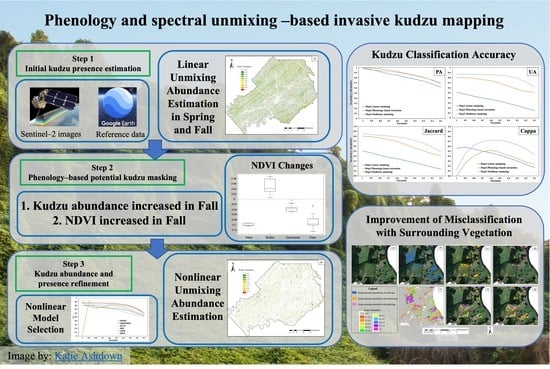
Phenology and Spectral Unmixing-Based Invasive Kudzu Mapping: A Case Study in Knox County, Tennessee
Abstract
:1. Introduction
2. Materials and Methods
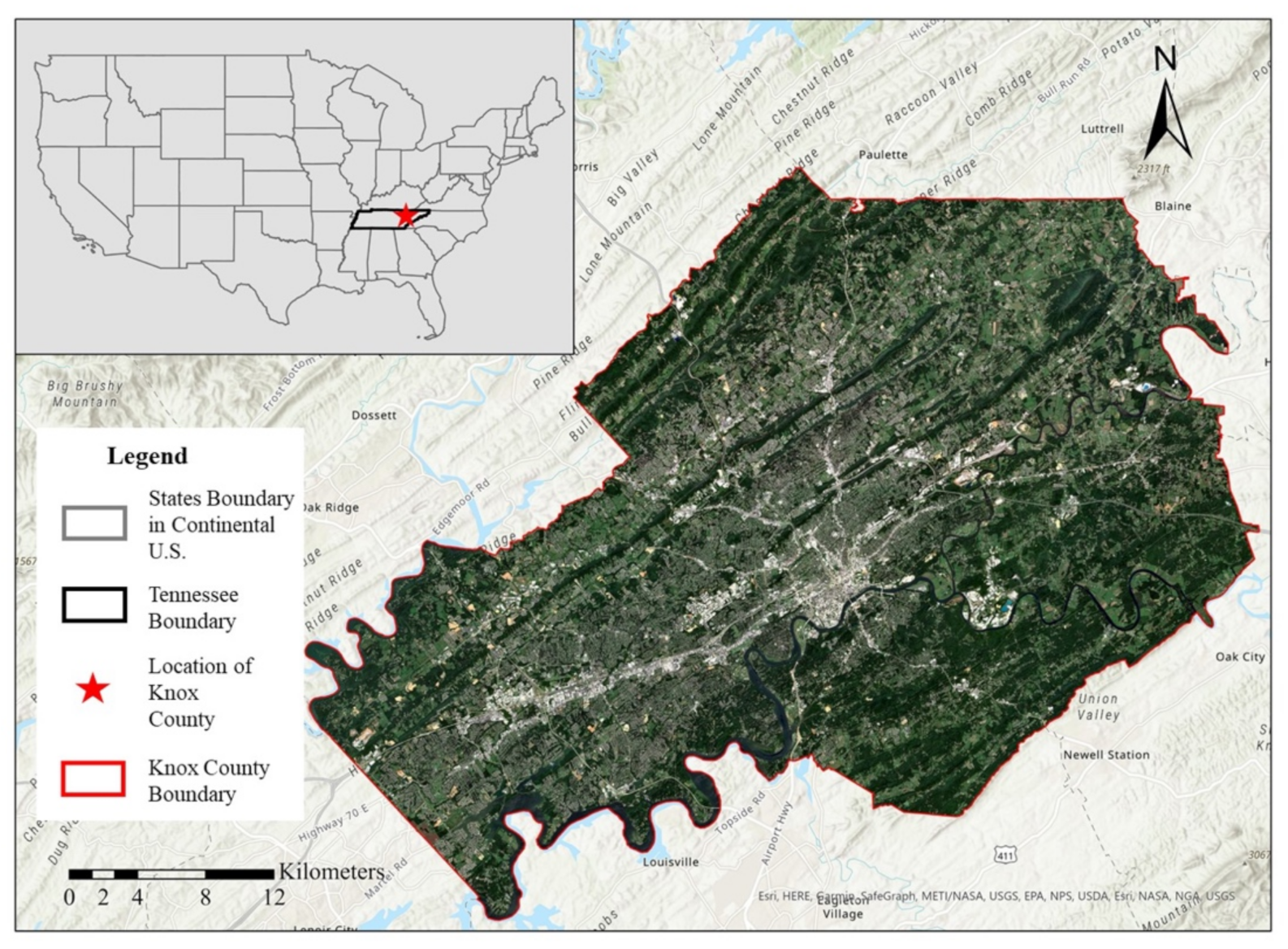
2.1. Study Area
2.2. Datasets
2.2.1. Remote Sensing Images and Pre-Processing
