1. Introduction
Fine-grained tree species classification is the basis of forest management planning and interference monitoring, which is conducive to the scientific management and effective use of forest resources. Numerous continuous narrow spectral bands and high spatial resolution of hyperspectral images (HSI) can provide a wealth of available spectral information for each pixel on the land cover mapping [
1]. Forest-type recognition is an important aspect of hyperspectral image application [
1,
2,
3], and it plays an important role in the fine-grained classification of tree species [
4].
Deep learning can extract high-level and semantic features [
5,
6]. Its objective function focuses directly on classification, and completes the process of data feature extraction and classifier training automatically. The complex feature extraction and selection process is replaced by a simple end-to-end deep workflow [
7,
8].
Recently, convolution neural networks (CNNs) for hyperspectral classification have made significant developments [
9,
10,
11,
12,
13,
14,
15]. For instance, Chen et al. [
10] addressed the classification problem of three hyperspectral public data sets (Indian Pines, University of Pavia, and Kennedy Space Center) by making use of 1D, 2D, and 3D CNNs and found that the 3D CNN can obtain the best classification effect, whereas the 2D CNN is superior to the 1D CNN. In [
16], Zhang et al. explored an improved 3D CNN, named 3D-1D CNN, for tree species classification, which converts the joint spatial-spectral feature extracted by the last Conv3D layer into a 1D feature. This model can shorten the training time of the 3D CNN model by 60%, although it loses some classification accuracy.
Deep learning has achieved inspiring results in classification applications, but when there are few training examples, its classification performance will still decrease [
17]. In the forest, sample collection will be hindered due to the complex terrain and stand structure. At the same time, the heterogeneity of structure and tree species composition and the similarity of image features make it difficult to label the samples when classifying forest tree species. Therefore, the problem of tree species classification based on a few-shot set of deep learning methods needs to be solved urgently [
18,
19].
Few-shot learning refers to performing related learning tasks when there are fewer training samples in categories. According to the different application scenarios, the number of categories and the sample size within a category show differences in the few-shot public data sets given at present. They mainly include Omniglot [
20], CIFAR-100 [
21], MiniImageNet [
19], Tiered-ImageNet [
22], and CUB-200 [
23]. The overall classification results show that the more categories and more samples in the categories, the more conducive to few-shot image classification [
24]. Hyperspectral images processing presents many challenges, including high data dimensionality and usually low numbers of training samples [
25]. Currently, public hyperspectral remote sensing image data sets include Indian Pines [
9,
10,
26,
27,
28,
29], Salinas [
9,
27], Pavia University [
9,
10,
26,
27,
28,
29,
30,
31], and the Kennedy Space Center (KSC) [
10,
30], etc.
Table 1 shows the detailed information of the four hyperspectral data sets. In general, these data sets have low spatial resolution, significant differences between categories, and regular boundaries, and are mainly used in the classification of urban ground objects and crops. When applied to the classification of forestry tree species, the accuracy often decreases because the spectral response of different plants of the same family and genus are very similar, especially under the fragmented species distribution, complex topography, and the occluded canopy [
16]. Therefore, establishing a hyperspectral data set suitable for forest tree classification is the primary problem of our research. In the complex forest stand structure, the uneven distribution of samples and the noise generated by the background pixels are difficult to identify directly through hyperspectral images. It is necessary to rely on data collected by ground plot surveys and forest sub-compartment surveys, etc., but the sample points obtained in this way are difficult and relatively scarce. Therefore, in the process of making samples, it is necessary to comprehensively consider the complexity of the hundreds of dimensional bands of hyperspectral remote sensing images and the limitations of obtaining prior knowledge in forest stands, and to establish a data set with the number of categories in line with the actual situation and the size of the samples within the category as sufficient as possible. The data set can be applied to the classification of tropical and subtropical forest species similar to those of this study area.
