1. Introduction
Crop modeling, which is a method of obtaining quantitative knowledge of how crops grow by interacting with the environment, is a useful tool for predicting the yield and quality to help with crop management [
1,
2]. Developed crop models include climatic variables such as the temperature, precipitation, and length of day [
3]. However, crop models are only applicable to normal patterns of the given climatic conditions, and all other conditions are assumed optimal [
1]. Therefore, crop models cannot provide realistic predictions of the crop yield or quality. Remote sensing (RS) technology has been combined with spectroscopy for nondestructive monitoring of the crop yield and quality under actual environmental conditions [
4]. Spectral reflectance data containing visible, red-edge, and near-infrared (NIR) spectral bands have tried to managing agriculture over a wide area [
5] by helping predict the growth status [
6]. This technology is expected to play a key role in integrated crop management systems [
7].
Rice (
Oryza sativa L.) is a major carbohydrate source for meeting energy and nutrient needs, and consumption is steadily increasing globally [
8]. It is an essential crop, especially in Asia, and its successful annual production has significant implications for the global community [
9]. Properties such as the color, flavor, and composition of the rice depend on the variety, storage conditions, and amylose content. Interactions among proteins, the breakdown products of lipid oxidation, and starch change the cell walls and proteins, which affect the rice quality [
10]. The rice quality is improved by reducing protein content and amylose [
11], which form by nitrogen uptake during the vegetative growth period, especially in the grain filling stage.
To recognize a specific rice field, a spatially precise RS system is required rather than a conventional spectrometer [
12]. Spectral imagery combines spectrometry and imaging, and it has been used to observe specific spatial variations [
13]. Spectral imagery can be used to assess the crop growth in individual areas while excluding external disturbances [
14]. It can be used to develop a crop model that includes various parameters, such as the biomass, crop quality, and physiological active substances of the crop, with a comprehensive range of crop–canopy attributes [
15].
Regression models have been developed to predict crop parameters during the cultivation period [
16]. Simple, multiple, and multivariate linear regression analyses have been used with RS spectral data for agricultural applications. Simple linear regression (SLR) has been used with satellite spectral imagery to predict rice yields where the yield is the dependent variable and the vegetation index (VI) is the independent variable [
17]. Multiple linear regression (MLR) and partial least squares regression (PLSR) have been used with airborne and satellite spectral imagery to predict the protein and nitrogen contents of rice [
18,
19]. Unmanned aerial vehicles (UAVs) are suitable for precise monitoring of crop parameters with a high spatial resolution by flying at low altitudes, and they are advantageous for acquiring time series data with unconstrained scheduling [
20]. The multispectral imagery acquired by UAVs has been used for developing linear regression models by combining VIs for predicting the rice yield or biomass depending on time series [
21]. Various approaches have been attempted to predict rice yield by labeling pixels in the ear region using K-means clustering on RGB imagery as well as spectral imagery [
22]. In the prediction of rice-protein content, which requires more spectroscopy than morphology, some studies have suggested that green normalization difference VI (GNDVI) calculated from NIR and green is the most advantageous [
23,
24].
Recently, machine learning (ML) has been used to predict rice parameters with a high level of accuracy [
25]. Among studies using ML with spectral imagery acquired by UAVs, linear ML-based ridge regression (RR) was used to predict the nitrogen content of rice in a single year [
26]. Additionally, a nonlinear ML-based artificial neural network (ANN) was used to predict the moisture content of rice to reduce economic losses regarding the yield, quality, and drying cost at harvest time in a single year [
27]. Among the various regression methods for the prediction of rice parameters, it is important to select a regression method that can be developed as a year-invariant model. The potential of developing a year-invariant model must be verified through cross-validation for different years via the collection of multiyear data [
28]. However, although the model is based on multiyear data, changes in the environmental conditions at various fields may make it difficult to reproduce the model [
18]. Therefore, cumulative climate data during growth periods in each year may be required as input variables besides the spectral imagery to realize a year-invariant model [
29]. In addition, incorporating three-dimensional (3D) technology may have a positive effect on predicting rice growth grown under various environmental conditions. A study showed the possibility of predicting the crop yields depending on different space-times using a 3D-convolution neural network in RGB imagery acquired by UAV [
30].