2.2.2. Reference Data
2.3. Spectral Unmixing and Phenology-Based Kudzu Identification Method
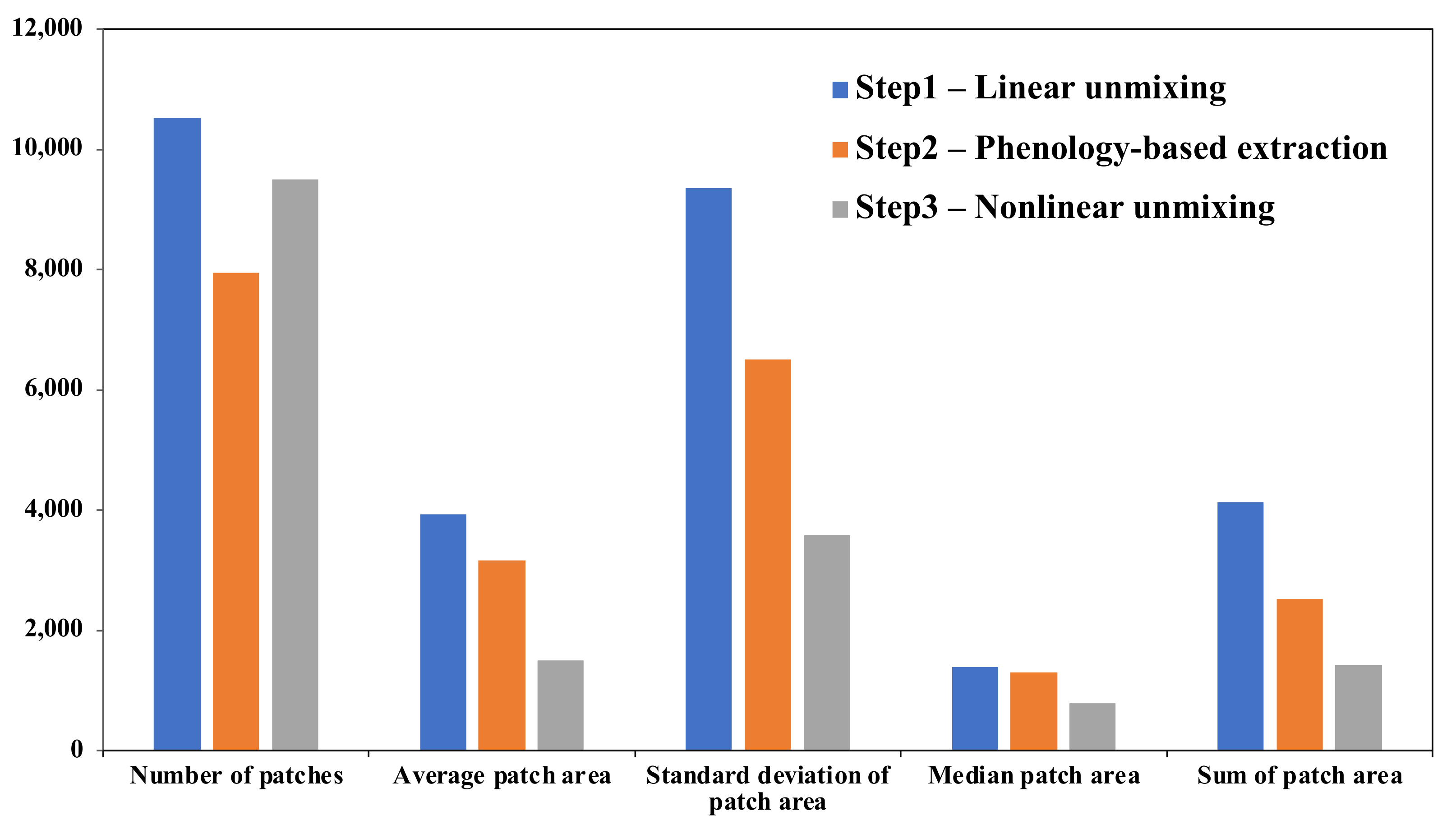
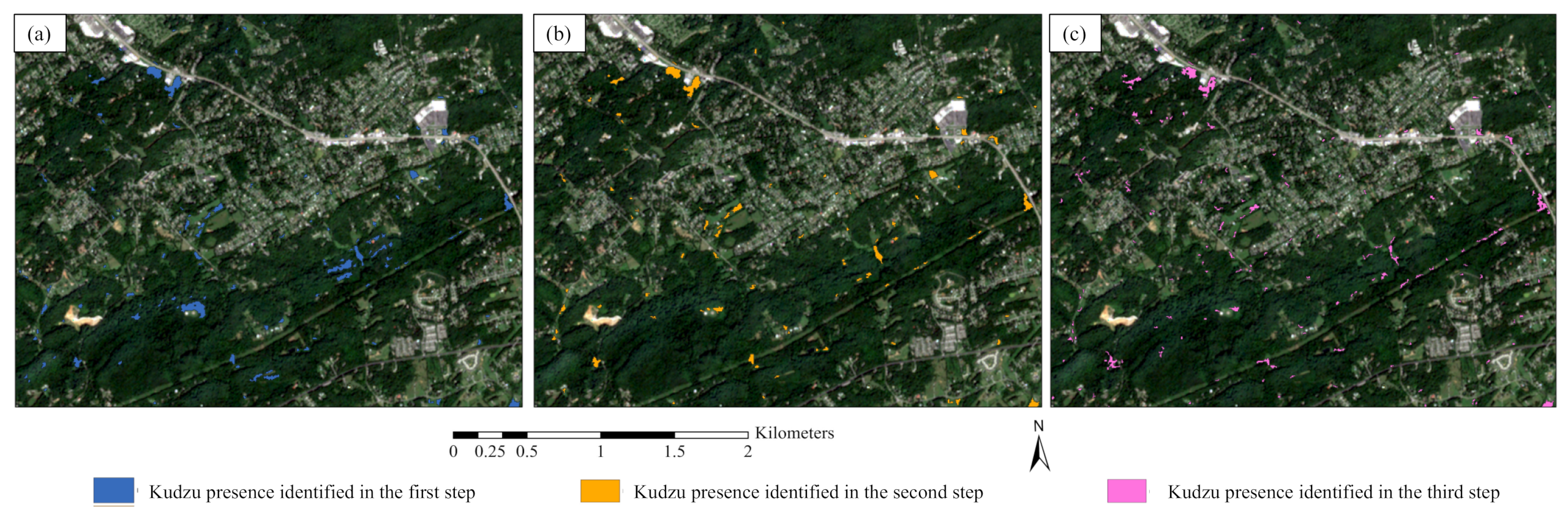
