Abstract
Remote sensing from optical radiometers in space offers a nondestructive approach to estimating above ground biomass (AGB) with high spatial and temporal resolution, but the application is challenged by cloud cover and differences in soil background and crop phenology. We present a framework based on Sentinel-2 imagery for relating the adjusted summed NDVI measurements to the AGB. The resulting R2 values for the measured and estimated AGB ranged from 0.79 to 0.98 for individual paddocks, and the R2 from a pooled dataset (multiple crops, years, and locations) was 0.86. Application of the pooled dataset model to a separate validation dataset resulted in an R2 of 0.88; however, there was a bias that resulted in the underestimation of the measured biomass. Analysis of the impacts of the gaps in the time series showed a decrease of 0.43% per gap day for the summed NDVI values. To address the impacts of clouds, we demonstrate the use of active optical and additional satellite imagery to fill the gaps due to clouds in the Sentinel-2 imagery. The framework presented results of the spatial daily estimates of the AGB and crop growth rates.
1. Introduction
Above ground biomass (AGB) is an important crop parameter, providing an indicator of yield, crop growth, and net primary productivity, while also supporting crop management decisions. Whereas traditional approaches to AGB measurement involve destructive sampling, remote sensing technologies provide a nondestructive approach to estimate the AGB at a range of spatial scales, providing a flexible means of measuring and monitoring crop growth and condition. With rapid advancements in remote sensing technology, it is now possible to have both high spatial resolution and high temporal frequency imagery. This makes paddock-specific analyses of temporal and spatial crop production dynamics, including AGB, possible, in addition to broader regional assessments.
Estimation of the AGB from satellite time series dates back to the early 1980s (e.g., [1,2]). Since then, the body of work utilizing remote sensing in crop studies has developed, encompassing approaches such as the development of empirical relationships [3,4,5], integration with crop and biophysical models [6,7,8], and the use of crop phenology metrics [9,10].
There is great diversity in the remote sensing platforms and sensors used to estimate crop AGB including multispectral [4,10], hyperspectral [3], LiDAR [11], and radar technology [12,13]. Vegetation indices such as the Normalised Vegetation Difference Index (NDVI) [14] are used extensively to derive the AGB from remotely sensed data.
The time series of vegetation indices, such as the NDVI, frequently provide the basis for calculating crop phenology metrics such as the length of the growing season and the start and the end of the growing season. Software packages such as TIMESAT [15] and CropPhenology [16] have been developed to aid the calculation of these metrics. Summed NDVI, also referred to as time-integrated NDVI, is a phenology metric demonstrated to predict AGB or net primary production. It represents the magnitude of vegetative growth within a season. At a broad scale, the summed NDVI has been used to represent regional productivity in animal and crop systems [17,18], as well as grassland systems [19], over multiple years. Araya et al. [9] used summed NDVI to examine the crop growth variability across paddocks using MODIS imagery. They found the summed NDVI to be sensitive to seasonal variability.
Several studies have reported strong correlations between the summed NDVI and the crop AGB or yield. The use of summed NDVI reduces issues associated with relating the AGB directly to the NDVI values due to the timing of AGB accumulation within crops and peak NDVI values [20]. Mirasi et al. [21] and Nakayama [18] found strong correlations between the summed NDVI and yield for a number of crops including wheat and rice. Perry et al. [10] found strong correlations between the wheat AGB and summed NDVI derived from MODIS imagery across a number of paddocks. At a smaller spatial scale (sub-paddock experimental plots), Tefera et al. [22] demonstrated a correlation between the summed NDVI and field pea biomass. Calera et al. [20] reported strong relationships between the AGB (irrigated barley and corn crops) and summed NDVI at multiple growth stages throughout the year. Mirasi et al. [21] also examined the variation in correlation through time, with a stronger correlation between the summed NDVI and crop yield at heading (half emerged) compared with the tillering and ripening growth stages. A similar increase in variance after anthesis was noted by Perry et al. [10].
Whilst these studies demonstrate the successful use of the summed NDVI for AGB estimation, there are multiple factors, which impact this, such as differences in sensors, variation in soil background reflectance, and spatial scale. Image preprocessing, including the derivation of a smoothed NDVI time series, and specific approaches to deriving a summed NDVI value also vary between studies.
One specific challenge for AGB estimation from satellites is gaps in the NDVI time series due to cloud coverage. There are a number of gap-filling approaches including Harmonic Analysis of Time Series (HANTS) and various curve fitting algorithms. The most appropriate technique is often application specific [23,24,25].
In this paper, we focus on the use of the public domain Sentinel-2 (S-2) imagery to estimate crop AGB within paddocks across multiple regions in Australia with varying climatic conditions and soil types. We use this diverse dataset to examine not only the estimation of the crop AGB from imagery but to investigate issues impacting the broader application of this approach, such as the transferability of the S-2 AGB relationships between crop types, regions, and paddocks, and the impact of image acquisition gaps on the underlying time series derivation and subsequent measurements of AGB. We examine approaches to normalize relationships across different paddocks and the integration of multiple sensors to overcome the impact of clouds on the NDVI time series. We demonstrate the impact of gaps in the image time series data and the timing of those gaps on the uncertainty in the resulting AGB estimates. The analyses are completed using real, as opposed to synthetic, data, presenting a robust analysis of the impacts for practical applications of these approaches. The framework we present to overcome these issues facilitates the ongoing development of robust approaches to AGB estimation and crop production monitoring from satellites.
2. Materials and Methods
In this section, we briefly describe the datasets used. The determination of the AGB from the NDVI time series, including the processing of the time series, is discussed in Section 3 in greater detail. The datasets evaluated consist of paired coincident measurements of AGB and cumulative NDVI from the time series from satellite imagery.
2.1. AGB Ground-Truth Datasets
The AGB data were acquired over field sites representing rain fed broad acre cereal cropping from southern and western Australia. The production areas are described as a Mediterranean climate, with an annual rainfall from <300 mm to >500 mm. The major soils are deep coarse-textured sands and sandy loams, duplex soils with coarse-textured sands over clay, and fine-textured red–brown earths of low hydraulic conductivity [26]. The crops are grown during the cooler wetter winter and mature during the spring months.
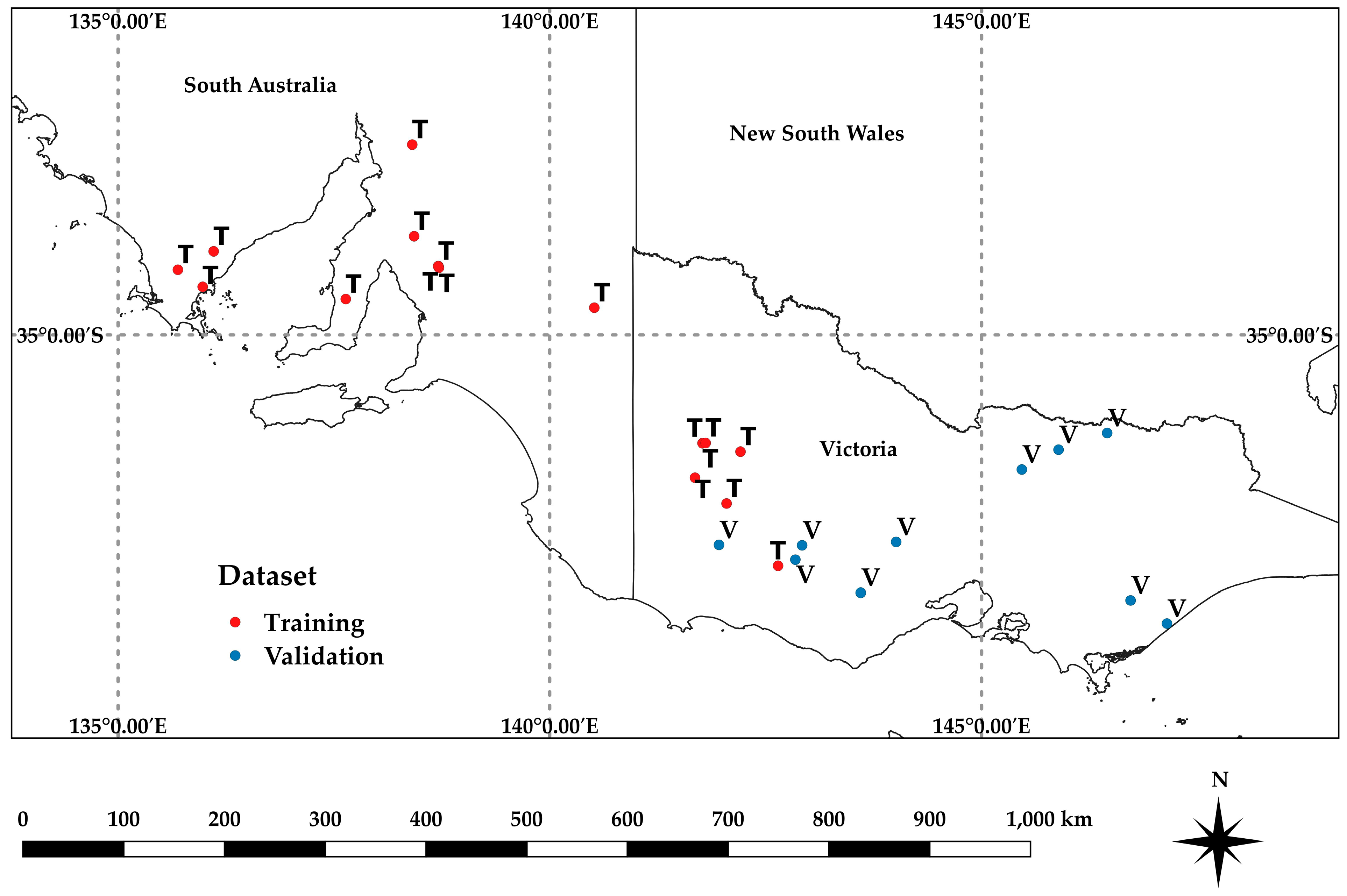
Two separate AGB datasets were used for the analysis (Table 1, Figure 1). A training dataset (‘Training’) was compiled with data pooled from 23 paddocks in South Australia and Victoria, across four growing seasons (2018–2021), multiple growth stages, and several crops representing grain farming systems. These data were divided into calibration and validation data as described in the Results. A separate validation dataset (‘Validation’), consisting of 13 paddocks, across multiple growth stages for several crops was acquired in Victoria in 2017 and 2018. The Training dataset was used to develop models of biomass estimates for individual paddocks and an overall model using all locations. The Validation dataset was used to assess the results of the overall model on completely separate data.

Table 1.
Crop biomass datasets.
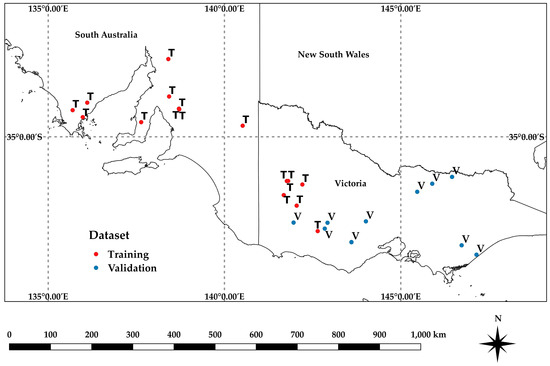
Figure 1.
Locations for the Training and Validation datasets across broad acre cropping regions in southern Australia, as described in Table 1.
Georeferenced biomass cuts were used to determine the AGB and used as ground truth. The crop biomass data were acquired by the researchers and their collaborators as part of agronomic research using standard protocols. For the Training dataset, standard methods using dried and weighed sample biomass cuts (e.g., 4 rows by 1 m length) were used to estimate the AGB. Biomass cuts were taken for each paddock at least once during the season and up to eight times, with samples taken at 9–60 sites across the paddock. The biomass cuts acquired for the Validation dataset were taken using a transect approach, with three 0.5 m cuts along one row along a 10 m transect representing one sample location.
2.2. Satellite Imagery Used
The primary imagery data source for this work was the public domain S-2 Level 2A image datasets [27]. Daily Planet imagery [28] was acquired for specific dates to supplement the S-2 time series. For both of the data sources, the surface reflectance products were used. NDVI was computed from the red and near-infrared bands of the reflectance imagery. Preprocessing was performed to detect and remove imagery impacted by clouds. This was completed in two ways: by visual inspection and using the cloud mask metadata. For the latter, the S-2 Scene Classification Layer (SCL) classes for vegetation and soil [29] were used to select cloud-free pixels. Likewise, the Planet Usable Data Map 2 (UDM2; [30]) metadata were used to select cloud-free pixels. A threshold (e.g., >98% cloud free pixels within the paddock area) was used to determine if a given image was included in the datasets.