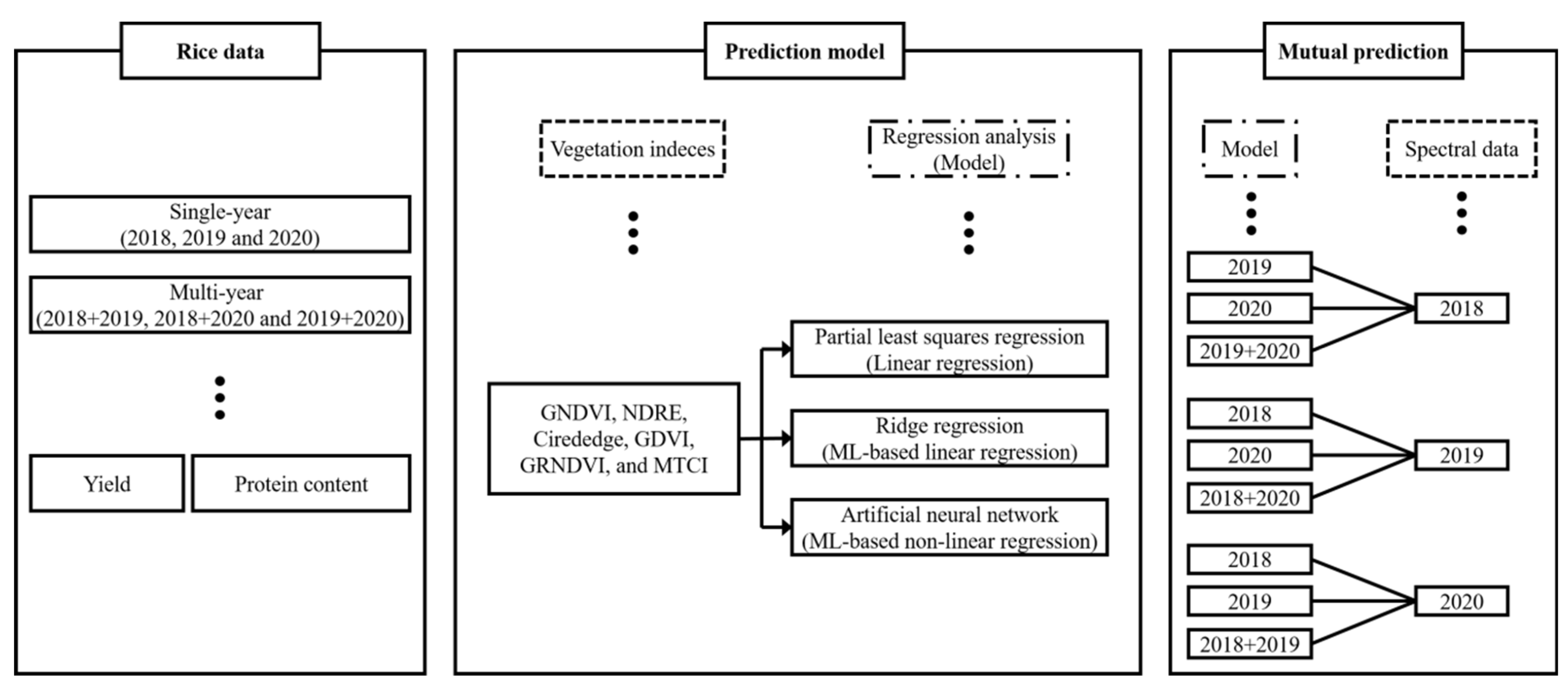
The objective of this study was to develop a regression model for predicting the rice yield and protein content and to select the most suitable regression analysis method for the year-invariant model. UAV-based multispectral imagery data for rice canopies at the ripening stage were collected in 2018, 2019, and 2020. Different types of regression models were developed for single years (2018, 2019, and 2020) and multiyears (2018 + 2019, 2018 + 2020, 2019 + 2020, and all years): PLSR (linear regression), RR (ML-based linear regression), and ANN (ML-based nonlinear regression). The regression models were evaluated by comparing the prediction performances for the rice yield and protein content. The reproducibility was verified via cross-validation of the regression models for single- and multi-year analyses in nonoverlapping years through mutual prediction. Finally, the regression model with the highest potential for the year-invariant model was selected.