2.3.1. Initial Estimation by Linear Unmixing
2.3.2. Phenology-Based Potential Kudzu Masking
2.3.3. Nonlinear Unmixing Refinement
2.3.4. Creating Kudzu Presence Maps
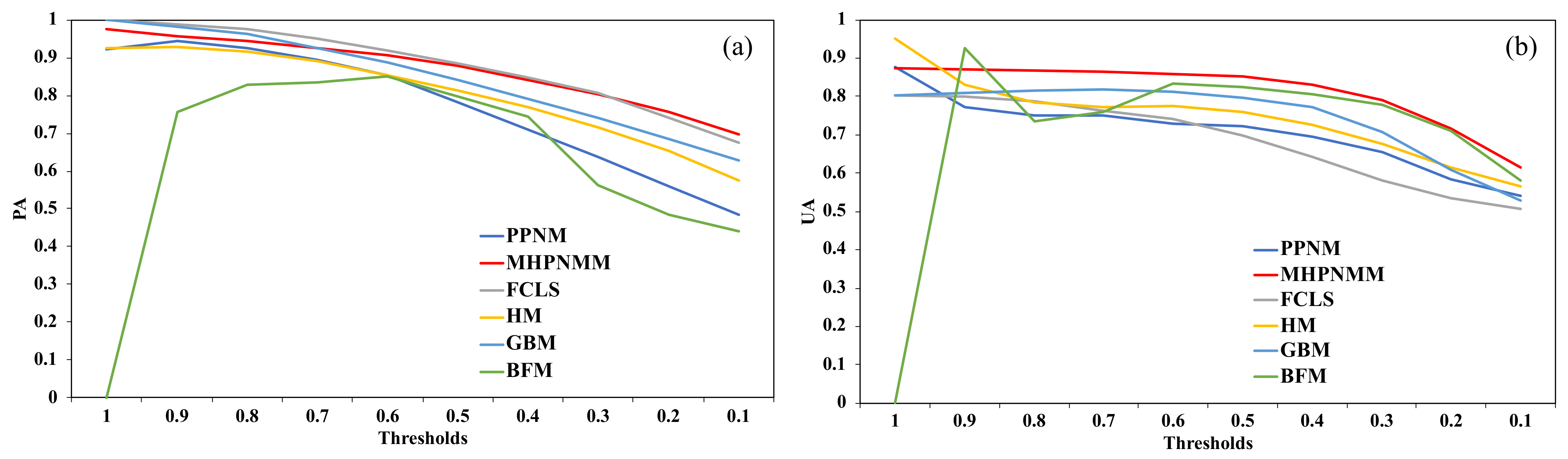
2.4. Accuracy Assessment
3. Results
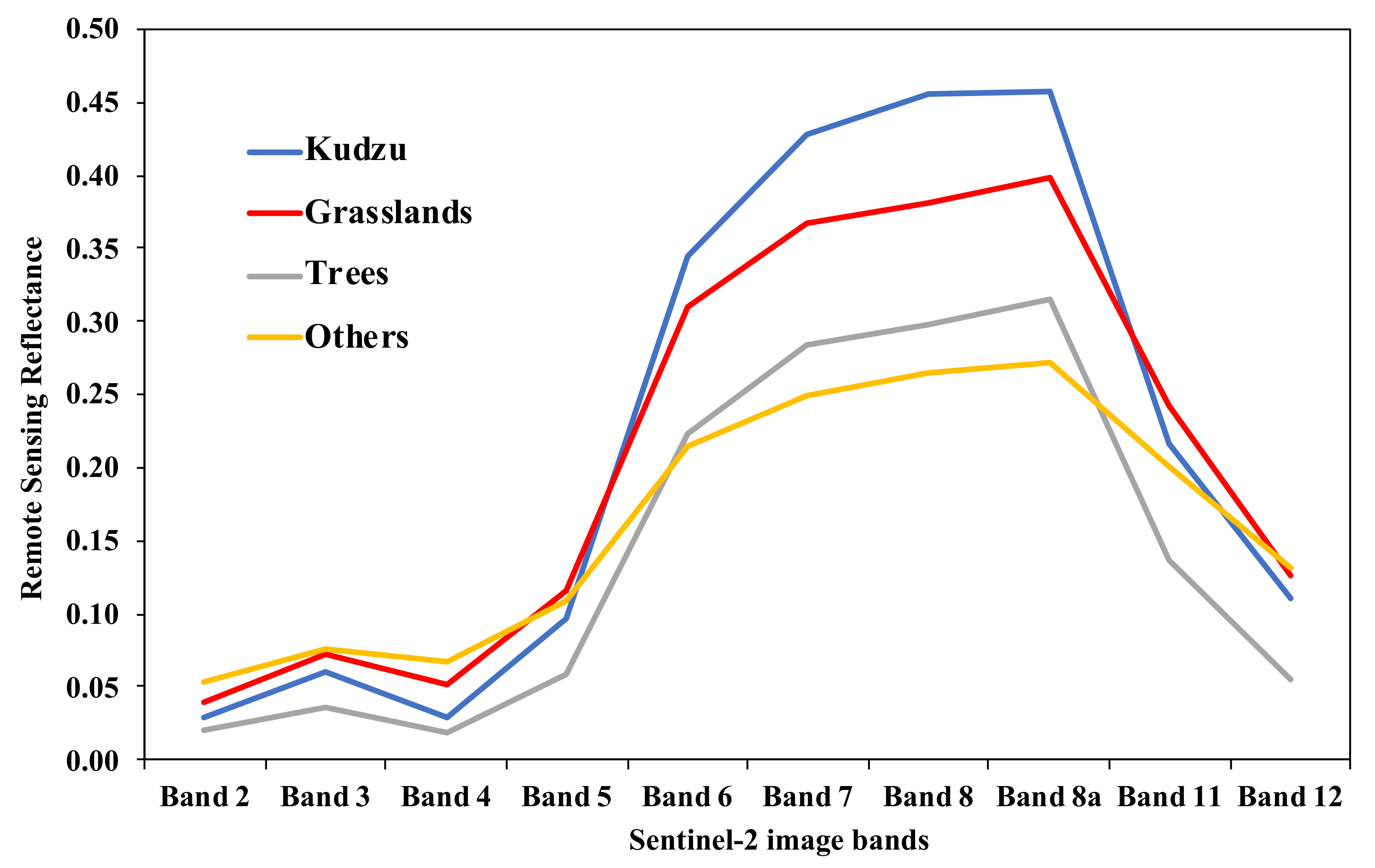