2.3. Ground-Based NDVI Measurements
Ground-based active optical sensing (Holland Scientific Crop Circle ACS-430; Lincoln, NE, USA) was used to create paddock-wide maps of NDVI for a small subset of the Training dataset. The sensor was deployed using all-terrain vehicles, with the sensor head positioned between 0.5 m and 1.0 m above the crop. Transects were driven throughout the paddocks, and the individual points were interpolated to spatial datasets with 5 m grid cells. The interpolation was performed using block kriging with 30 m block sizes, and the interpolated cells were aligned to the S-2 pixels’ footprints. These data were used to help backfill gaps in the S-2 NDVI time series, as is detailed in Section 3.
2.4. Determining the Small Integral of the NDVI Time Series
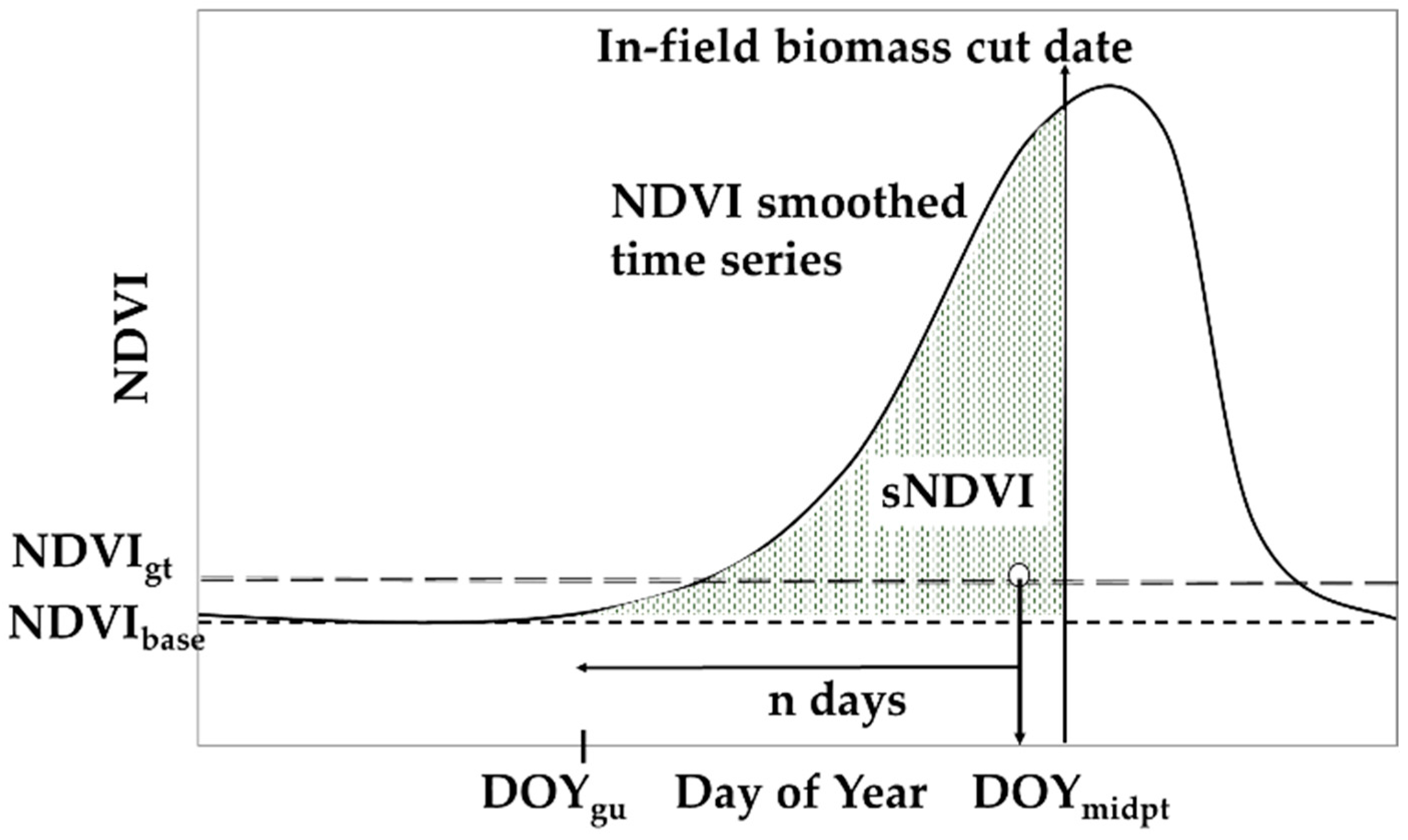
S-2 was acquired through the year for each site, and additional NDVI data were added as required from the Planet imagery and ground-based NDVI. Note that the determination of the small integral (sNDVI) is implemented per pixel; that is, each pixel represents a complete and independent time series. The following steps assume that the time series are cloud free, and that the gaps are small (e.g., 14 days or fewer) or have been filled with auxiliary data (e.g., Planet imagery or ground-based NDVI). The key parameters described are also shown in Figure 2. Note that the values selected for the key parameters were determined through experience as the technique was developed.
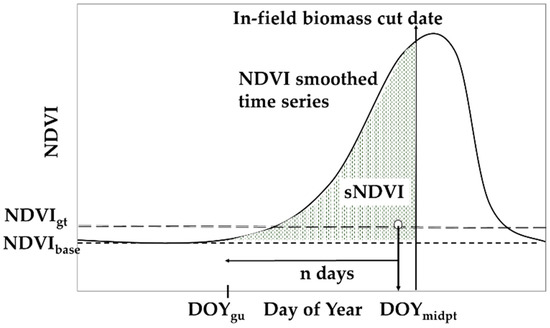
Figure 2.
Key parameters used to determine sNDVI. See text for the description of the methodology.
- Step 1:
- Interpolate the time series
This step involved determining the pre- and post-cropping season NDVI values and interpolating the time series to daily time steps. First, the baseline NDVI was defined as:
NDVIbase = min(NDVI > 0)
The NDVI for day of year (DOY) one through d was set to NDVIbase to eliminate the effects of summer crops and weeds on the interpolation. For this paper, d equal to 137 was used. Next the time series was fitted using the LOESS function, interpolating the time series to a daily time step. Note that the LOESS function uses a parameter (SPAN), which defines the interpolation window. For this paper, we used a SPAN value of 0.25.
- Step 2:
- Determine the start, end, and mid-point of the season
In this step, the crop phenological stages were determined based on the NDVI time series. The greenness threshold, NDVIgt, was determined as:
NDVIgt = a(NDVImax − NDVIbase) + NDVIbase.
This parameter was used to determine the beginning of the crop growth. For this paper, the parameter a was set to 0.10, based on the results from previous datasets (unpublished) used in the development of the technique. The start, end, and mid-point of the season were then determined based on the NDVIgt:
DOYstart = first DOY where NDVI > NDVIgt
DOYend = last DOY where NDVI > NDVIgt
DOYmidpt = INT(0.5(DOYstart + DOYend) + DOYstart).
- Step 3:
- Calculate the small integral (summed NDVI, sNDVI)
The crop greenup date, DOYgu, was determined from the mid-season date:
DOYgu = DOYmidpt − n days.
The parameter n was set to 100 in this paper and was based on the earliest expected or observed emergence of the crop. The small integral was based on the DOYgu, as the NDVI values prior to DOYgu were not included in the summation. The summation was also adjusted for the differences in pre- and post-season NDVI values across sites by subtracting the value of NDVIbase. This results in a small integral as indicated by the shaded area under the curve shown in Figure 2. The integral (sNDVI) was determined by summing the adjusted interpolated time series using daily time steps starting at DOYgu and ending at the desired DOY (e.g., a date corresponding to plant biomass sampling).
For the combined datasets utilized in this paper, the summation sNDVI was computed on a pixel basis for each crop biomass measurement date. Each georeferenced AGB measurement was paired with the corresponding sNDVI for that spatial location and measurement date.
2.5. Software Used
The S-2 level 2A surface reflectance imagery was accessed and processed to create NDVI time series using Google Earth Engine [31]. The R statistical software [32] was used extensively in this work, for both the image processing (computing the small integral or summed NDVI) and the statistical analysis. Selected data analysis and visualization was performed using QGIS [33]. The QGIS plug in Precision Agriculture Tools ([34]) was used to process the active optical data. Some of the S-2 imagery was also processed using ENVI version 5.5.2 and IDL version 8.7.2 (Harris Geospatial Solutions, Inc).
3. Results
In this section, we present the results evaluating the use of the NDVI time series to estimate the AGB. The estimated values of the AGB were compared with the measurements (ground truth) after fitting for individual paddocks and pooled across sites and years. The impact of gaps in the time series was also evaluated, and we describe the use of the supplemental satellite and ground-based data.
