Salehi, B. Deep Convolutional Neural Network for Complex Wetland Classification Using Optical Remote Sensing Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3030–3039. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Mountrakis, G.; Li, J.; Lu, X.; Hellwich, O. Deep learning for remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2018, 145, 1–2. [Google Scholar] [CrossRef]
- Griffiths, P.; van der Linden, S.; Kuemmerle, T.; Hostert, P. A Pixel-Based Landsat Compositing Algorithm for Large Area Land Cover Mapping. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 2088–2101. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.; Hobart, G.; Luther, J.; Hermosilla, T.; Griffiths, P.; Coops, N.; Hall, R.J.; Hostert, P.; Dyk, A.; et al. Pixel-Based Image Compositing for Large-Area Dense Time Series Applications and Science. Can. J. Remote Sens. 2014, 40, 192–212. [Google Scholar] [CrossRef]
- Corbane, C.; Politis, P.; Kempeneers, P.; Simonetti, D.; Soille, P.; Burger, A.; Pesaresi, M.; Sabo, F.; Syrris, V.; Kemper, T. A global cloud free pixel- based image composite from Sentinel-2 data. Data Brief 2020, 31, 105737. [Google Scholar] [CrossRef]
- Lucas, R.; Mueller, N.; Siggins, A.; Owers, C.; Clewley, D.; Bunting, P.; Kooymans, C.; Tissott, B.; Lewis, B.; Lymburner, L.; et al. Land Cover Mapping using Digital Earth Australia. Data 2019, 4, 143. [Google Scholar] [CrossRef]
- Geoscience Australia. Digital Earth Australia—Public Data—Surface Reflectance 25m Geomedian v2.1.0. 2019. Available online: https://data.dea.ga.gov.au/?prefix=geomedian-australia/v2.1.0/ (accessed on 17 May 2022).
- Roberts, D.; Mueller, N.; Mcintyre, A. High-Dimensional Pixel Composites from Earth Observation Time Series. IEEE Trans. Geosci. Remote Sens. 2017, 55, 6254–6264. [Google Scholar] [CrossRef]
- Roberts, D.; Dunn, B.; Mueller, N. Open Data Cube Products Using High-Dimensional Statistics of Time Series. In Proceedings of the IGARSS 2018. 2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 8647–8650. [Google Scholar] [CrossRef]
- Van Der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef]
- ABARES. Catchment Scale Land Use of Australia—Update December 2018. 2019. Available online: https://www.agriculture.gov.au/abares/aclump/land-use/catchment-scale-land-use-of-australia-update-december-2018 (accessed on 17 May 2022).
- ACLUMP. Land Use Mapping Technical Specifications. 2016. Available online: http://www.agriculture.gov.au/abares/aclump/land-use/mapping-technical-specifications (accessed on 17 May 2022).
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A Survey on Deep Transfer Learning. In Artificial Neural Networks and Machine Learning—ICANN 2018. ICANN 2018; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11141. [Google Scholar] [CrossRef]
- Alshehhi, R.; Marpu, P.R.; Woon, W.L.; Mura, M.D. Simultaneous extraction of roads and buildings in remote sensing imagery with convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2017, 130, 139–149. [Google Scholar] [CrossRef]
- Marmanis, D.; Datcu, M.; Esch, T.; Stilla, U. Deep Learning Earth Observation Classification Using ImageNet Pretrained Networks. IEEE Geosci. Remote. Sens. Lett. 2016, 13, 105–109. [Google Scholar] [CrossRef]
- Penatti, O.A.B.; Nogueira, K.; Dos Santos, J.A. Do deep features generalize from everyday objects to remote sensing and aerial scenes domains? In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; pp. 44–51. [Google Scholar] [CrossRef]
- Yakubovskiy, P. Segmentation Models. GitHub Repository. 2019. Available online: https://github.com/qubvel/segmentation_models (accessed on 17 May 2022).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. Available online: https://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html (accessed on 17 May 2022).