2. Materials and Methods
2.1. Experimental Design
2.1.1. Field Site and History
The experiment was conducted in a rice field of the Agricultural Research and Extension Services in Jinju-si, Gyeongsangnam-do, Republic of Korea for a 3 year period (2018–2020), as shown in
Figure 1. The rice-field area was 1550 m
2. The primary soil type of the rice field was sandy loam, and the rice cultivar was Yeonghojinmi. This is a high-quality cultivar artificially derived from crossing the high-quality Hitomebre with Junam, which has high disease resistance. Yeonghojinmi is a mid-late maturing ecotype that is mainly cultivated in the southern part of the Republic of Korea [
31]. Rice seedlings were mechanically transplanted at a density of 30 × 14 cm.
Table 1 presents the rice-cultivation schedule, which includes seedling management, water management, fertilizer prescription, and harvest time. The fertilizer prescription was divided into six levels of nitrogen (N) fertilization: 0, 5, 7, 9, 11, and 17 kg/1000 m
2. Additionally, each level received 4.5 kg/1000 m
2 of phosphorous (P) and 5.7 kg/1000 m
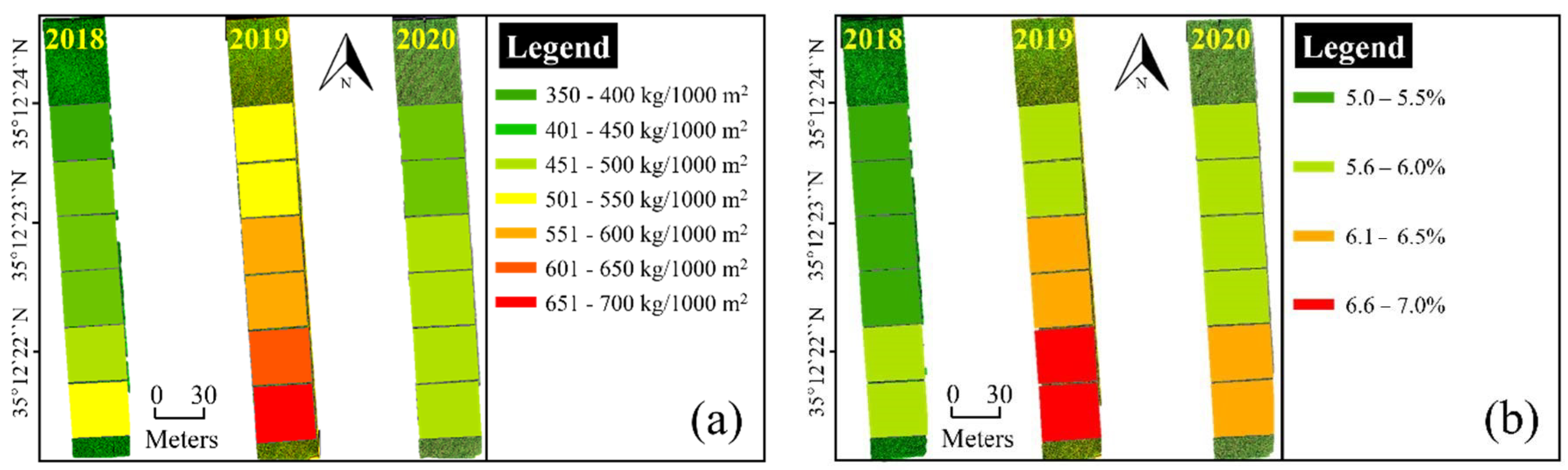
2 of potassium (K) for all years. N fertilization was applied three times at a ratio of 50%, 20%, and 30%, corresponding to the basal dressing before seedling transplantation, topdressing in the tillering stage, and topdressing in the panicle initiation stage. All P fertilization was applied to the basal dressing, and K fertilization was applied at a ratio of 70%, 0%, and 30%. The climatic conditions, such as the growing degree days (GDD) and accumulated sunlight hours (ASH), were collected from a weather station located in the Agricultural Research and Extension Services. The GDD and ASH were 1046 °C and 884 h, respectively, for 2018; 1118 °C and 873 h, respectively, for 2019; and 1108 °C and 790 h, respectively, for 2020. The multispectral images were acquired at midday on 4 October 2018; 1 October 2019; and 5 October 2020. Immediately after image acquisition, sampling was performed three times in 2018 and four times in 2019 and 2020, depending on each nitrogen treatment.