Attention plays an important role in human perception. A significant characteristic of the human visual system is that it does not try to process the entire scene immediately, but selectively focuses on the salient parts in order to better capture the visual structure. Attention can be directed to the focal point, and the expression ability can be improved by using the attention mechanism, that is, focusing on important features and suppressing unnecessary features. Convolutional block attention module (CBAM) is a simple and effective attention module for feedforward convolutional neural networks [
32]. Given an intermediate feature map, CBAM will sequentially infer the attention map along two separate dimensions (channel and spatial) [
32] and then multiply the attention map by the input feature map for adaptive feature refinement [
33]. Since CBAM is a lightweight general-purpose module, it can be seamlessly integrated into any convolutional neural network (CNN) architecture [
25,
34], while the overhead is negligible, and it can engage in end-to-end training together with the basic CNN [
35,
36,
37]. Applying CBAM to the tree species classification of hyperspectral images aims to overcome the dimensional dilemma, adaptively reduce the impact of redundant bands on classification, and achieve a precise and efficient feature extraction so as to improve classification performance.
Matching networks uses the latest advances in attention to achieve fast learning. It is a weighted nearest-neighbor classifier applied within an embedding space. During the training process, the model imitates the test scenario of the few-shot task by sub-sampling the class labels and samples [
19]. The training process of the network is to establish the relationship or mapping between labels and samples in the training set, and directly apply it to the test set in the same way. Prototypical networks is part of special matching networks and has a simple network structure, which does not require complex hyper-parameters, guiding the learning of new tasks using past prior knowledge and experience [
38]. Moreover, it has great potential for solving few-shot classification problems. Compared with matching networks, it has fewer parameters and is more convenient to train. However, for the classification of hyperspectral images, the general prototypical networks structure is simple, and the problem of weak model generalization is prone to occur [
39].
Considering the large and fine-grained spatial and spectral characteristics of airborne hyperspectral images, we are faced with two major challenges:
Extracting effective features for classification based on a large amount of spatial and spectral information of hyperspectral images.
Obtaining a high-precision forest tree species classification model with limited samples.