3.1. Relating Biomass to the Small Integral
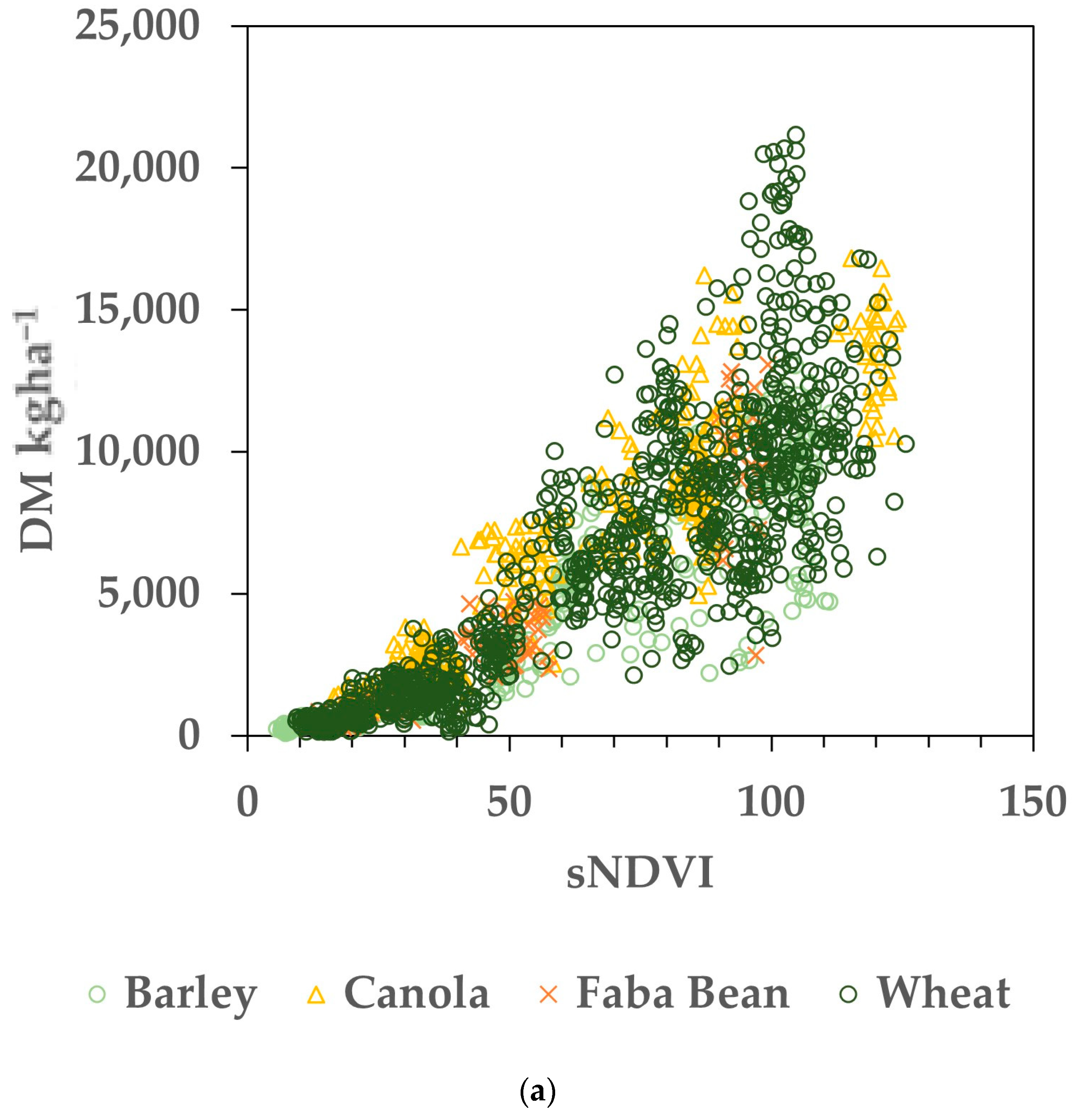
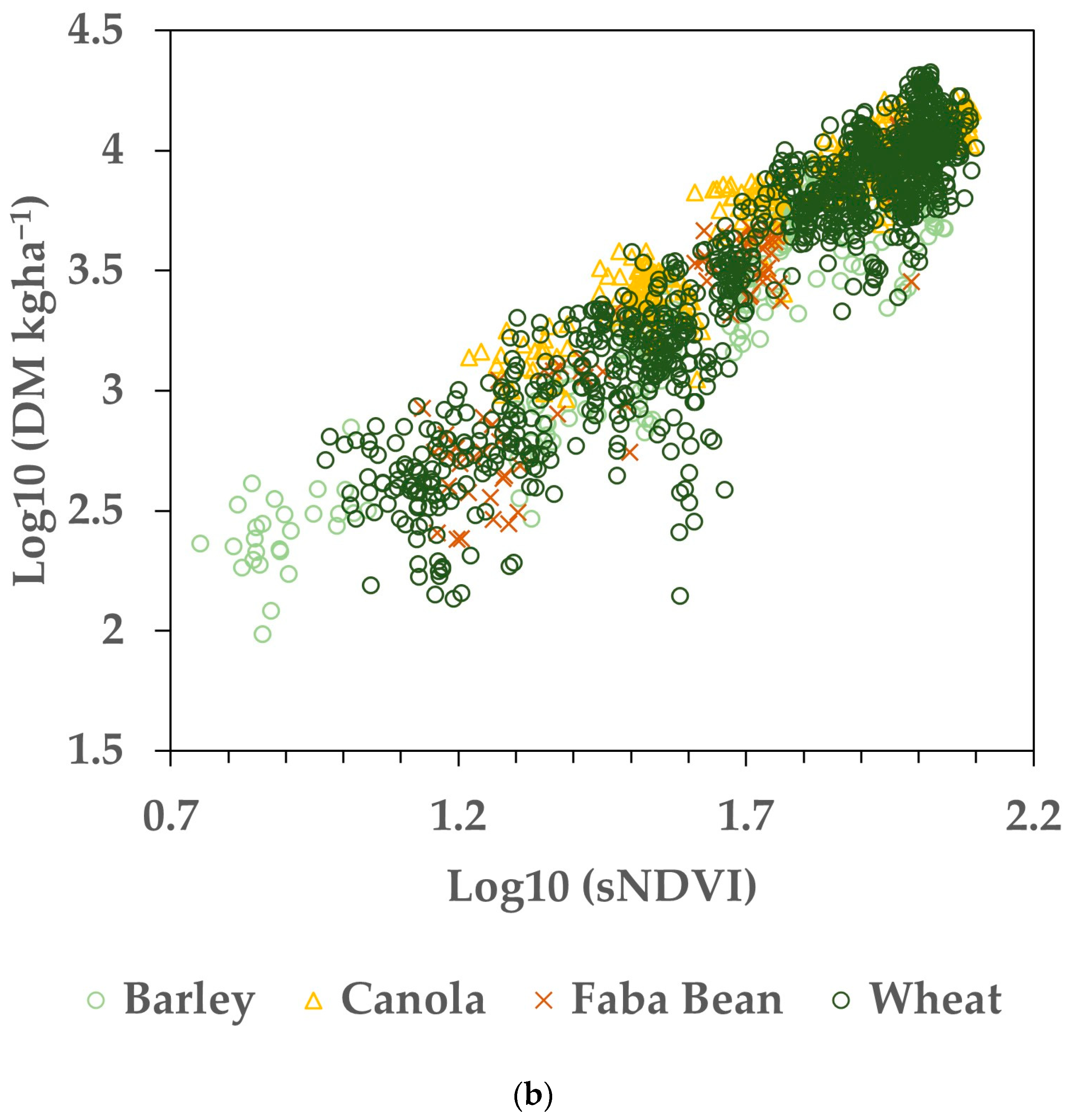
The datasets were compiled by pairing the AGB measurements (from biomass cuts) to the small integral of the NDVI time series (sNDVI) value for the same date, as described in the Section 2 (Figure 2). Regressions were performed on the Training dataset for individual paddocks and the pooled data. Eighty percent (selected randomly) of the entire dataset was used to generate the models of AGB as a function of sNDVI. Figure 3a shows the resulting relationship between the AGB (kg ha−1) and the sNDVI for 80% of the Training dataset, used as calibration. The same data, with log10 transformation, are shown in Figure 3b. The random error was more consistent across the range of sNDVI and AGB values using the log10 transformation (Figure 3b), while the errors increased with the magnitude of sNDVI and AGB (Figure 3a). To facilitate the error analysis, the datasets were transformed using log10 prior to fitting.
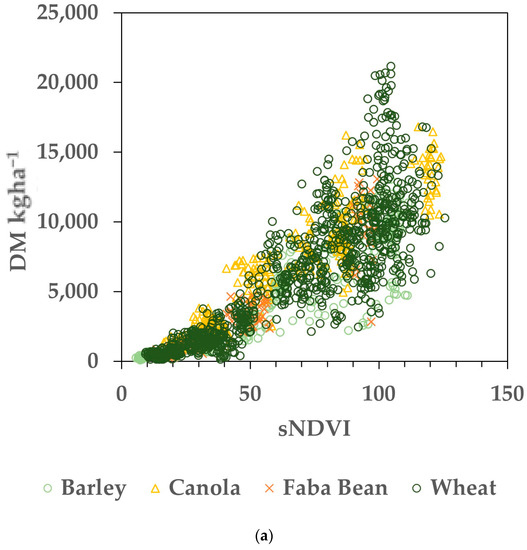
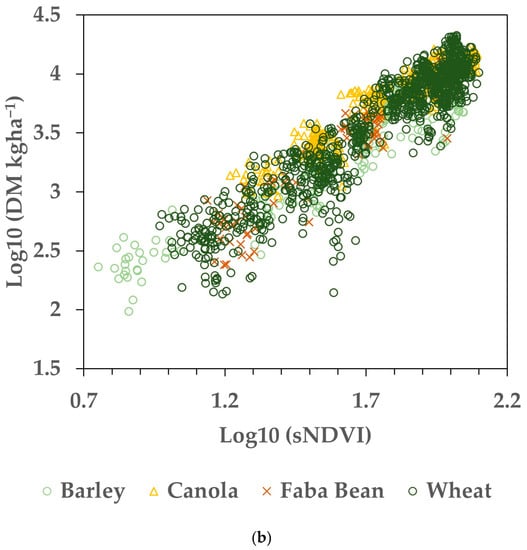
Figure 3.
Calibration data representing 80% (N = 1664) of the Training dataset. (a) Data before log transformation. (b) Crop biomass and sNDVI data transformed with both using log10. Note that the remaining 20% of the dataset was used to evaluate the estimated versus measured biomass values.
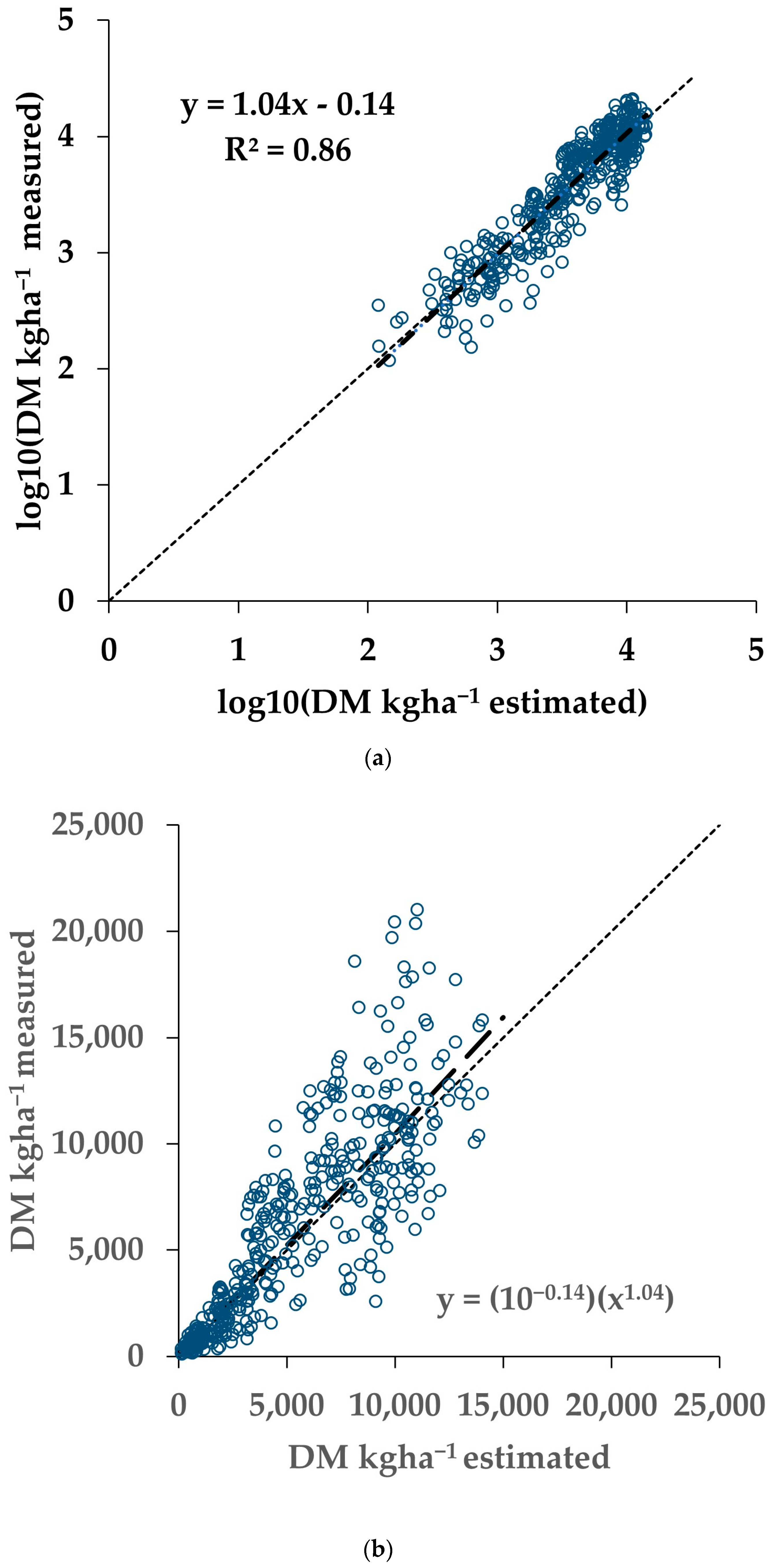
The remaining 20% of the Training dataset was used to assess the fit between the measured AGB and the AGB estimated from the sNDVI. The results are shown in Table 2. Note that some paddocks included in the total were not reported individually, as the number of samples was not sufficient. Figure 4 shows the measured AGB graphed versus the estimated values for the 20% validation data in log10 space (Figure 4a) and transformed back to the original units (Figure 4b).

Table 2.
Calibration results for the estimated AGB on the sNDVI. The full Training dataset was split into calibration (80%) and validation (20%).
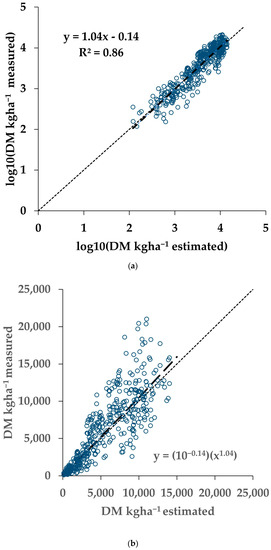
Figure 4.
Validation data representing 20% (N = 416) of the Training dataset. Both the biomass and sNDVI data were transformed using log10 prior to fitting. (a) Measured versus estimated AGB in log10 transformation. (b) Measured versus estimated AGB transformed back from log10.