2.1.2. Measurement of the Rice Yield and Protein Content
To measure the rice yield and protein content, 100 and 15 rice plants were respectively sampled from each N fertilization treatment field just before harvest (18 October 2018; 17 October 2019; and 17 October 2020). The rice yield was measured by multiplying the number of rice ears per square meter (N), percent ripened grain (PRG), and unit seed weight (SW):
The protein content of the polished rice was measured with a rice taste analyzer (Infratec 1241, FOSS Tecator, Hoganas, Sweden).
2.2. Acquisition of Multispectral Images
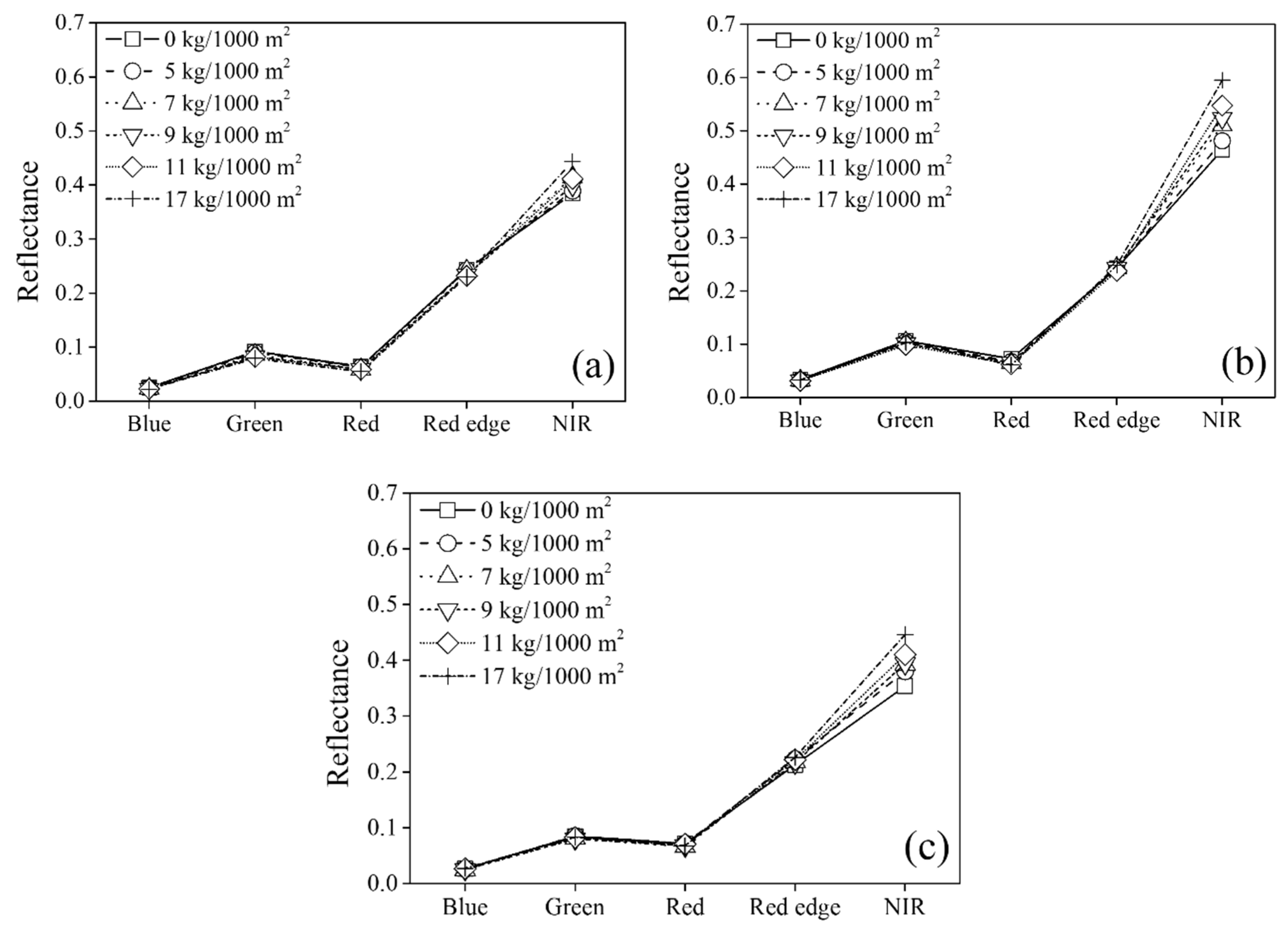
Multispectral images were acquired with a Red-edge-M (Micasense, Inc., Seattle, WA, USA) mounted on UAVs: a 3DR Solo (3D Robotics, Berkeley, CA, USA) in 2018 and a M600 (DJI Technology Co., Ltd., Shenzhen, China) in 2019 and 2020. The 3DR Solo and M600 are quadcopters weighing 1.5 and 9.1 kg, respectively, with dimensions of 0.26 m × 0.26 m × 0.25 m and 1.67 m × 1.52 m × 0.76 m, respectively. The flight plan software for the UAVs were Mission Planner (ArduPilot Dev Team, New York, NY, USA) for the 3DR Solo and DJI Pilot (DJI Technology Co., Ltd., China) for the M600. Red-edge-M was equipped with a sunshine sensor to correct the reflectance data for each spectral image depending on the sunlight conditions at the time of image capture. The multispectral image and sunshine sensors were configured to measure the reflectance of the central wavelength ± full width at half maximum for five spectral bands: blue (475 ± 32 nm), green (560 ± 27 nm), red (668 ± 14 nm), red-edge (717 ± 12 nm), and NIR (842 ± 57 nm). The image sensor provided 12 bits/pixel images of each spectral band geotagged with the GPS information and pitch, roll, and yaw information from the inertial measurement unit sensor.
Because different UAVs were used each year, the flight conditions varied. For the 3DR Solo used in 2018, the spatial resolution, flight speed, front overlap rate, and side overlap rate were about 1.4 cm at an altitude of 20 m, 2 m/s, 80%, and 80%, respectively. For the M600 used in 2019 and 2020, these were about 3.5 cm at an altitude of 50 m, 3 m/s, 70%, and 70%, respectively. Multiple spectral images per flight were acquired under different flight conditions. The multispectral images were acquired 10–16 days (4 October 2018; 1 October 2019; and 5 October 2020) before harvest.
2.3. Image Processing and Analysis
The geotagged images were mosaicked with geometric and radiometric correction in Pix4d Mapper Pro (Pix4d S.A., Prilly, Switzerland). The mosaicked multispectral images were converted into GNDVI images with the equation in