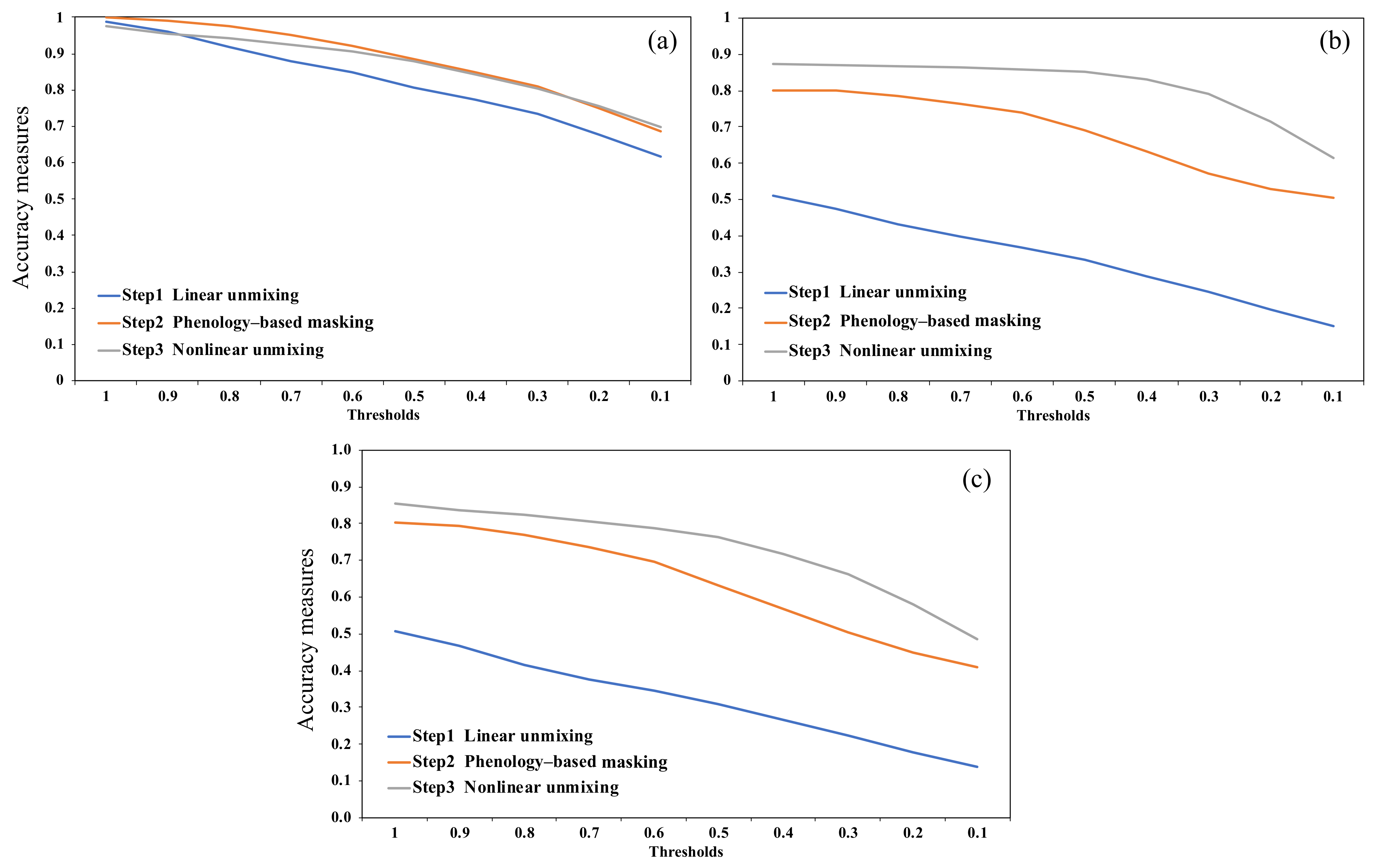
3.1. Performance of the Initial Linear Unmixing Estimation for Kudzu Mapping
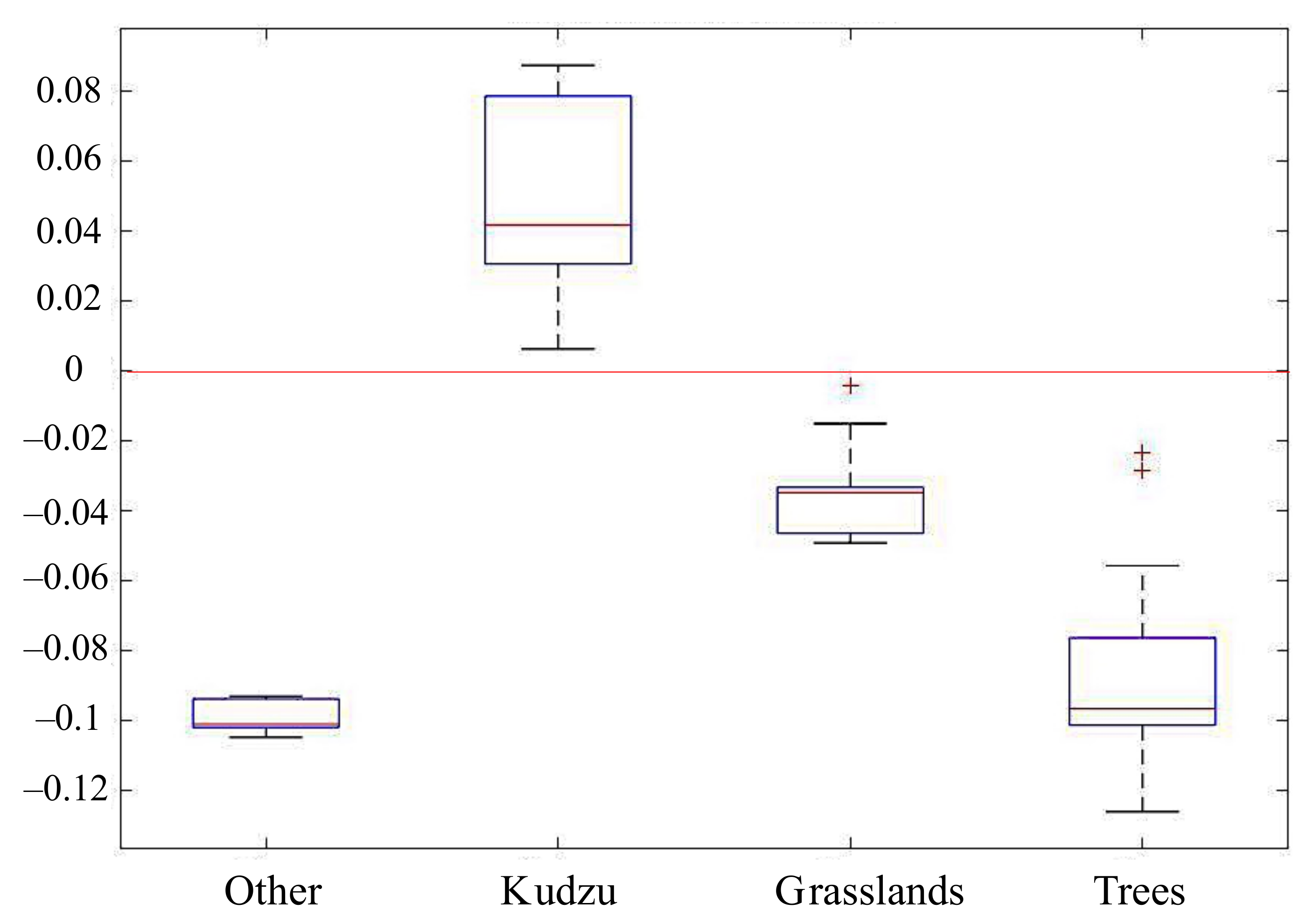