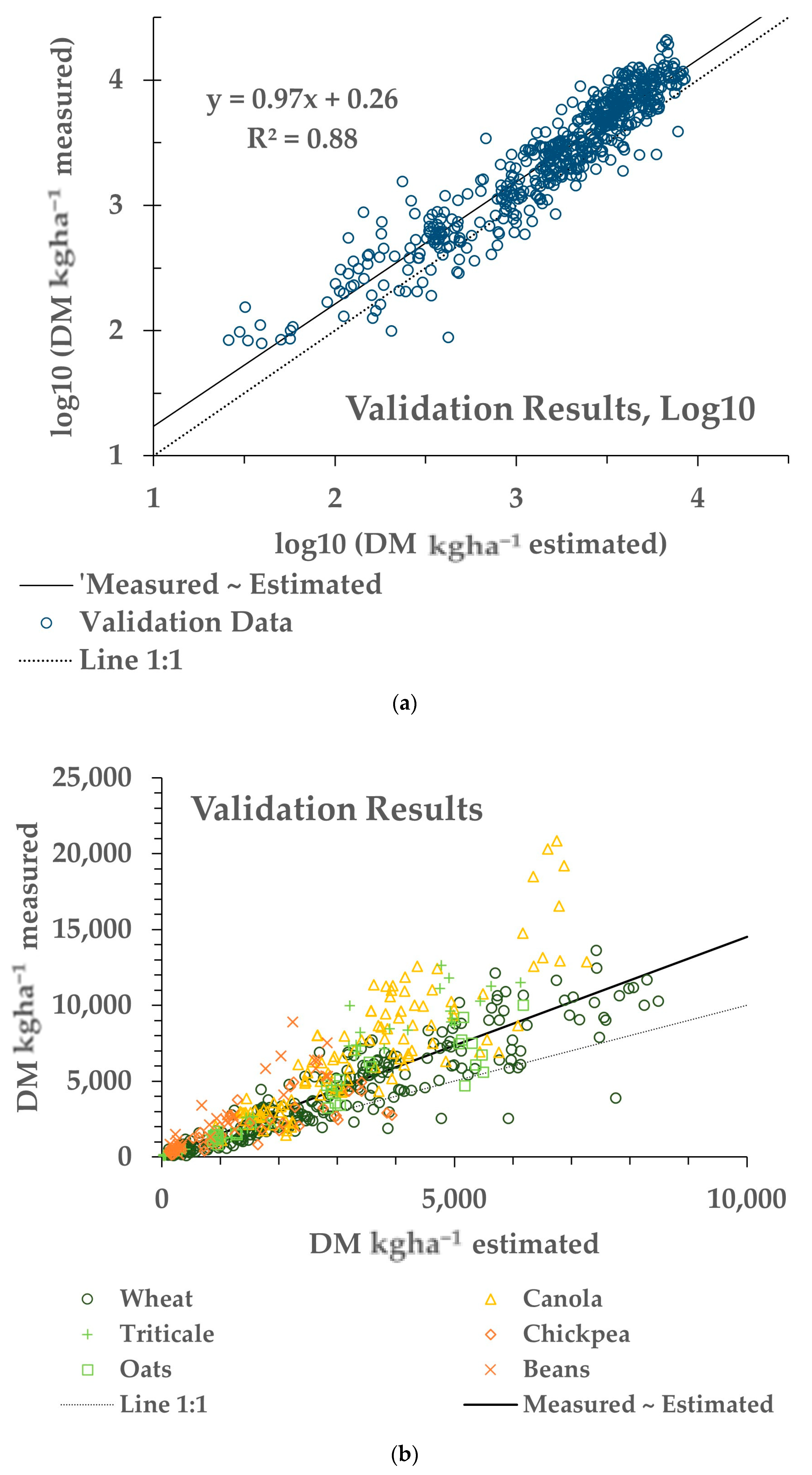
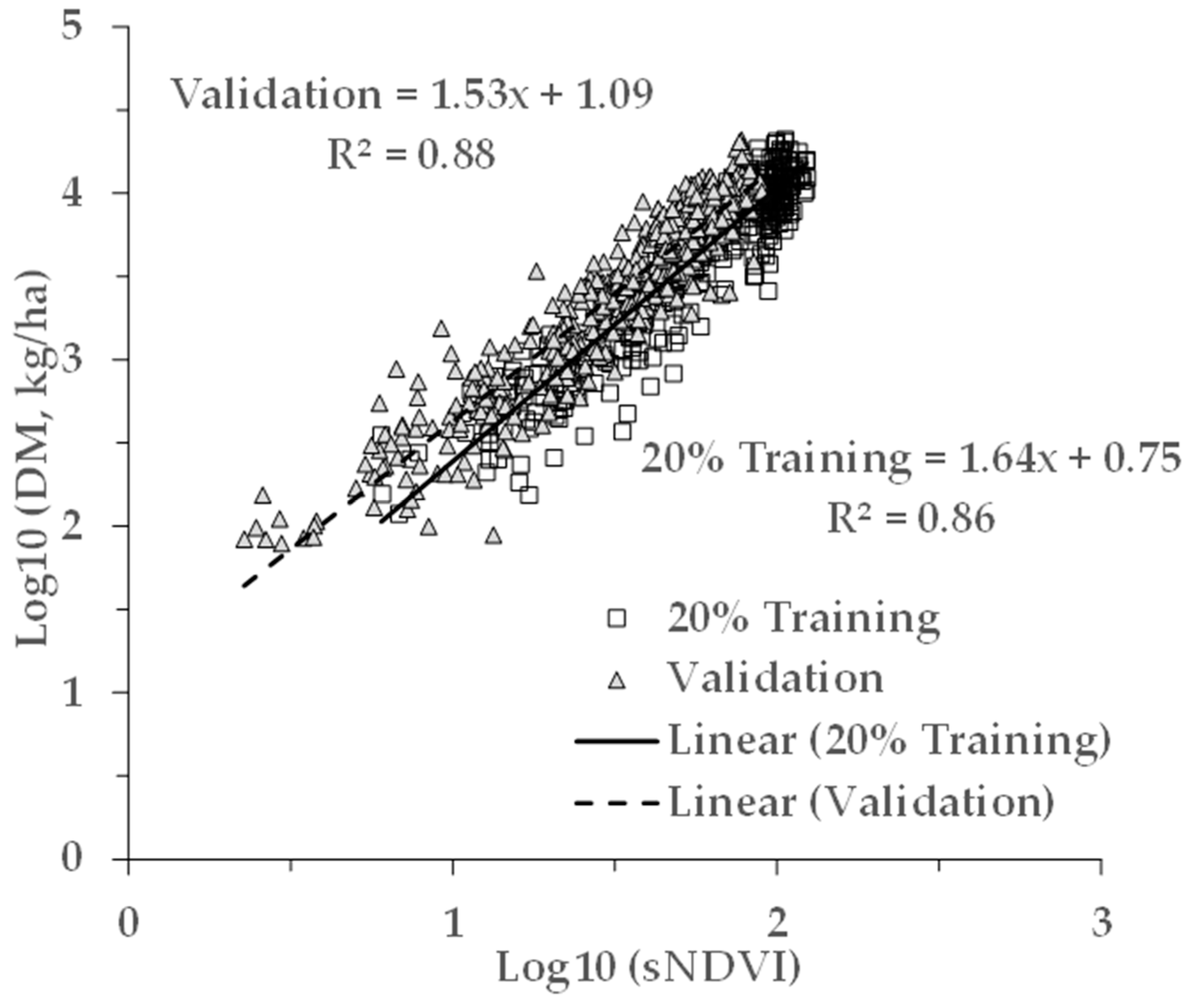
A validation of the overall regression model (last row of Table 2) was performed using the Validation dataset (Table 1), a separate dataset than used for the results in Table 2. The model parameters were applied to the sNDVI values of the Validation dataset to generate the estimated AGB values. The results of the measured and estimated AGB for the Validation dataset are shown in Figure 5.
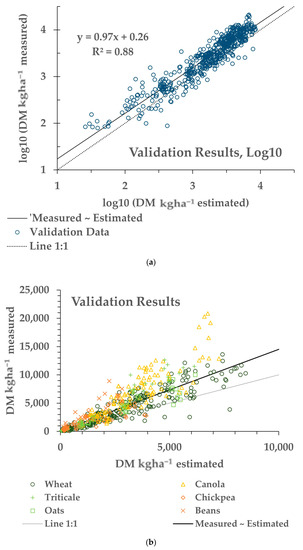
Figure 5.
Results from a validation dataset (N = 550), not utilised for the training results shown in Table 2. (a) Measured versus estimated AGB in log10 transformation. The solid line indicates the fitted relationship, and the small dashed line indicates the 1:1 relationship between the modelled and measured AGB. (b) The same results transformed back to the original space. The solid and dashed lines indicate the fitted and 1:1 relationships, respectively. The symbols indicate the crop type.
3.2. Evaluating the Effects of Gaps in the Time Series
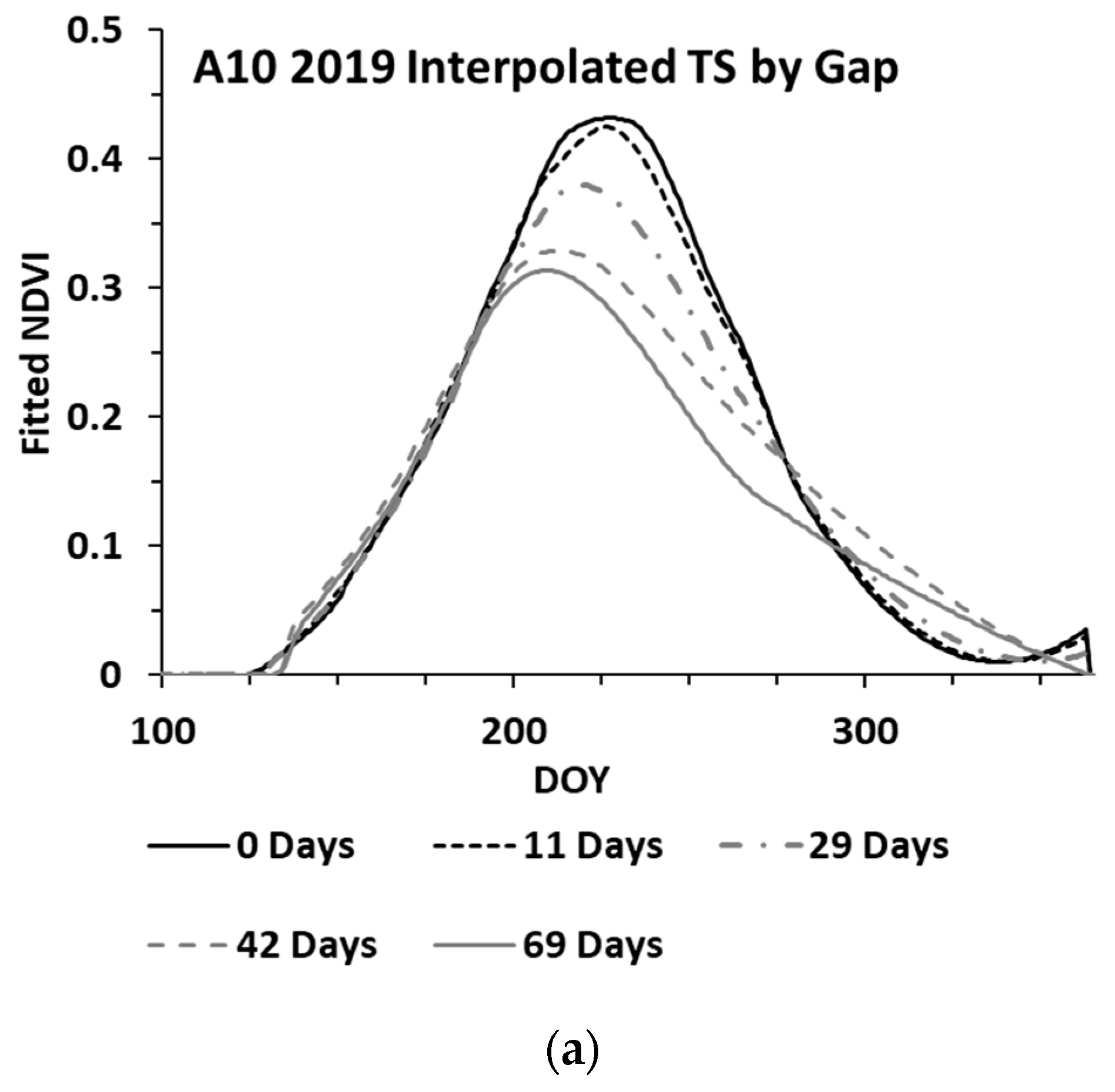
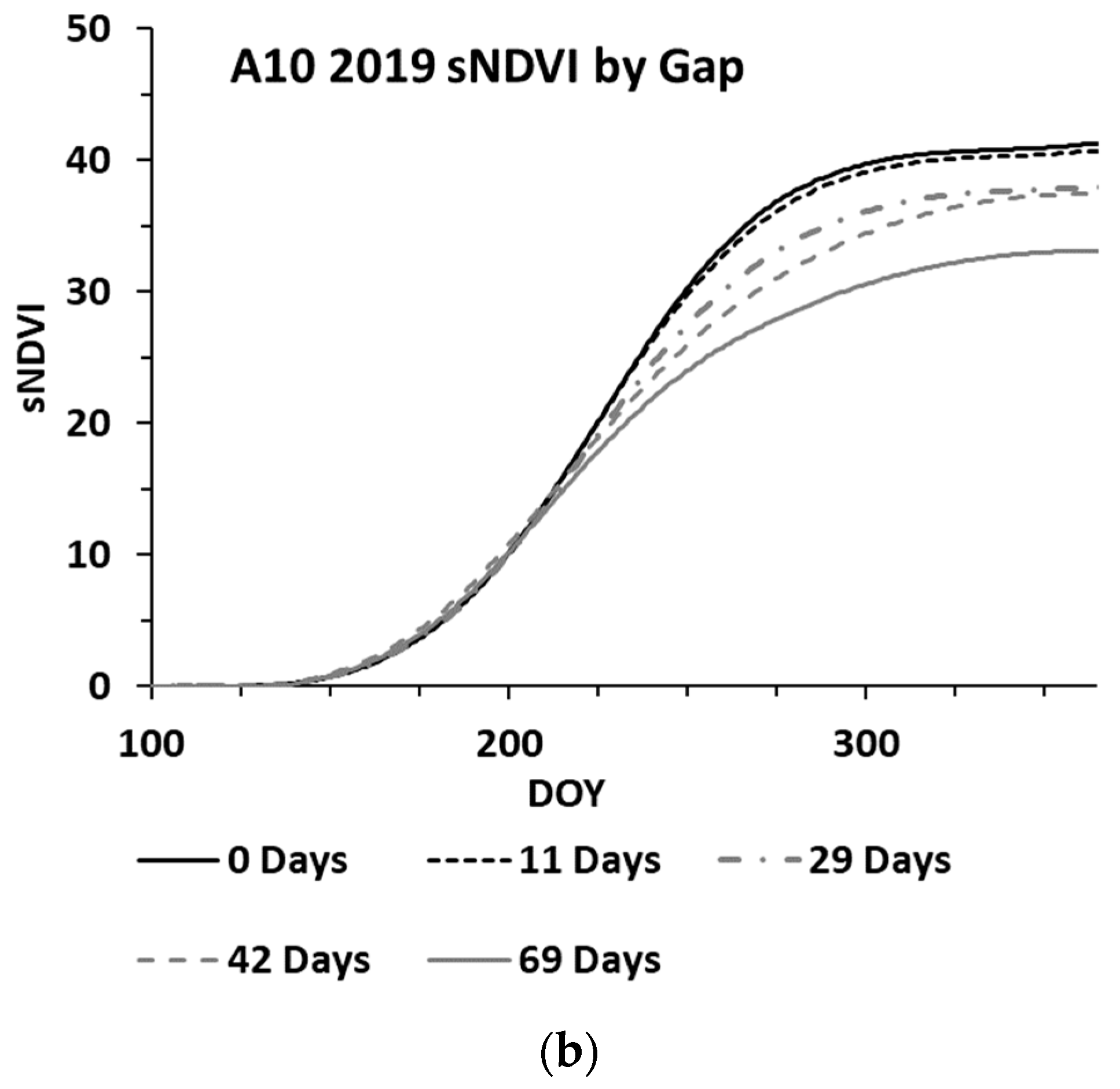
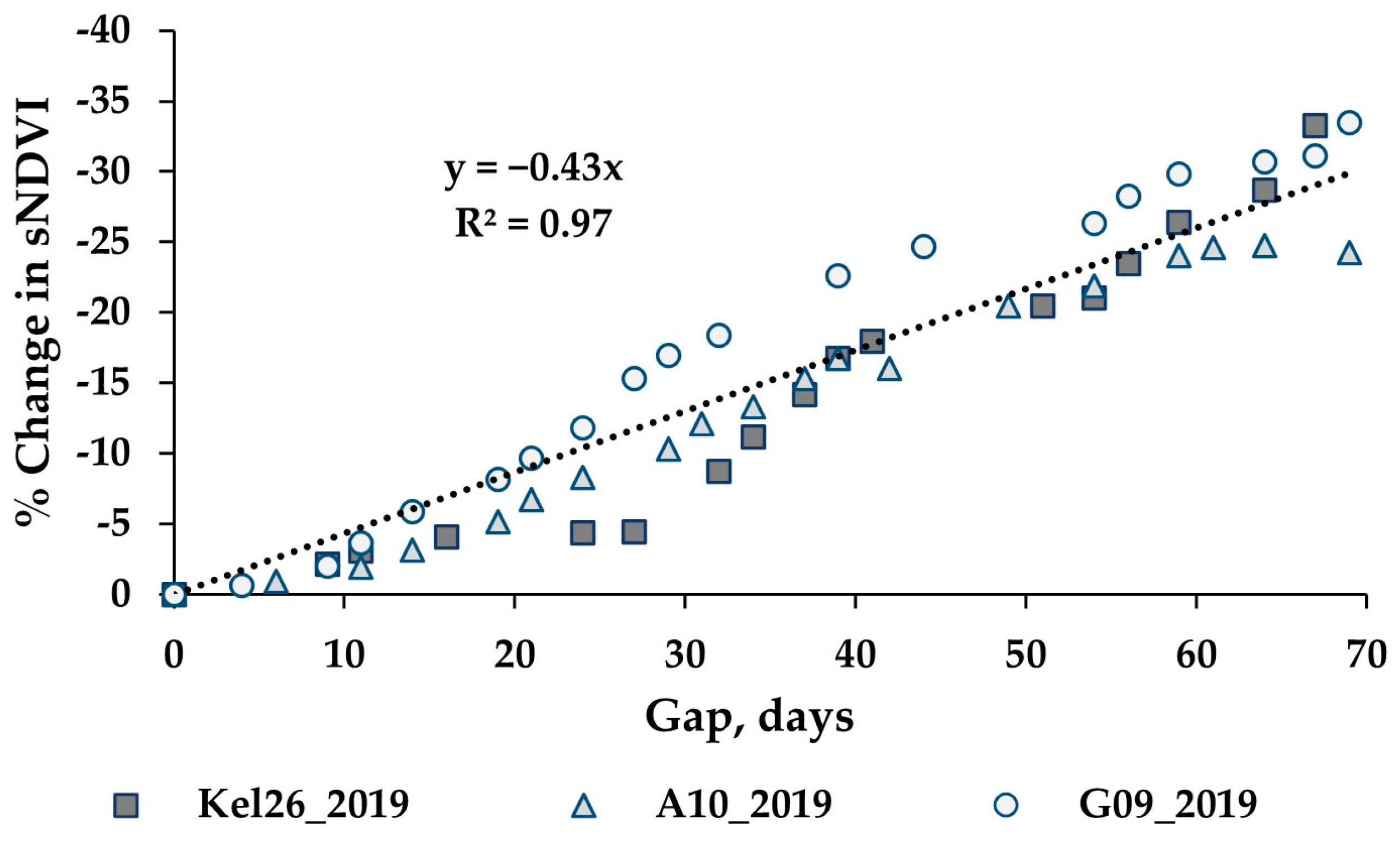
While the methods used were developed to work on an irregular time series, cloud cover can lead to significant gaps (e.g., 30 days or more between clear sky image acquisitions). An analysis was performed to assess the impacts of increasing gap sizes on the estimated biomass. The analysis utilized three wheat paddocks in Western Australia, with clear sky conditions throughout 2019 providing an almost continuous time series for the year. The S-2 SLC classes 4 and 5 were used to verify that the paddock was comprised of ≥99% cloud free pixels for each date. One hundred pixels were randomly selected from each paddock, each representing a time series for the winter crop growing season. These time series were analysed by iteratively removing dates to simulate cloud gaps, with the synthetic gap centred at mid-season. The gap sizes graduated