In this study, we proposed a CBAM-P-Net model by embedding a CBAM module into the prototypical networks, analyzed the influence of the convolutional attention module on the network operation efficiency and results, optimized the prototypical networks structure and tuning parameters, proposed a training sample size and method suitable for tree species classification based on airborne hyperspectral data, and discussed the classification performance of CBAM-P-Net on hyperspectral images under the condition of few-shot. The results show that our method improved the efficiency of feature extraction, although it makes the network more complex and increases the computational cost to a certain extent.
2. Materials and Methods
2.1. Study Area
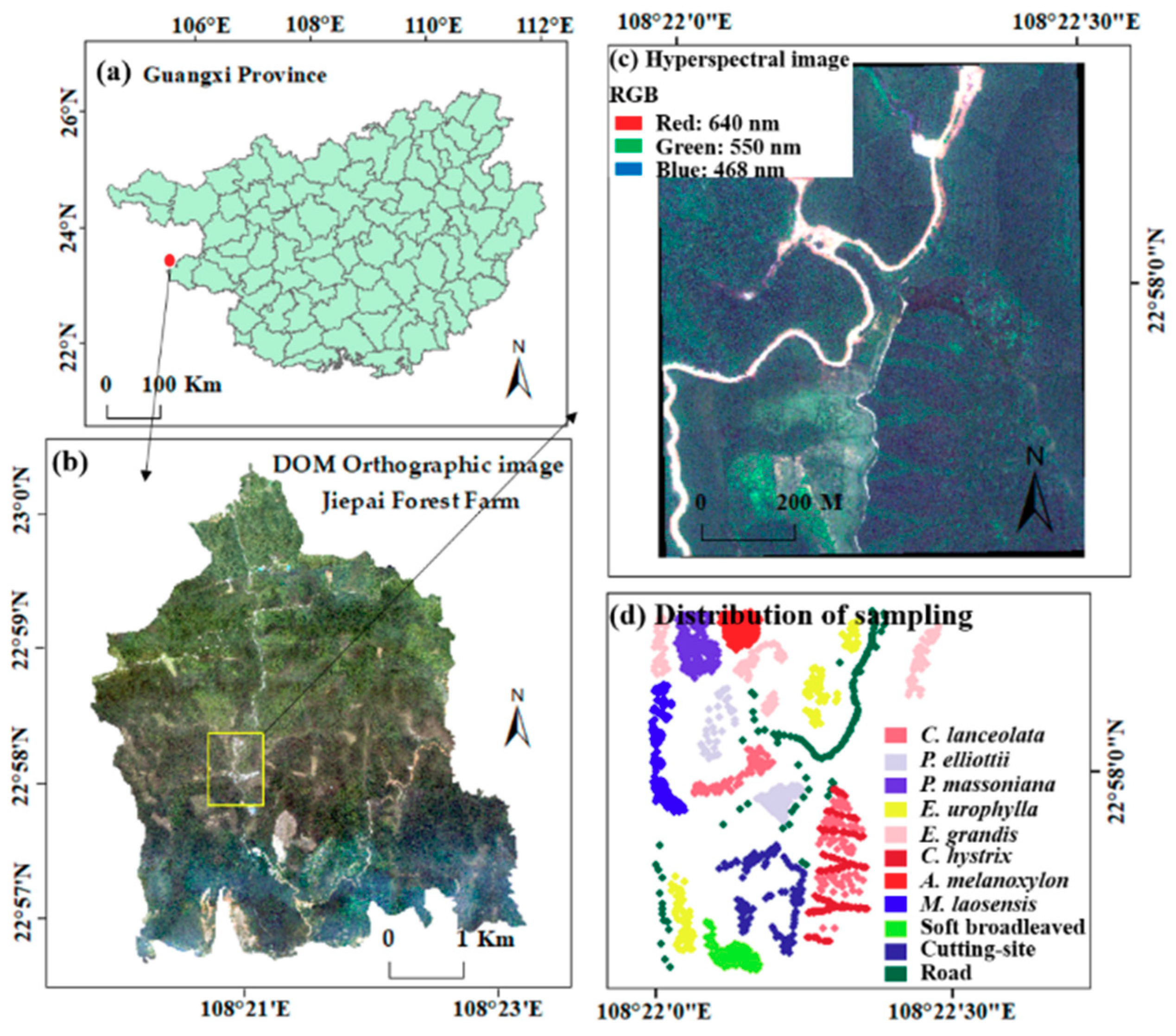
The study area is a sub-area of the Jiepai branch of Gaofeng Forest Farm in Nanning City, Guangxi Province, China (108°22′1″~108°22′30″ E, 22°57′42″~22°58′13″ N), belonging to the subtropical monsoon climate, with an area of 74.1 hm2. The average temperature is 21.6 °C, the average annual precipitation is 1200–1500 mm, and the average relative humidity is 79%. It is a hilly landform with an altitude of 149–263 m and a slope of 6~35°. The forest composition and structure in the study area have the typical characteristics of subtropical forests, with diverse tree species, fragmented and irregular distribution, complex tree structure, and a varied and luxuriant understory vegetation, which brings challenges to the classification of tree species. This paper classifies 11 categories in the study area, including 9 tree species, cutover land, and road. Of these, coniferous species include Cunninghamia lanceolate (C. lanceolate, CL), Pinus elliottii (P. elliottii, PE), and Pinus massoniana (P. massoniana, PM), and broadleaf species include Eucalyptus urophylla (E. urophylla, EU), Eucalyptus grandis (E. grandis, EG), Castanopsis hystrix (C. hystrix, CH), Acacia melanoxylon (A. melanoxylon, AM), Mytilaria laosensis (M. laosensis, ML), and other soft broadleaf species (SB). C. lanceolate, P. elliottii, P. massoniana, A. melanoxylon, and M. laosensis are mixed forests, and the remains are pure forests. Exploring the high-precision classification method of tree species in the study area has important guiding significance for the classification and mapping of forest stands with complex structures and composition.
2.2. Data Collection and Processing
2.2.1. Airborne Hyperspectral Data
The hyperspectral data acquisition was conducted on 13 January and 30 January 2019, under cloudless conditions, at noon. The hyperspectral equipment was equipped with the CAF’s (Chinese Academy of Forestry) LiCHy (LiDAR, CCD and Hyperspectral) system integrated by the German IGI company, including LiDAR sensors (LMS-Q680i, produced by RIEGL company), CCD cameras (DigiCAM-60), AISA Eagle II hyperspectral sensors (produced by Finland SPECIM company), and inertial navigation units (IMU). The aircraft had a flying speed of 180 km/h, a relative altitude of 750 m, an absolute altitude of 1000 m, and a course spacing of 400 m. The hyperspectral data is the radiance data after radiometric calibration and geometric correction. It contained 125 bands with a wavelength range of 400–987 nm, a spectral resolution of 3.3 nm, and a spatial resolution of 1 m.