- Ulmas, P.; Liiv, I. Segmentation of Satellite Imagery using U-Net Models for Land Cover Classification. arXiv 2020, arXiv:2003.02899. [Google Scholar]
- Benbahria, Z.; Smiej, M.F.; Sebari, I.; Hajji, H. Land cover intelligent mapping using transfer learning and semantic segmentation. In Proceedings of the 2019 7th Mediterranean Congress of Telecommunications (CMT), Fès, Morocco, 24–25 October 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Walsh, E.; Bessardon, G.; Gleeson, E.; Ulmas, P. Using machine learning to produce a very high resolution land-cover map for Ireland. Adv. Sci. Res. 2021, 18, 65–87. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med. Phys. 2019, 46, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Chollet, F. Keras: The Python Deep Learning Library. 2015. Available online: https://keras.io/ (accessed on 17 May 2022).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Foody, G.M. Status of land cover classification accuracy assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Spatial Services, NSW Department of Finance and Services. Spatial Information eXchange (SIX) Maps. 2022. Available online: http://maps.six.nsw.gov.au/ (accessed on 17 May 2022).
- Thomlinson, J.R.; Bolstad, P.V.; Cohen, W.B. Coordinating Methodologies for Scaling Landcover Classifications from Site-Specific to Global: Steps toward Validating Global Map Products. Remote Sens. Environ. 1999, 70, 16–28. [Google Scholar] [CrossRef]
- Stoian, A.; Poulain, V.; Inglada, J.; Poughon, V.; Derksen, D. Land Cover Maps Production with High Resolution Satellite Image Time Series and Convolutional Neural Networks: Adaptations and Limits for Operational Systems. Remote Sens. 2019, 11, 1986. [Google Scholar] [CrossRef]
- Pollatos, V.; Kouvaras, L.; Charou, E. Land Cover Semantic Segmentation Using ResUNet. In Proceedings of the Workshops of the 11th EETN Conference on Artificial Intelligence 2020, Athens, Greece, 2–4 September 2020; pp. 55–65. Available online: http://ceur-ws.org/Vol-2844/ainst2.pdf (accessed on 17 May 2022).
- Syrris, V.; Hasenohr, P.; Delipetrev, B.; Kotsev, A.; Kempeneers, P.; Soille, P. Evaluation of the Potential of Convolutional Neural Networks and Random Forests for Multi-Class Segmentation of Sentinel-2 Imagery. Remote Sens. 2019, 11, 907. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar] [CrossRef]
- Reina, G.A.; Panchumarthy, R.; Thakur, S.P.; Bastidas, A.; Bakas, S. Systematic Evaluation of Image Tiling Adverse Effects on Deep Learning Semantic Segmentation. Front. Neurosci. 2020, 14, 65. [Google Scholar] [CrossRef]


| Dataset Splits | Count | Percent |
|---|---|---|
| Train | 19,086 | 81% |
| Validation | 3532 | 15% |
| Test | 1024 | 4% |
| TOTAL | 23,642 | 100% |
| Class Count | Label Class | Producer’s Acc. (Recall) | Omission Errors | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vis. Id. Class | Forest | Grassland | Horticulture | Crop | Plantation | Bare | Water | Built-Up | Total | Total | ||
| Forest | 93 | 8 | 5 | 2 | 4 | 12 | 6 | 130 | 71.5% | 28.5% | 100.0% | |
| Grassland | 5 | 80 | 9 | 5 | 5 | 9 | 25 | 33 | 171 | 46.8% | 53.2% | 100.0% |
| Horticulture | 1 | 76 | 77 | 98.7% | 1.3% | 100.0% | ||||||
| Crop | 1 | 10 | 6 | 92 | 1 | 2 | 3 | 115 | 80.0% | 20.0% | 100.0% | |
| Plantation | 1 | 1 | 92 | 2 | 1 | 97 | 94.8% | 5.2% | 100.0% | |||
| Bare | 1 | 84 | 2 | 87 | 96.6% | 3.4% | 100.0% | |||||
| Water | 2 | 57 | 59 | 96.6% | 3.4% | 100.0% | ||||||