from four days to a maximum of 69 days. With each gap iteration, the time series was fitted to compute the sNDVI for daily time steps. Figure 6 shows the mean of the 100 time-series (pixels) from one paddock, showing the effect of the increasing gap size on the interpolated NDVI values (Figure 6a) and the summed NDVI time series (Figure 6b). The impact of the increasing gap size on the sNDVI values for the three paddocks is shown in Figure 7. The percent change represents the relative change from the minimum gap sNDVI and is based on the average of 100 time series (pixels) from each of the three paddocks. These differences were computed at daily time steps. Each point in the graph represents the greatest percent change (minimum value) from DOY 200 to DOY 365, which represented the growing season.
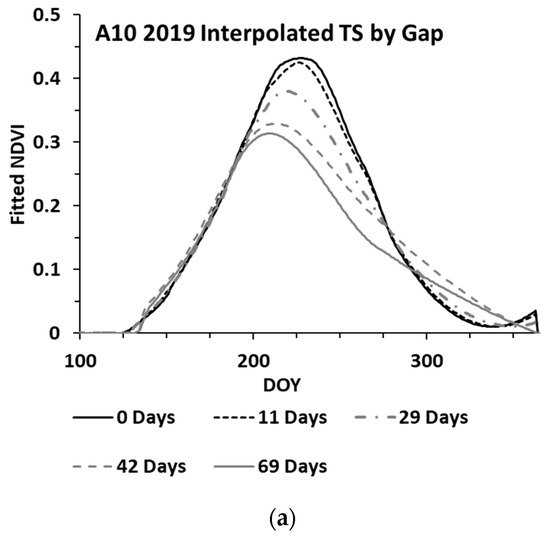
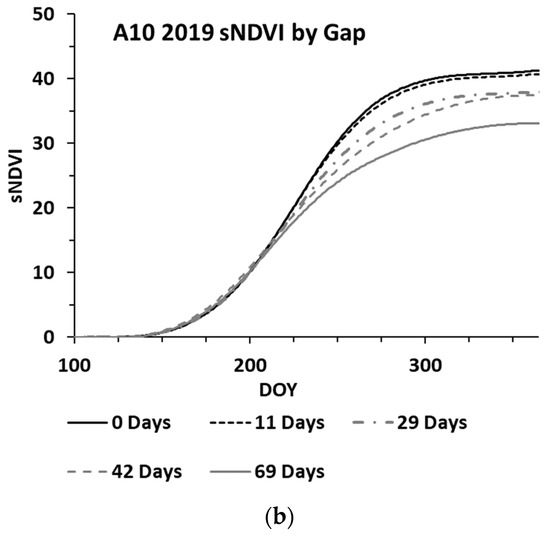
Figure 6.
Mean of 100 time series (pixels), showing the effect of the increasing gap size on the (a) interpolated and (b) sNDVI time series.
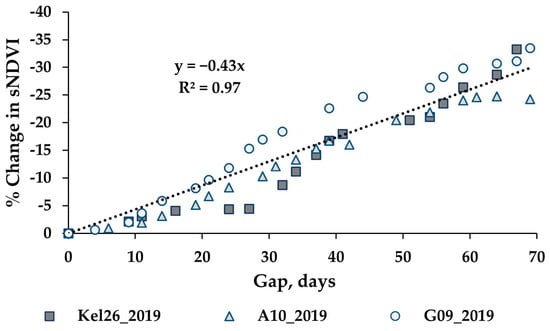
Figure 7.
Evaluation of three wheat paddocks with near complete time series for the effect of the gap on the sNDVI. The increasing gap size is centred near mid-season. The percent change represents the relative change from a gap near zero and is based on the average time series of 100 pixels from each paddock. Each point represents the greatest percent change from DOY 200 to 365.
The error in sNDVI due to gaps was translated to the estimated AGB, but the magnitude depended on the size of the gap and the date of the fitted sNDVI. Table 3 shows the impact of the increasing gaps on the estimated biomass for the three paddocks. The model parameters from Table 2 (last row) were used to translate the sNDVI values fitted with gaps of about 31 days (one month) and 67 days (maximum gap). The change in AGB due to the gap was expressed as the difference between the AGB estimated with no gap and with the gap, divided by the AGB without the gap effect.