Table 2. The reflectance of the rice canopy area for each spectral image was extracted at each sampling position based on the optimal threshold value for minimizing the effects of water and soil from the GNDVI image.
2.4. Regression Analysis
2.4.1. Partial Least Squares Regression
PLSR is a multivariate linear regression method where the least squares method is applied to linear combinations of independent and dependent variables to derive latent variables (LVs) with high covariance. In other words, PLSR proceeds in the direction that describes both dependent and independent variables. PLSR establishes the optimal LVs that maximize the explained variance in the dependent variable from independent variables and that minimize the predicted residual sum of squares (RSS) and mean square error (MSE) [
32].
2.4.2. Ridge Regression
RR is a regularized linear regression method that limits the regression coefficient while minimizing RSS and MSE. The advantage of RR over the least squares method is the tradeoff between the bias and variance [
33]. If RR is set to a penalty of zero, it simply produces an unbiased least squares predictor, and the variance for the predictor is large. Increasing the penalty decreases the RR coefficient, which causes a slight bias but sharply decreases the variance and MSE. Above a certain penalty, the decrease in variance with increasing penalty slows down, and the bias significantly increases as the coefficient approaches zero [
34]. In other words, the penalty determines the strength of the constraint. As the penalty increases, the flexibility of RR decreases, which decreases the variance but increases the bias.
2.4.3. Artificial Neural Network
ANN is a type of ML that mimics the central nervous system. The perceptron is a simple ANN algorithm that receives multiple signals, such as independent variables, and outputs one signal [
35]:
The weight,
, acts like neurons in the brain transferring information through electrical signals. Each input signal is an independent variable that is given a unique
. When the sum of the signals exceeds a predetermined threshold in the activation function, the output is 1; otherwise, the output is 0 or −1. The bias,
, determines the strength of the perceptron activation. However, a perceptron cannot solve nonlinear problems [
36]. This difficulty can be overcome by constructing an ANN, which stacks perceptron layers. The ANN comprises an input layer, a hidden layer, and an output layer. The hidden layer (i.e., black box) can produce the optimal predictor by learning the input and output.