| Built-up | 1 | 3 | 1 | 1 | 58 | 64 | 90.6% | 9.4% | 100.0% | |||
| Total | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 800 | 79.0% | Overall Accuracy | |
| User’s Acc. (Precision) | 93.0% | 80.0% | 76.0% | 92.0% | 92.0% | 84.0% | 57.0% | 58.0% | ||||
| Commission Errors | 7.0% | 20.0% | 24.0% | 8.0% | 8.0% | 16.0% | 43.0% | 42.0% | ||||
| Total | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 76.0% | Kappa | ||
| F1 | 80.9% | 59.0% | 85.9% | 85.6% | 93.4% | 89.8% | 71.7% | 70.7% | 74.7% | Weighted-Mean F1 | ||
| Class Count | Model Prediction | Producer’s Acc. (Recall) | Omission Errors | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vis. Id. class | Forest | Grassland | Horticulture | Crop | Plantation | Bare | Water | Built-Up | Total | Total | ||
| Forest | 101 | 9 | 2 | 3 | 4 | 3 | 2 | 6 | 130 | 77.7% | 22.3% | 100.0% |
| Grassland | 4 | 144 | 3 | 11 | 2 | 1 | 1 | 5 | 171 | 84.2% | 15.8% | 100.0% |
| Horticulture | 1 | 10 | 52 | 12 | 2 | 77 | 67.5% | 32.5% | 100.0% | |||
| Crop | 6 | 109 | 115 | 94.8% | 5.2% | 100.0% | ||||||
| Plantation | 5 | 6 | 1 | 1 | 82 | 1 | 1 | 97 | 84.5% | 15.5% | 100.0% | |
| Bare | 2 | 2 | 76 | 1 | 6 | 87 | 87.4% | 12.6% | 100.0% | |||
| Water | 3 | 2 | 5 | 3 | 46 | 59 | 78.0% | 22.0% | 100.0% | |||
| Built-up | 1 | 4 | 4 | 1 | 54 | 64 | 84.4% | 15.6% | 100.0% | |||
| Total | 115 | 183 | 58 | 147 | 88 | 84 | 51 | 74 | 800 | 83.0% | Overall Accuracy | |
| User’s Acc. (Precision) | 87.8% | 78.7% | 89.7% | 74.1% | 93.2% | 90.5% | 90.2% | 73.0% | ||||
| Commission Errors | 12.2% | 21.3% | 10.3% | 25.9% | 6.8% | 9.5% | 9.8% | 27.0% | ||||
| Total | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 80.2% | Kappa | ||
| F1 | 82.4% | 81.4% | 77.0% | 83.2% | 88.6% | 88.9% | 83.6% | 78.3% | 82.3% | Weighted-Mean F1 | ||
| Land Cover | Vis. Id. Class (A) | Label Correct (B) | Class Accuracy (B/A) | Model Correct (C) | Class Accuracy (C/A) | Model Correct When Label Correct (D) | Class Accuracy (D/B) |
|---|---|---|---|---|---|---|---|
| Forest | 130 | 93 | 71.5% | 101 | 77.7% | 83 | 89.2% |
| Grassland | 171 | 80 | 46.8% | 144 | 84.2% | 76 | 95.0% |
| Horticulture | 77 | 76 | 98.7% | 52 | 67.5% | 51 | 67.1% |
| Crop | 115 | 92 | 80.0% | 109 | 94.8% | 90 | 97.8% |
| Plantation | 97 | 92 | 94.8% | 82 | 84.5% | 78 | 84.8% |
| Bare | 87 | 84 | 96.6% | 76 | 87.4% | 73 | 86.9% |
| Water | 59 | 57 | 96.6% | 46 | 78.0% | 44 | 77.2% |
| Built-up | 64 | 58 | 90.6% | 54 | 84.4% | 51 | 87.9% |
| Total | 800 | 632 | 664 | 546 | |||
| Weighted-Mean Class Accuracy | 67.1% | 85.4% | 93.4% | ||||
| Overall Accuracy | (B/A): | 79.0% | (C/A): | 83.0% | (D/B): | 86.4% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boston, T.; Van Dijk, A.; Larraondo, P.R.; Thackway, R. Comparing CNNs and Random Forests for Landsat Image Segmentation Trained on a Large Proxy Land Cover Dataset. Remote Sens. 2022, 14, 3396. https://doi.org/10.3390/rs14143396
Boston T, Van Dijk A, Larraondo PR, Thackway R. Comparing CNNs and Random Forests for Landsat Image Segmentation Trained on a Large Proxy Land Cover Dataset. Remote Sensing. 2022; 14(14):3396. https://doi.org/10.3390/rs14143396
Chicago/Turabian StyleBoston, Tony, Albert Van Dijk, Pablo Rozas Larraondo, and Richard Thackway. 2022. "Comparing CNNs and Random Forests for Landsat Image Segmentation Trained on a Large Proxy Land Cover Dataset" Remote Sensing 14, no. 14: 3396. https://doi.org/10.3390/rs14143396