Table 3.
Effect of the time-series gap centred at mid-season on estimated AGB.
3.3. Filling of Gaps in the Time Series
While gaps due to cloud cover may be unavoidable, other satellite and/or ground based measurements may be used to fill these gaps. Four of the 2021 paddocks in the Training dataset had frequent cloud cover, and Planet imagery and ground based active optical measurements were used to fill between the S-2 observations. Planet NDVI values were used to estimate the S-2 NDVI based on relationships developed with coincident imagery. One hundred fifty image pairs of coincident S-2 and Planet imagery were acquired during 2019–2021, for the same four paddocks. A regression model was developed on the coincident imagery, randomly selecting 200 pixels from each pair (total N > 28,000). Eighty percent of the data was used for calibration, and the remaining 20% was used to validate the model. The results are shown in Table 4. The calibration results indicate an uncertainty of 0.06 NDVI of the estimated S-2 values.

Table 4.
Estimating the S-2 NDVI values using Planet imagery 1.
Likewise, ground-based measurements of the NDVI using an active optical sensor (Model ACS430, Holland Scientific, Lincoln NE USA) were related to the S-2 NDVI based on coincident sets of measurements. The ground-based NDVI measurements, made along tracks throughout the paddocks, were interpolated to 5 m grid cells. Nine pairs of coincident S-2 imagery and active optical NDVI maps were acquired, and approximately 200 pixels were randomly selected from the set. Note that the active optical NDVI does not ‘saturate’ at high green biomass values as quickly S-2 or Planet NDVI, so the relationship was fitted with a second order polynomial. The resulting model is shown in Table 5.

Table 5.
Estimating the S-2 NDVI values using active optical measurements 1.
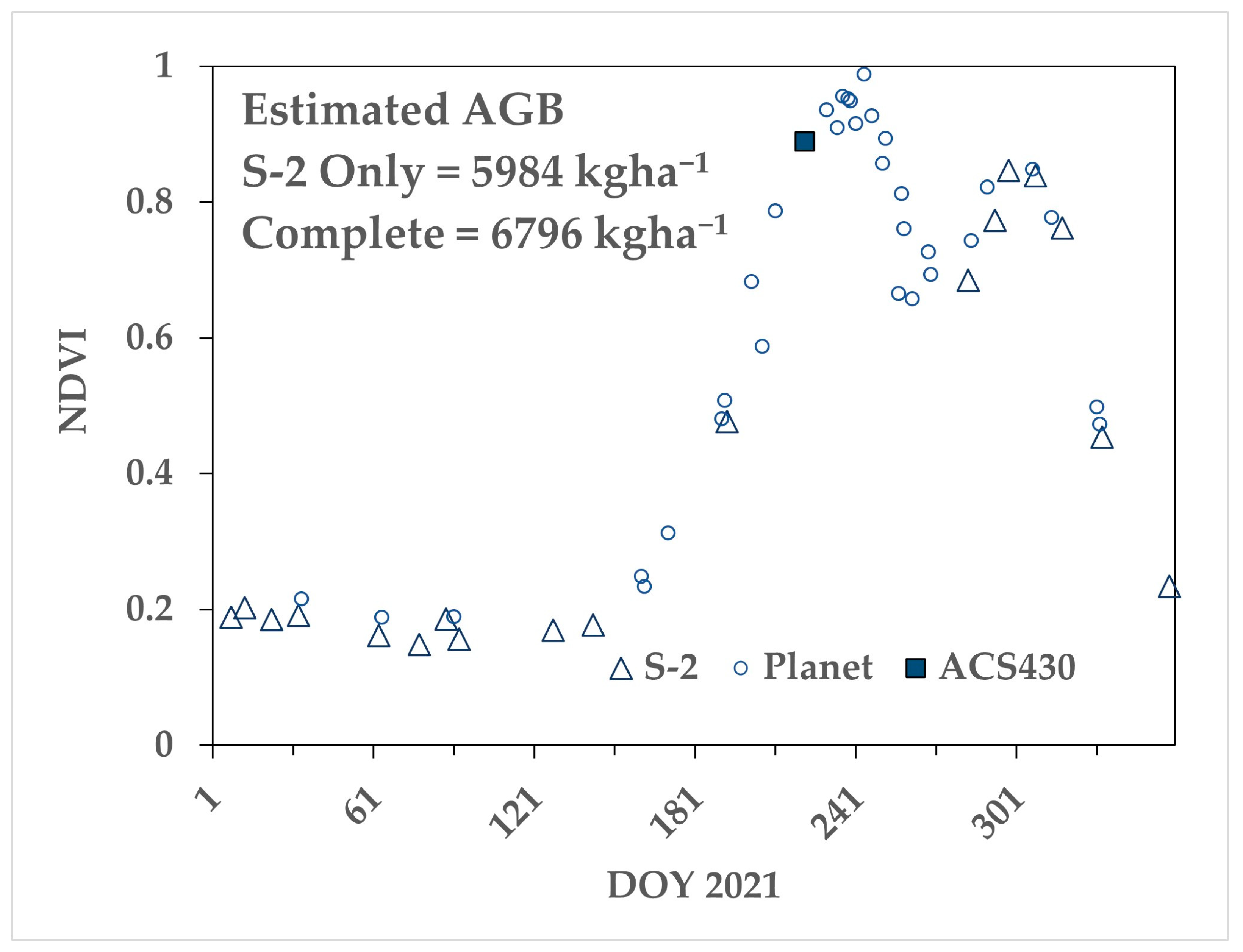
Figure 8 shows an example time series from one of the four 2021 paddocks, sown to canola. Note that the S-2 observations (open triangles) completely missed the first NDVI peak, which was defined by the modelled S-2 pixels based on Planet and active optical NDVI values. The estimated AGB at DOY 266 (near mid-season) increased from 5984 kg ha−1, using only S-2, to 6796 kg ha−1, with the complete time series.
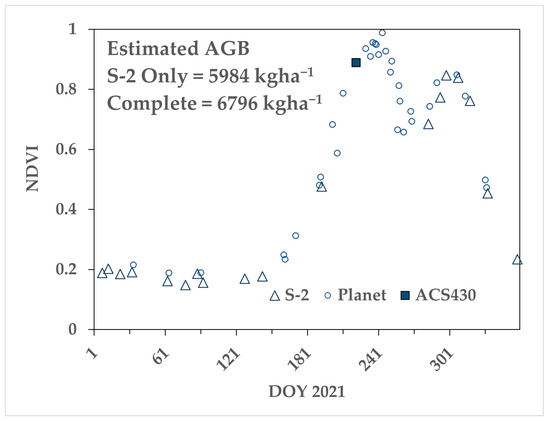
Figure 8.
Example AGB estimates for a canola crop grown in 2021 near Nurrabiel Victoria with and without a complete time series. The S-2 time series from the original pixels (open triangle) had a gap of 92 days, which resulted in an AGB estimate at DOY 266 of 5984 kg ha−1. Supplementing the time series with Planet imagery (open circles) and active optical (filled square) increased the estimated biomass to 6796 kg ha−1.
4. Discussion
In the previous section, a framework was presented to estimate the AGB, relating the NDVI time series to measured AGB. Accuracy results were presented using local calibrations, calibration from pooled data, and applying the calibration from one dataset to another. Validation using 20% of the Training dataset generated models for the individual paddocks with R2 values ranging from 0.79 to 0.98, while the pooled data resulted in an R2 of 0.86 (Table 2). Eight of the twelve paddocks had models with validation R2 values greater than the pooled data. The slope for the pooled data (1.04) was within the range of the individual paddocks (0.86 to 1.09). The intercept value for the pooled data (−0.14) was also within the range of the individual paddocks (−0.44 to 0.55). These parameters suggest that the individual model fitting did not always result in more accurate estimations of the AGB.
Applying the model from the pooled data (last row of Table 2) on the separate Validation dataset resulted in an apparent bias in the fitting of the Validation dataset. This can be seen in Figure 4 noting the 1:1 line (dotted). While the slope was close to 1 (0.97), and the R2 was 0.88, the model underestimated the measured AGB (intercept = 0.26). This difference is explained by the apparent differences in the relationships between the AGB and sNDVI for the Training and Validation data (Figure 9). This may be due in part to the ground footprint of the sNDVI in the Validation dataset. Each data point represents an average of seven to nine S-2 pixels, rather than one or two as with the Training dataset. Taking the mean NDVI of several pixels might have the effect of lowering the NDVI and sNDVI values for the same measured AGB value. The results showed that differences in the relationship between the measured AGB and the remotely-sensed NDVI can translate through to the AGB estimates.
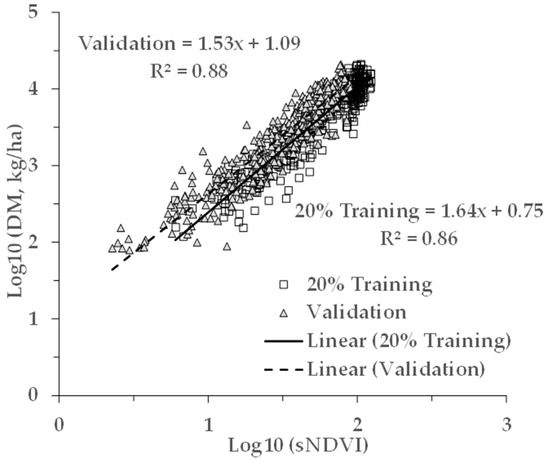
Figure 9.
Comparison of the AGB and sNDVI relationships between the Validation and Training datasets. For a given value of sNDVI, the DM values for the Validation dataset appear to be greater.
In addition to the appropriate calibration data, filling gaps in the time series is important to the estimation of AGB. The results from the gap analysis indicate that increasing the mid-season gap resulted in a reduction in the peak NDVI and sNDVI values, as well as a change in the time series (Figure 6). The analysis of three wheat paddocks indicated a change in the sNDVI of −0.43% for each additional gap day (Figure 7). The translation of this error to the estimated AGB depends on the timing of the AGB estimates as well as the size of the gaps. However, the results shown in Table 3 indicate there can be substantial (e.g., >75%) decreases in the AGB with gaps exceeding 60 days. Figure 8 shows an example time series and the resulting AGB from one of the 2021 paddocks in the Training datasets, where a gap of 92 days in the S-2 imagery resulted in a 12% decrease in the estimated AGB.
The results support the use of the framework as an approach to estimate the AGB directly from the NDVI. Other approaches utilize the relationship between the AGB and the accumulation of absorbed photosynthetically active radiation (PAR), as presented by Monteith [35]. Meng et al. [36] modelled the fraction of PAR utilized by the canopy (fPAR) from the NDVI to estimate the biomass for winter wheat, using MODIS imagery downscaled to 30 m. They performed a validation with 40 data points of direct biomass measurements, reporting an average relative error of about 17%. Their results also indicated the increase in uncertainty with the biomass amount (as seen in Figure 5 from that paper). Relating the NDVI time series to fPAR to model biomass takes advantage of the differences in fPAR and light use efficiency but requires measurements and/or assumptions to compute this quantity (e.g., air temperature and evapotranspiration).
Implementation of the presented framework requires the selection or development of a calibration model and the availability of a suitable time series. The calibration between the sNDVI and the measured AGB can be developed for specific crops (paddock and year), across multiple sites, or taken from the model presented within this paper. The most direct use of the framework is to develop a model for a given paddock and crop (growing season). When planning biomass cuts, the range of biomass values (e.g., <1000 kg ha−1 to >5000 kg ha−1) is more important than the phenological stages; so, a sampling strategy of regular intervals (e.g., every 2–3 weeks) is advised. Ideally, there should be sufficient data (e.g., >100 data points for the season) to provide calibration and validation for a model. The results demonstrate the successful use of pooled data using similar sites, with the same sampling techniques and image processing to calculate the sNDVI. If applying a model that was derived from other datasets, this may produce increased errors in the AGB estimates (as exemplified by the Validation dataset results presented). If applying a model derived from separate data, any available measured AGB should be checked against the calibration dataset used. Regardless of the calibration model used, it is important to start with a cloud free time series with minimal gaps. To build the NDVI time series, S-2 surface reflectance data offer an excellent source of public domain imagery. Large gaps in the time series (e.g., >30 days) should be filled with supplemental data, as shown in the results. If active optical sensing is available, these data can be utilized during prolonged cloudy periods. An alternative is to ‘back fill’ using other imagery sources (e.g., daily Planet) if cloud-free imagery is available.