2.5. Development of the Regression Models and Mutual Prediction
PLSR-, RR-, and ANN-based prediction models for the rice yield and protein content were developed with six VIs (green normalization difference vegetation index (GNDVI), normalization difference red-edge index (NDRE), chlorophyll index red edge (CIrededge), difference NIR/Green green difference vegetation index (GDVI), green-red NDVI (GRNDVI), and medium resolution imaging spectrometer terrestrial chlorophyll index (MTCI)) in
Table 2 in Python (Python Software Foundation, USA), as shown in
Figure 2. The model performances for single-year data (2018, 2019, and 2020) and multiyear data (2018 + 2019, 2018 + 2020, 2019 + 2020, and all years) were evaluated according to the coefficient of determination (R
2), root-mean-square error (RMSE), and relative error (RE).
Table 2.
Vegetation indices calculated from the reflectances of five spectral bands of rice canopies extracted from multispectral images.
Table 2.
Vegetation indices calculated from the reflectances of five spectral bands of rice canopies extracted from multispectral images.
| Vegetation Index | Equation | Reference |
|---|
| Green normalization difference vegetation index (GNDVI) | | [37] |
| Normalization difference red-edge index (NDRE) | | [38] |
| Chlorophyll index red edge (CIrededge) | | [39] |
| Difference NIR/Green green difference vegetation index (GDVI) | | [40] |
| Green-red NDVI (GRNDVI) | | [41] |
| Medium resolution imaging spectrometer terrestrial chlorophyll index (MTCI) | | [42] |