Table 2 summarizes the detailed parameters of the hyperspectral sensors.
The Quick Atmospheric Correction (QUAC) method was used to perform atmospheric correction on hyperspectral images to eliminate the interference of light and the atmosphere on the reflectivity of ground objects. Due to the complex terrain of the study area, the brightness value of the images was uneven, so the hyperspectral image was corrected by the DEM based on the synchronously acquired LiDAR data, which eliminated the changes in the image radiance value caused by the undulation of the terrain. Savitzky-GOlay (SG) filtering [
40] was used to smooth the spectral data and effectively remove the noise caused by various factors.
2.2.2. Field Survey Data
The field data survey was conducted at the Jiepai branch of Gaofeng Forest Farm from 16 January 16 to 5 February 2019. First, through the visual interpretation of GF-2 satellite images with a resolution of 1 m, the sample plots were set up with uniform distribution. Ten plots with the size of 25 m × 25 m and nine plots with 25 m × 50 m were laid out, of which seven were C. lanceolate pure forest, three were E. urophylla pure forests, three were E. grandis pure forests, and the remaining six were other forest stands and mixed forests. The tree species mainly included C. lanceolate and P. massoniana, E. urophylla, E. grandis, C. hystrix, etc., with a total of 1657 trees. In the plot, each tree was positioned using the China Sanding STS-752 Series Total Station and measured including tree species, tree height, crown width, branch height, diameter at breast height, and other factors. At the same time, for areas where it was not possible to set up plots due to the complex terrain, a field positioning survey was conducted by handheld GPS, with 10–20 points for each tree species.
In order to keep the number of sample points of each feature category consistent and evenly distributed, for categories with too many sample points, the points located at plot edges and that were too densely distributed were deleted. For categories where the number of sample points was too few, based on field survey GPS location points and sample site survey data, combined with a 0.2 m resolution digital orthophoto map (DOM) and forest sub-compartments survey data, the sample points were manually marked on the image of the study area. In this way, 112 sample points were obtained for each category, a total of 1232 sample points (
Figure 1).
2.3. Sample Data and Prototypical Networks Construction
In the previous research, we have produced a complete set of sample data and constructed the classification framework of the prototypical networks [
39]. The sample data set is based on hyperspectral images, as the data source, centered on the screen coordinate representation of the actual measured point’s latitude and longitude, and clipped with different window sizes through the open source framework GDAL. The window size starts from 5 × 5, with a step length of 2 m, and then clips the sample data until 31 × 31, when the clipping area exceeds the study area. Finally, a sample data set (11 classes, 112 samples in each class, a total of 1232 samples) consistent with the number of sample points was obtained in different window sizes. The sample data were divided into training samples and test samples according to the ratio of 80% and 20%.