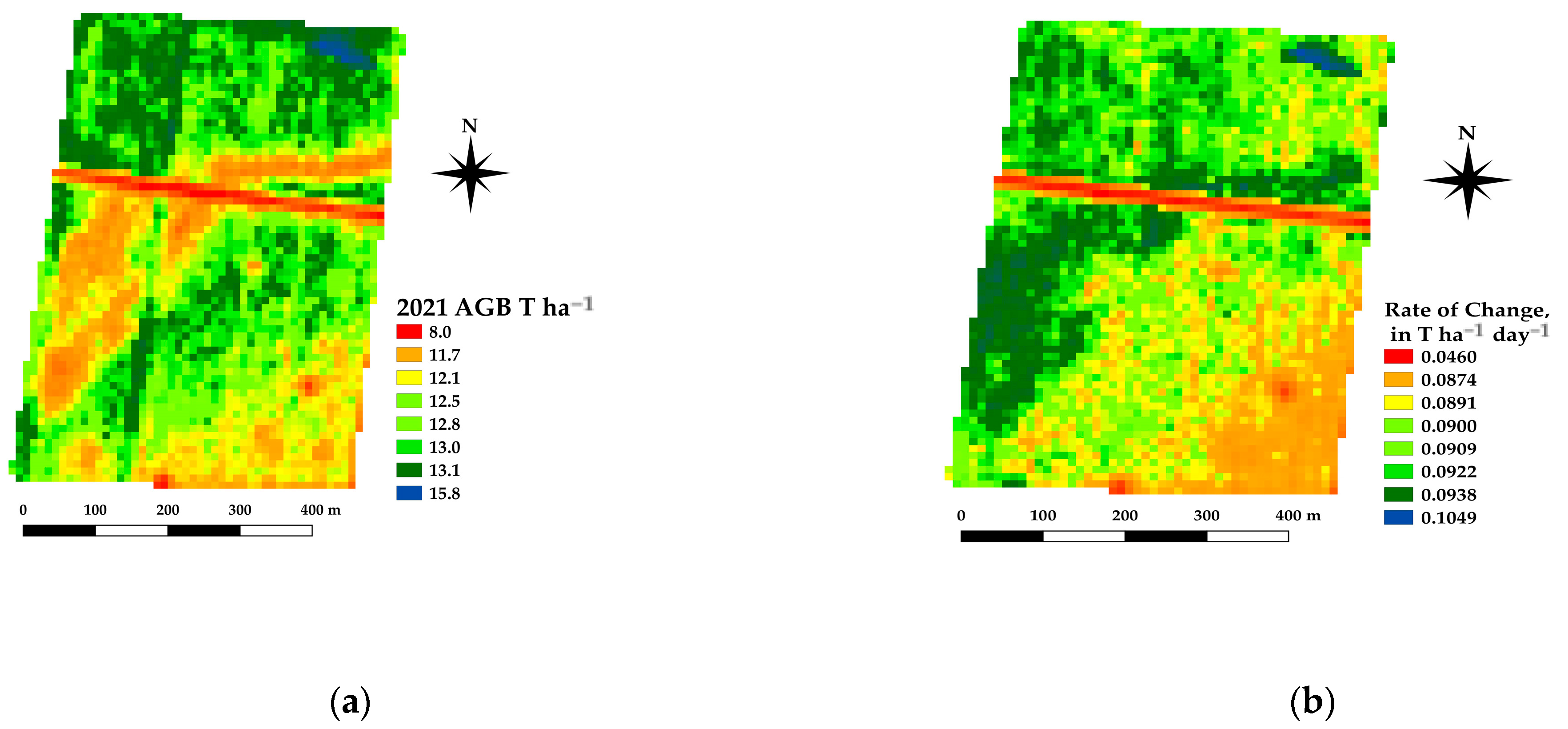
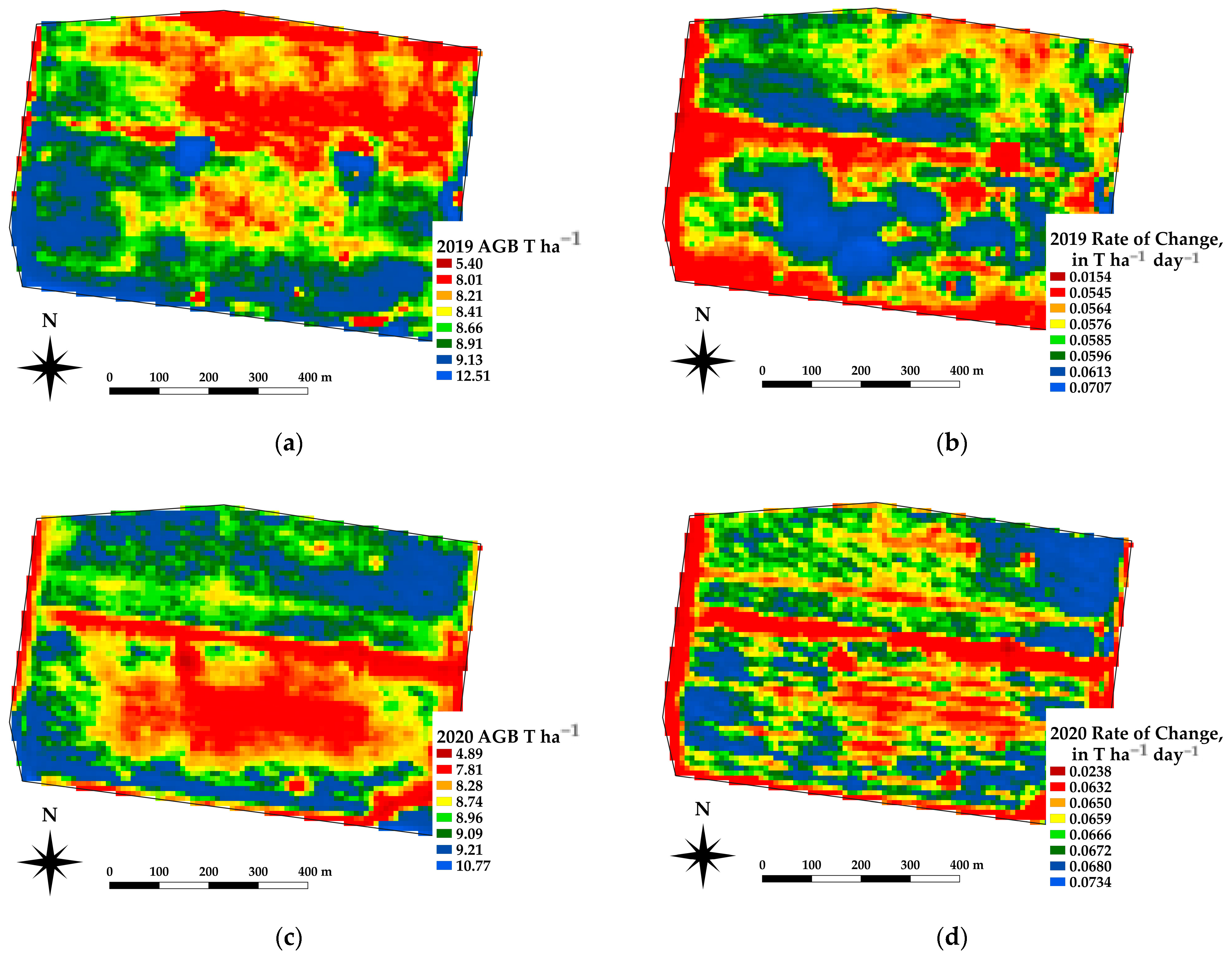
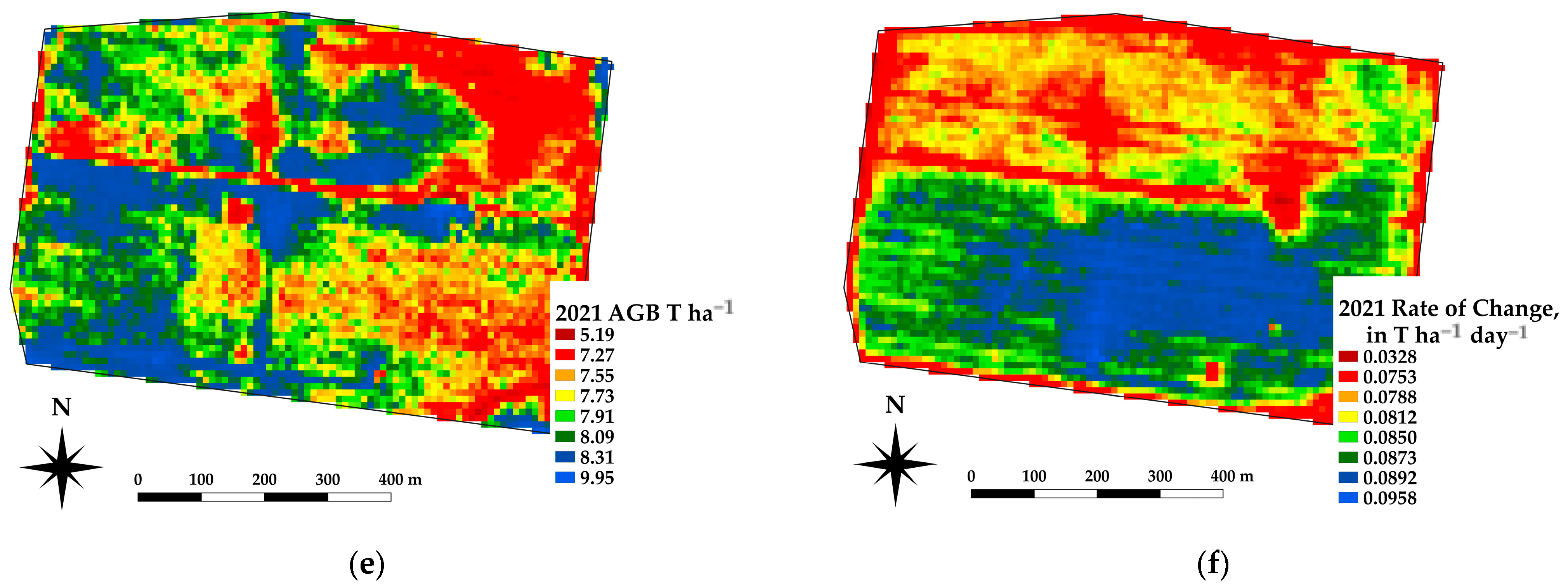
The framework presented offers the ability to estimate the spatial estimates of the AGB at daily time steps, which in turn allows for daily rates of change in crop growth. Spatial differences in the AGB and crop growth rates across a paddock are being used as an indicator for soil constraints [37]. Figure 10 shows an example of the AGB and rate of change for a canola crop grown near Nurrabiel Victoria, for DOY 300 in 2021. The AGB appeared to vary between 11.7 and 13.8 T ha−1 for most of the paddock area (note the red linear area is an access road and has low AGB values). The rate of change was determined by fitting the AGB as a function of the DOY using a second order polynomial for each pixel independently. The first derivative of the fitted equation was then used to indicate the rate of change (ROC) in T ha−1 day−1. The AGB and ROC datasets can be compared within the growing season and across paddocks, crops, and years. Figure 11 shows an example of the AGB and ROC for a paddock near Nurcoung, Victoria. The maps shown compare AGB and ROC for a canola crop grown in 2019, a wheat crop grown in 2020, and a faba bean crop grown in 2021. These data represent the inherent variability in the soil, as manifested in the corresponding crop growth, as well as variability between different crop types and seasonal conditions. These resulting spatial and temporal datasets are being incorporated into machine learning approaches to relate change in the crop growth to other spatial and temporal datasets characterizing soils, leading to a better understanding of crop growth restraints, the effects of amelioration activities, and more efficient crop and paddock management.
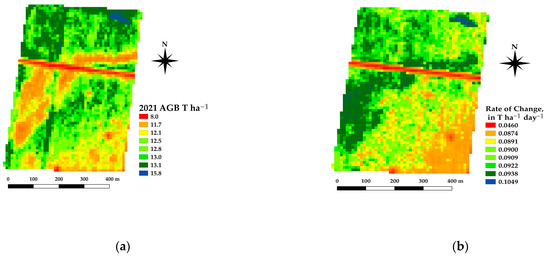
Figure 10.