The grid search method was used to identify the best LVs for PLSR, penalty for RR, and epoch for ANN that minimized the MSE. This means that the model exhibited the best performance with the selected tuning parameters. The ANN structure comprised five neurons matching the number of spectral bands as independent variables in a single hidden layer; this was based on the results of a previous study showing that a single hidden layer is sufficient for predicting the rice-protein content [
43]. A rectified linear unit was used to overcome the vanishing gradient problem of sigmoids and serve as the activity function for the ANN [
44]. The prediction performances of PLSR, RR, and ANN for the rice yield and protein content were evaluated.
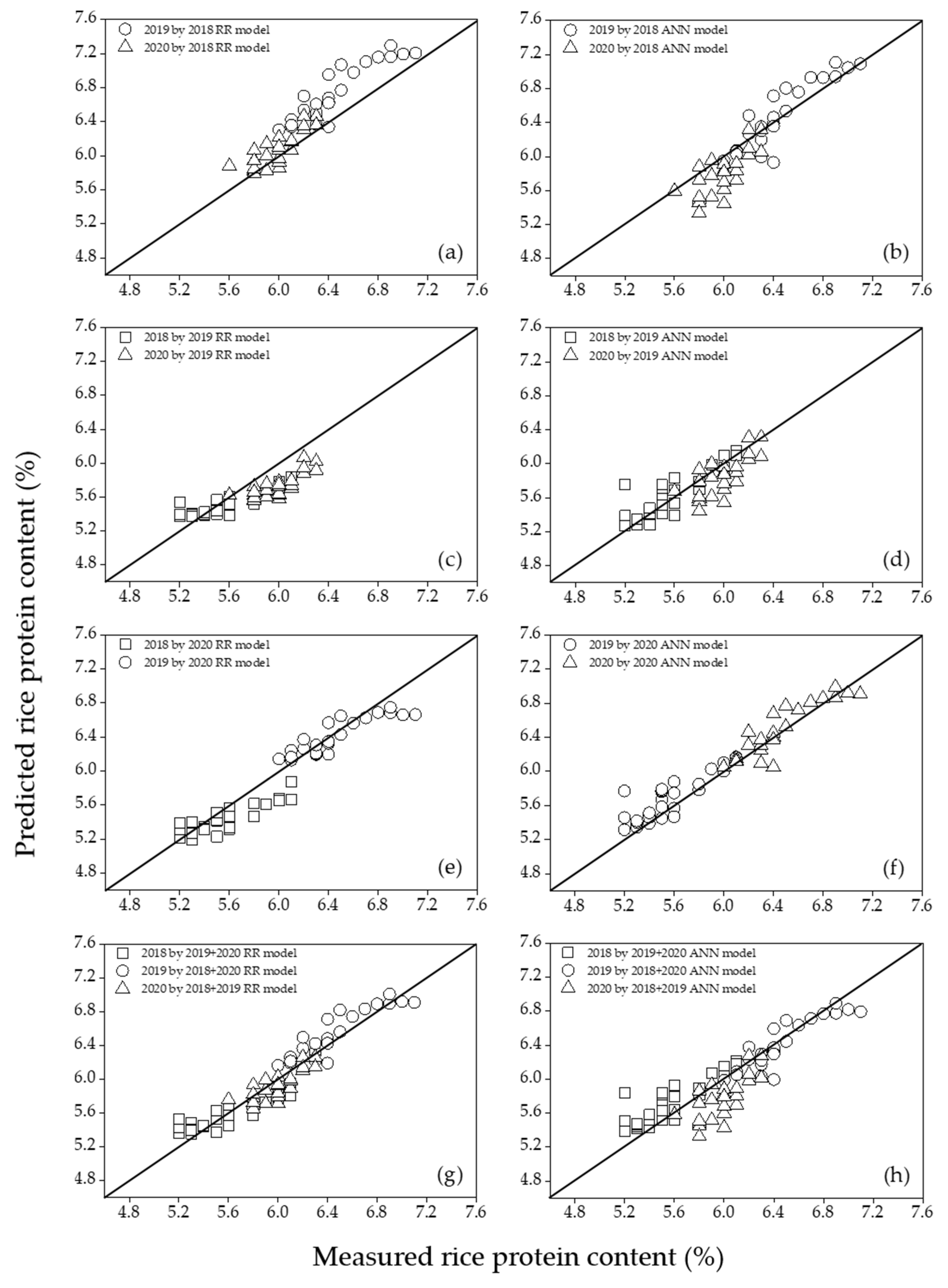
The regression models were cross-validated for single- and multi-year data in nonoverlapping years through mutual prediction, as shown in
Figure 2. Cross-validation was used to verify that the model could predict the rice yield and protein content in nonoverlapping years and was evaluated according to the RMSE of prediction (RMSEP) and RE calculated from the 1:1 reference line. Finally, by evaluating the impact of each input VI, the regression model with the highest potential for the year-invariant model was selected.
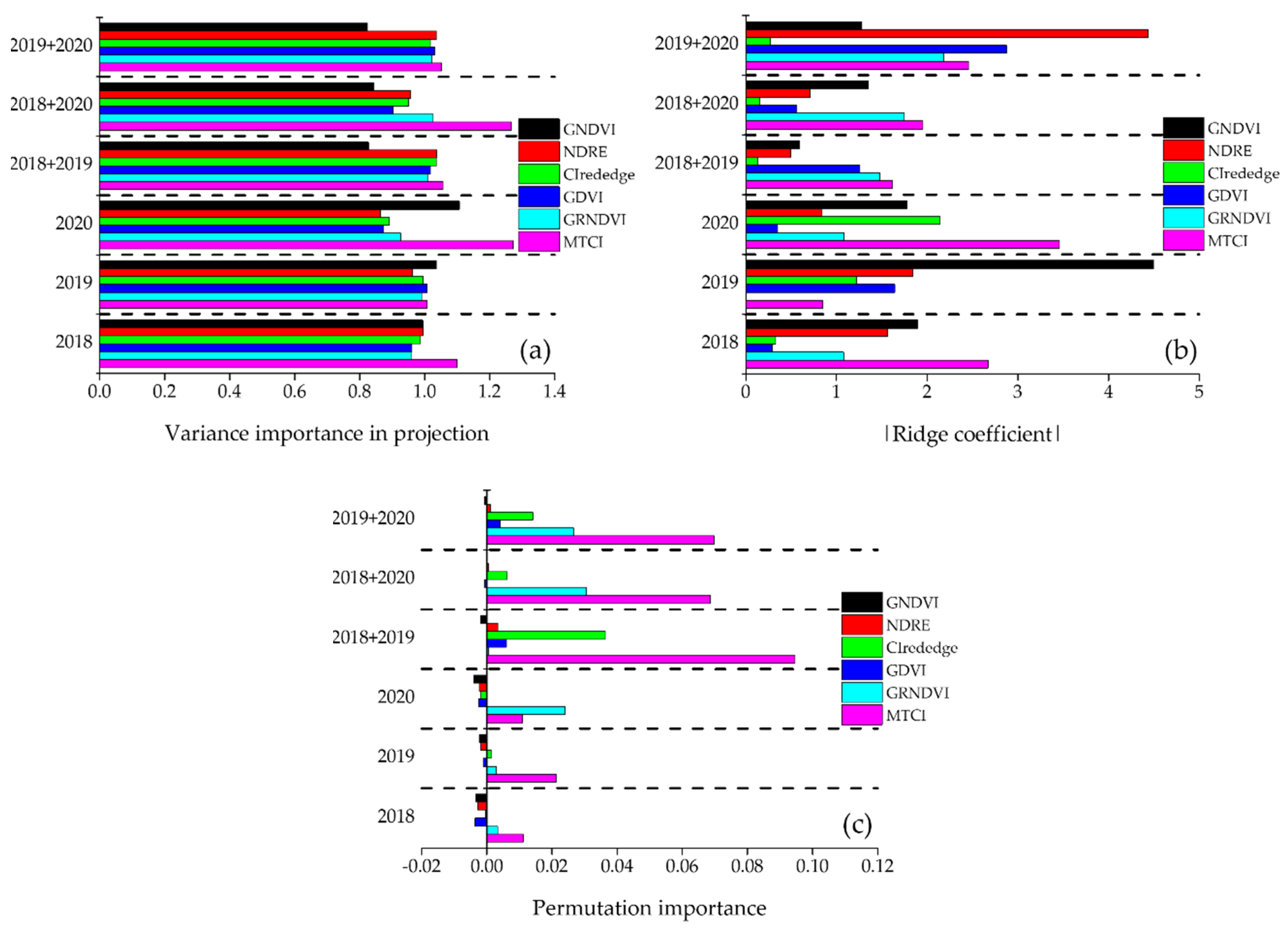
5. Conclusions
PLSR, RR, and ANN were applied to UAV-based multispectral imagery of rice canopies in the ripening stage to develop prediction models for the rice yield and protein content. ANN exhibited a poorer prediction performance (R2 ≥ 0.71, RMSE ≤ 29.0 kg/1000 m2, and RE ≤ 5.68% in the case of rice yield and R2 ≥ 0.57, RMSE ≤ 0.16%, and RE ≤ 2.88% in the case of rice-protein content) than PLSR and RR. However, for an accurate prediction of both rice yield and rice-protein content, the ANN model yielded more stable prediction