The classification principle of the prototypical networks is that the points of each class are clustered around a prototype. Specifically, the neural network learns the nonlinear mapping of the input to the embedding space and uses the average value of the support set as the prototype of its class in the embedding space. Next, the nearest class prototype is found to classify the embedded query points [
39]. The classification framework of the prototypical networks is shown in
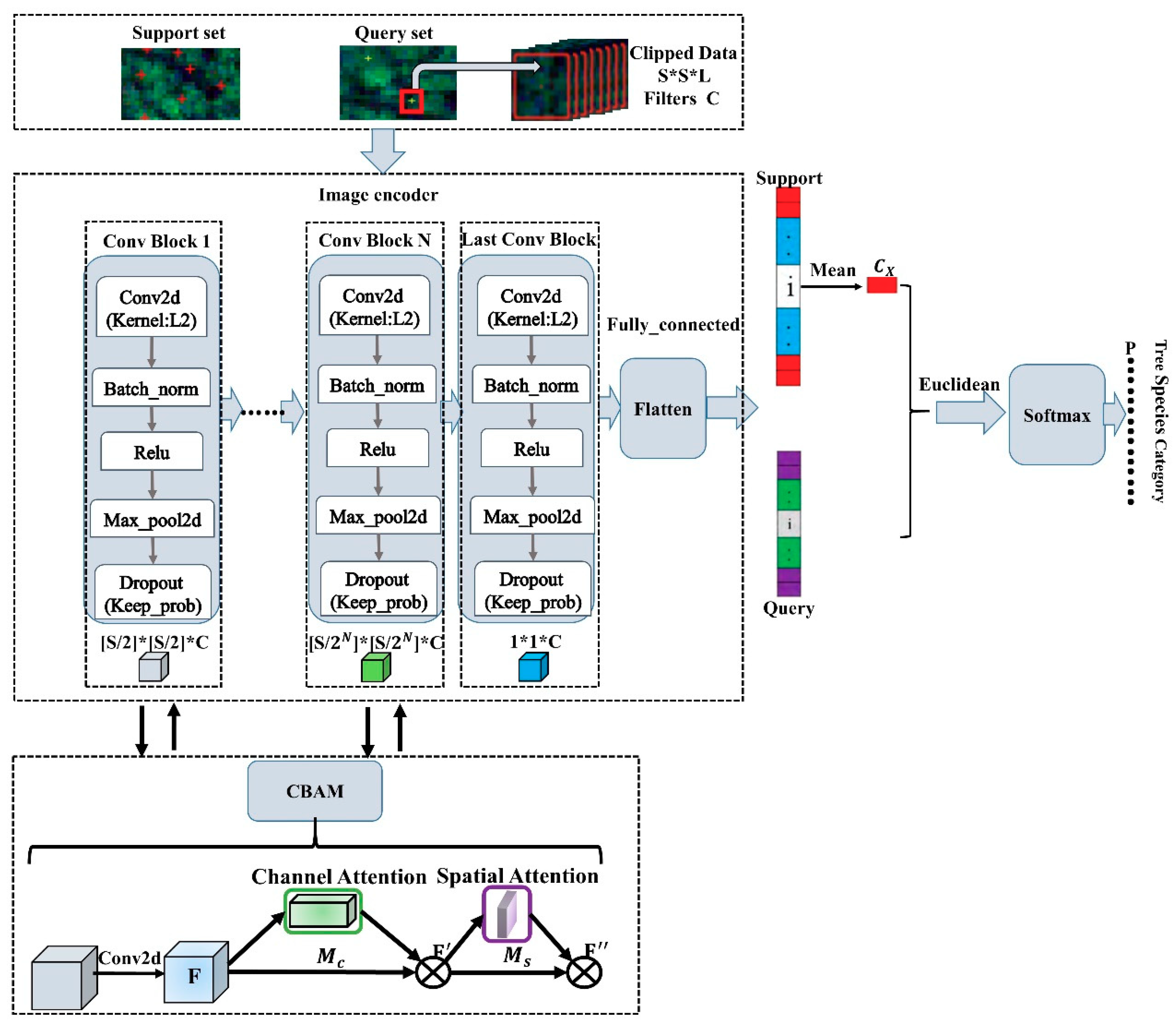
Figure 2, which mainly includes three parts: sample data input, image feature extraction, distance measurement and classification. In the prototypical networks, the sample data are divided into a support set and a query set. The support set is used to calculate the prototype, and the query set is used to optimize the prototype. If there are A classes and B samples in each class as the support set, it is A-way-B-shot. The image feature extraction part is to construct the embedding function (
,
is the learning parameter) to calculate the M-dimensional representation of each sample, that is, the image feature, and each class prototype (
) is the mean value of the feature vector obtained by the embedding function of the support set samples of its class. The square of the Euclidean distance is used to construct a linear classifier. From the projection of the sample to the embedding space, prototypical networks uses the distance function to calculate the distance from the query set x to the prototype, and then uses the softmax to calculate the probability of belonging to the category.