Example estimates of AGB (a) and Rate of Change (b) for a canola crop at DOY 300, 2021.
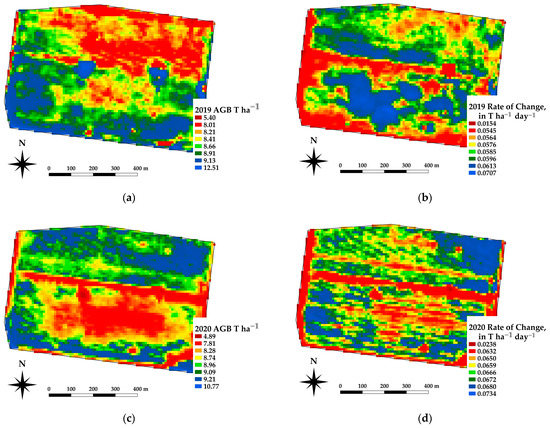
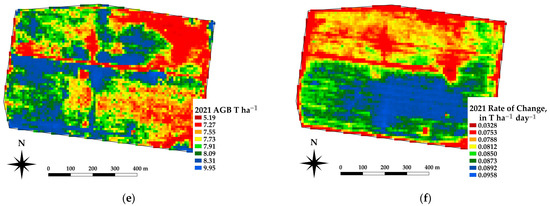
Figure 11.
Example estimates of AGB and Rate of Change for the same paddock sown to different crops. (a,b) AGB and ROC for canola at DOY 300, 2019. (c,d) AGB and ROC for wheat at DOY 300, 2020. (e,f) AGB and ROC for faba beans at DOY 300, 2021.
5. Conclusions
The framework presented offers the ability to estimate the spatial estimates of the AGB at daily time steps and spatial resolutions well suited to characterise in-field biomass and crop growth. The resulting R2 values for the measured and estimated AGB ranged from 0.79 to 0.98 for individual paddocks, and the R2 from a dataset pooled (multiple crops, years, and locations) was 0.86. The AGB and crop growth rate datasets produced may be quite valuable for agricultural applications such as agronomic research, crop management (e.g., fertiliser requirements), and input and validation for the refinement of crop biophysical models. The results presented in this paper highlight the importance of frequent image capture to support these spatial AGB estimates, with solutions given to reduce the impact of this issue.
The results presented show that the AGB can be estimated with a model developed on a separate dataset. However, any inherent differences in the relationship between the measured biomass and the sNDVI will impact the accuracy of the estimated AGB. Further development to reduce the impacts of differences across the datasets (crops, local soil and weather conditions, and sensor used) could include establishing more robust and universal models based on normalised values of sNDVI and AGB. Further testing of parameters used to calculate sNDVI such as the green-up date and the assumptions used to estimate the greenness threshold would be valuable in expanding the broader applicability of these models, as well as potentially reducing the error associated with these estimates.
Author Contributions
Conceptualization, E.P. and K.S.; methodology, E.P., K.S. and R.C.; software, E.P. and K.S.; validation, E.P.; formal analysis, E.P.; investigation, all authors; resources, all authors; data curation, all authors; writing—original draft preparation, E.P. and K.S.; writing—review and editing, E.P. and K.S.; project administration, E.P., K.S. and D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part through the Victorian Grains Innovation Partnership project 2A “Cereals: Minimising multiple soil constraints”, co-funded by the Grains Research and Development Corporation (GRDC) and Agriculture Victoria Research (AVR). Some of the data used in this project were from the GRDC-funded “Future Farm: Improving farmer confidence in targeted N management through automated sensing and decision support” (project number 9176493) and “Spatial variability of soil acidity and response to liming in cropped lands of the Victorian High Rainfall Zone” (DAV00152) with co-funding by AVR.
Data Availability Statement
The data associated with this research may be available upon request to the authors. However, much of these data are owned by the farming businesses with which we collaborated and so are not available for public access.
Acknowledgments
Dongryeol Ryu (University of Melbourne), Garry O’Leary, and Thabo Thayalakumaran (Agriculture Victoria) provided valuable discussions on the manuscript. Elizabeth Morse-McNabb (Agriculture Victoria) managed previous related work. We are grateful to all the grower (farmer) collaborators who kindly allowed access to their paddocks.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Tucker, C.J.; Holben, B.N.; Elgin, J.H.; McMurtrey, J.E. Remote sensing of total dry-matter accumulation in winter wheat. Remote Sens. Environ. 1981, 11, 171–189. [Google Scholar] [CrossRef] [Green Version]
- Asrar, G.; Kanemasu, E.T.; Jackson, R.D.; Pinter, P.J. Estimation of total above-ground phytomass production using remotely sensed data. Remote Sens. Environ. 1985, 17, 211–220. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Wang, J.; Song, X.; Feng, H. Winter wheat biomass estimation based on spectral indices, band depth analysis and partial least squares regression using hyperspectral measurements. Comput. Electron. Agric. 2014, 100, 51–59. [Google Scholar] [CrossRef]
- Ghosh, P.; Mandal, D.; Bhattacharya, A.; Nanda, M.K.; Bera, S. Assessing crop monitoring potential of Sentinal-2 in a spatio-temporal scale. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Volume XLII-5, ISPRS TC V Mid-Term Symposium “Geospatial Technology-Pixel to People”, Dehradun, India, 20–23 November 2018. [Google Scholar]
- Li, C.; Li, H.; Li, J.; Lei, Y.; Li, C.; Manevski, K.; Shen, Y. Using NDVI percentiles to monitor real-time crop growth. Comput. Electron. Agric. 2019, 162, 357–363. [Google Scholar] [CrossRef]
- Battude, M.; Al Bitar, A.; Morin, D.; Cros, J.; Huc, M.; Marais Sicre, C.; Le Dantec, V.; Demarez, V. Estimating maize biomass and yield over large areas using high spatial and temporal resolution Sentinel-2 like remote sensing data. Remote Sens. Environ. 2016, 184, 668–681. [Google Scholar] [CrossRef]
- Campos, I.; González-Gómez, L.; Villodre, J.; González-Piqueras, J.; Suyker, A.E.; Calera, A. Remote sensing-based crop biomass with water or light-driven crop growth models in wheat commercial fields. Field Crop. Res. 2018, 216, 175–188. [Google Scholar] [CrossRef]
- Liao, C.; Wang, J.; Dong, T.; Shang, J.; Liu, J.; Song, Y. Using spatio-temporal fusion of Landsat-8 and MODIS data to derive phenology, biomass and yield estimates for corn and soybean. Sci. Total Environ. 2019, 650, 1707–1721. [Google Scholar] [CrossRef]
- Araya, S.; Ostendorf, B.; Lyle, G.; Lewis, M. Remote Sensing Derived Phenological Metrics to Assess the Spatio-Temporal growth Variability in Cropping Fields. Adv. Remote Sens. 2017, 6, 212–228. [Google Scholar] [CrossRef] [Green Version]
- Perry, E.M.; Morse-McNabb, E.M.; Nuttal, J.G.; O’Leary, G.J.; Clark, R. Managing Wheat From Space: Linking MODIS NDVI and Crop Models for Predicting Australian Dryland Wheat Biomass. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3724–3731. [Google Scholar] [CrossRef]
- Li, W.; Niu, Z.; Huang, N.; Wang, C.; Gao, S.; Wu, C. Airborne LiDAR technique for estimating biomass components of maize: A case study in Zhangye City, Northwest China. Ecol. Indic. 2015, 57, 486–496. [Google Scholar] [CrossRef]
- Periasamy, S. Significance of dual polarimetric synthetic aperture radar in biomass retrieval: An attempt on Sentinel-1. Remote Sens. Environ. 2018, 217, 537–549. [Google Scholar] [CrossRef]
- Wigneron, J.P.; Ferrazzoli, P.; Olioso, A.; Bertuzzi, P.; Chanzy, A. A Simple Approach To Monitor Crop Biomass from C-Band Radar Data. Remote Sens. Environ. 1999, 69, 179–188. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.; Deering, D. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; Texas A&M University, Remote Sensing Center: College Station, TX, USA, 1973. [Google Scholar]
- Jönsson, P.; Eklundh, L. TIMESAT—A program for analyzing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef] [Green Version]
- Araya, S.; Ostendorf, B.; Lyle, G.; Lewis, M. CropPhenology: An R package for extracting crop phenology from time series remotely sensed vegetation index imagery. Ecol. Inform. 2018, 46, 45–56. [Google Scholar] [CrossRef]
- Hill, M.J.; Donald, G.E. Estimating spatio-temporal patterns of agricultural productivity in fragmented landscapes using AVHRR NDVI time series. Remote Sens. Environ. 2003, 84, 367–384. [Google Scholar] [CrossRef]
- Nakayama, T. Impact of anthropogenic activity on eco-hydrological process in continental scales. Procedia Environ. Sci. 2012, 13, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Zhang, G.; Ling, H.; Han, F. Comparison of time-integrated NDVI and annual maximum NDVI for assessing grassland dynamics. Ecol. Indic. 2022, 136, 108611. [Google Scholar] [CrossRef]
- Calera, A.; González-Piqueras, J.; Melia, J. Monitoring barley and corn growth from remote sensing data at field scale. Int. J. Remote Sens. 2004, 25, 97–109. [Google Scholar] [CrossRef]
- Mirasi, A.; Mahmoudi, A.; Navid, H.; Valizadeh Kamran, K.; Asoodar, M.A. Evaluation of sum-NDVI values to estimate wheat grain yields using multi-temporal Landsat OLI data. Geocarto Int. 2021, 36, 1309–1324. [Google Scholar] [CrossRef]
- Tefera, A.T.; Banerjee, B.P.; Pandey, B.R.; James, L.; Puri, R.R.; Cooray, O.; Marsh, J.; Richards, M.; Kant, S.; Fitzgerald, G.J.; et al. Estimating early season growth and biomass of field pea for selection of divergent ideotypes using proximal sensing. Field Crop. Res. 2022, 277, 108407. [Google Scholar] [CrossRef]
- Julien, Y.; Sobrino, J.A. Comparison of cloud-reconstruction methods for time series of composite NDVI data. Remote Sens. Environ. 2010, 114, 618–625. [Google Scholar] [CrossRef]
- Julien, Y.; Sobrino, J.A. Optimizing and comparing gap-filling techniques using simulated NDVI time series from remotely sensed global data. Int. J. Appl. Earth Obs. Geoinf. 2019, 76, 93–111. [Google Scholar] [CrossRef]
- Kandasamy, S.; Baret, F.; Verger, A.; Neveux, P.; Weiss, M. A comparison of methods for smoothing and gap filling time series of remote sensing observations–application to MODIS LAI products. Biogeosciences 2013, 10, 4055–4071. [Google Scholar] [CrossRef] [Green Version]
- Rovira, A. Dryland mediterranean farming systems in Australia. Aust. J. Exp. Agric. 1992, 32, 801–809. [Google Scholar] [CrossRef]
- European Space Agency. Copernicus Sentinel-2 (Processed by ESA); MSI Level-2A BOA Reflectance Product; Collection 0; European Space Agency: Paris, France, 2021. [Google Scholar] [CrossRef]
- Planet Team. Planet Application Program Interface: In Space for Life on Earth; Planet Team: San Francisco, CA, USA, 2017. [Google Scholar]
- Sentinel-2 MSI Technical Guide Level-2A Algorithm Overview. Available online: https://sentinels.copernicus.eu/web/sentinel/technical-guides/sentinel-2-msi/level-2a/algorithm (accessed on 27 April 2022).
- UDM 2. Available online: https://developers.planet.com/docs/data/udm-2/ (accessed on 27 April 2022).
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- QGIS.org. QGIS Geographic Information System. QGIS Association. 2022. Available online: http://www.qgis.org (accessed on 20 June 2022).
- Ratcliff, C.; Gobbett, D.; Bramley, R. PAT-Precision Agriculture Tools; Software Collection; CSIRO: Canberra, Australia, 2019.
- Monteith, J.L. Solar Radiation and Productivity in Tropical Ecosystems. J. Appl. Ecol. 1972, 9, 747–766. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Du, X.; Wu, B. Generation of high spatial and temporal resolution NDVI and its application in crop biomass estimation. Int. J. Digit. Earth 2013, 6, 203–218. [Google Scholar] [CrossRef]
- Collis, C. 3D mapping profiles soil-based constraints. In GRDC Groundcover; Grains Research Development Corporation: Canberra, Australia, 2022. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).