errors (24.2 kg/1000 m2 ≤ RMSEP ≤ 59.1 kg/1000 m2 in the case of rice yield, and 0.14% ≤ RMSEP ≤ 0.28% in the case of rice-protein content) than PLSR and RR (23.7 kg/1000 m2 ≤ RMSEP ≤ 83.3 kg/1000 m2 in the case of rice yield and 0.12% ≤ RMSEP ≤ 0.75% in the case of rice-protein content) in all single- and multi- analyses. In each analysis, the ANN model gave each VI the same ranking of importance. This consistency may have maintained the RMSEPs in each single-year and multiyear mutual prediction by ANN. For this reason, it was selected as the prediction model that best explained the rice yield and protein content, and as the most promising method for developing a year-invariant model under different cultivation and environmental conditions in the future. The MTCI was commonly the most important variable for predicting rice yield and protein content, followed by CIrededge for rice yield and GRNDVI for rice-protein content. This methodology, which select the optimal model by comparing the ranking of important variables for each year using other regression analysis and by evaluating error stability through mutual prediction, will be useful for the development of available prediction models in the field of agricultural remote sensing. The ANN model proposed in this study can verify the potential of a year-invariant model by collecting data from the same rice field under similar environmental conditions; however, it cannot verify the reproducibility of the model under different environmental conditions. Therefore, evaluation and reproduction of the ANN model is required for yield and protein content analysis in various environment conditions such as major rice-cultivation complexes.