This research uses slice data of H × W × C (Height × Width × Channels) as the input of prototypical networks. The image feature extraction architecture is composed of different numbers of convolution blocks (Conv Block 1...Conv Block N, Last Conv Block) according to the size of the clipped data window. Each convolution block includes a convolution layer (Conv2d, output dimension F is 64, convolution kernel is 3 × 3), a batch normalization layer (Batch_norm), a non-linear activation function (ReLU), and a maximum Pooling layer (Max_pool2d, pooling kernel is 2 × 2). At the same time, in order to avoid model overfitting, the L2 regularization ( = 0.001) of the convolution kernel is added to the convolution layer, and Dropout is added after the maximum pooling layer (Keep_prob is 0.7). Feature values are processed through the fully connected layer (Flatten) and softmax as the basis for classification. The same embedding function is taken to operate the support set and the query set, then using them it as input parameters for the loss and precision calculations. All models are trained through Adam-SGD. The initial learning rate is 10−4, halving the learning rate every 2000 training sessions. Euclidean distance is used as the measurement function, and the loss function is a negative log likelihood function to train the prototypical networks.
2.4. Convolutional Block Attention Module
Convolutional Block Attention Module (CBAM) is an attention module that combines the spatial and channel dimensions (
Figure 3). CBAM can achieve better results compared with SENet [
41] because the attention mechanism adopted by the latter only focuses on channels. In addition, MaxPool is added to the network structure of CBAM, which makes up for the information lost by AvgPool to a certain extent. It can be seen from
Figure 3 that taking the feature map (
) extracted by CNN as input, CBAM sequentially obtains a one-dimensional Channel Attention feature map (
) and a two-dimensional Spatial Attention feature map (
). The entire attention process can be summarized as:
The symbol means multiply by element, and represents a new feature obtained through channel attention, which is used as the input of spatial attention. Finally, the feature of the entire CBAM output is obtained.
The soft mask mechanism proposed by Wang et al. [
42] can guarantee a better performance of the attention module. The equation is modified as:
2.4.1. Channel Attention
Producing a channel attention map by exploiting the inter-channel relationship of the features (
Figure 4). In order to effectively calculate channel attention, the spatial size of the input feature map needs to be compressed, and average pooling and maximum pooling are commonly used. It can be seen from
Figure 4 that the module takes the feature map as input, and obtains the features (
, C represents the number of channels) through spatial-based global maximum pooling and global average pooling. Then, through a multi-layer perceptron (MLP) composed of two dense layers, the features output by the MLP are element-wise summation multiplication, and then, through the sigmoid activation function, the channel attention feature map (
) is generated. The feature map will multiply the input feature by the element to obtain a new feature (
). The calculation equation is indicated by Formula (5).
where
represents the sigmoid activation function,
is the weight of the first hidden layer in the MLP,
r is the feature compression rate, and
is the weight of the second hidden layer in the MLP.
2.4.2. Spatial Attention
Generating a spatial attention map by utilizing the inter-spatial relationship of the features (
Figure 5). First, average pooling and maximum pooling operations along the channel axis are applied to generate the corresponding feature vectors, which are connected according to the channel axis to form an effective feature descriptor. On this basis, the convolutional layer is applied to generate the spatial attention feature map (
), which will be multiplied by the input feature to obtain a new feature (
). The calculation equation is indicated in Formula (6).
where
represents the sigmoid activation function, and
is a convolution operation with a kernel size of 7 × 7.
As shown in
Figure 6, this study inserts CBAM between the convolution blocks of the prototypical networks to construct CBAM-P-Net. CBAM focuses on channel and spatial features. In addition, the soft mask mechanism is used in each convolution block attention sub-module to ensure the performance of the model.
2.5. Accuracy Verification
The classification accuracy of CBAM-P-Net includes training accuracy and testing accuracy. The training accuracy is expressed by last epoch accuracy (LEA). The testing accuracy is expressed by average accuracy (AA, Equation (7)), overall accuracy (OA, Equation (8)), and Kappa coefficient (Kappa, Equation (9)).
where
n is the number of categories,
Xii is the number of correct classifications of a category in the error matrix,
X+i is the total number of true reference samples of that category,
Xi+ is the total number of categories classified into this category, and
M is the total number of samples.
2.6. Experiments Design
In this study, we designed four experimental schemes, as shown in
Table 3.