3.2. Performance of Phenology-Based Kudzu Masking
3.3. Performance of the Nonlinear Unmixing Refinements for Kudzu Mapping
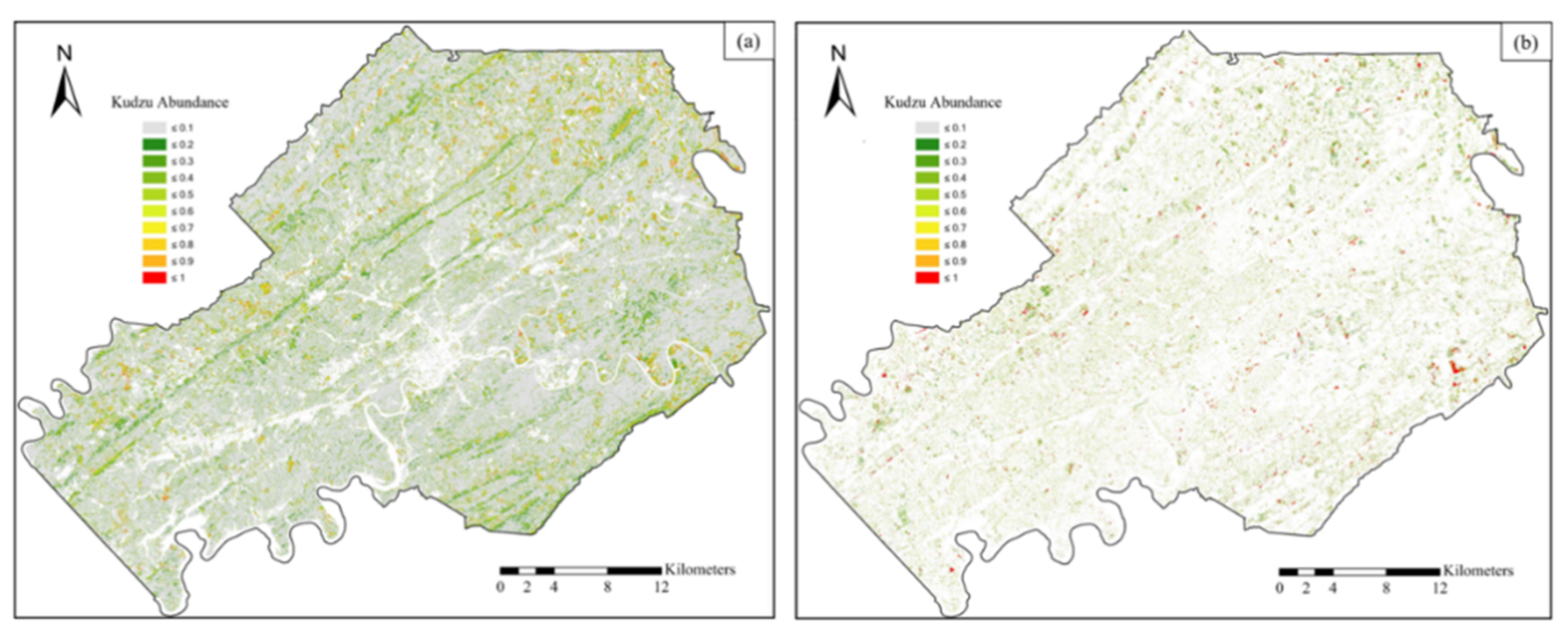
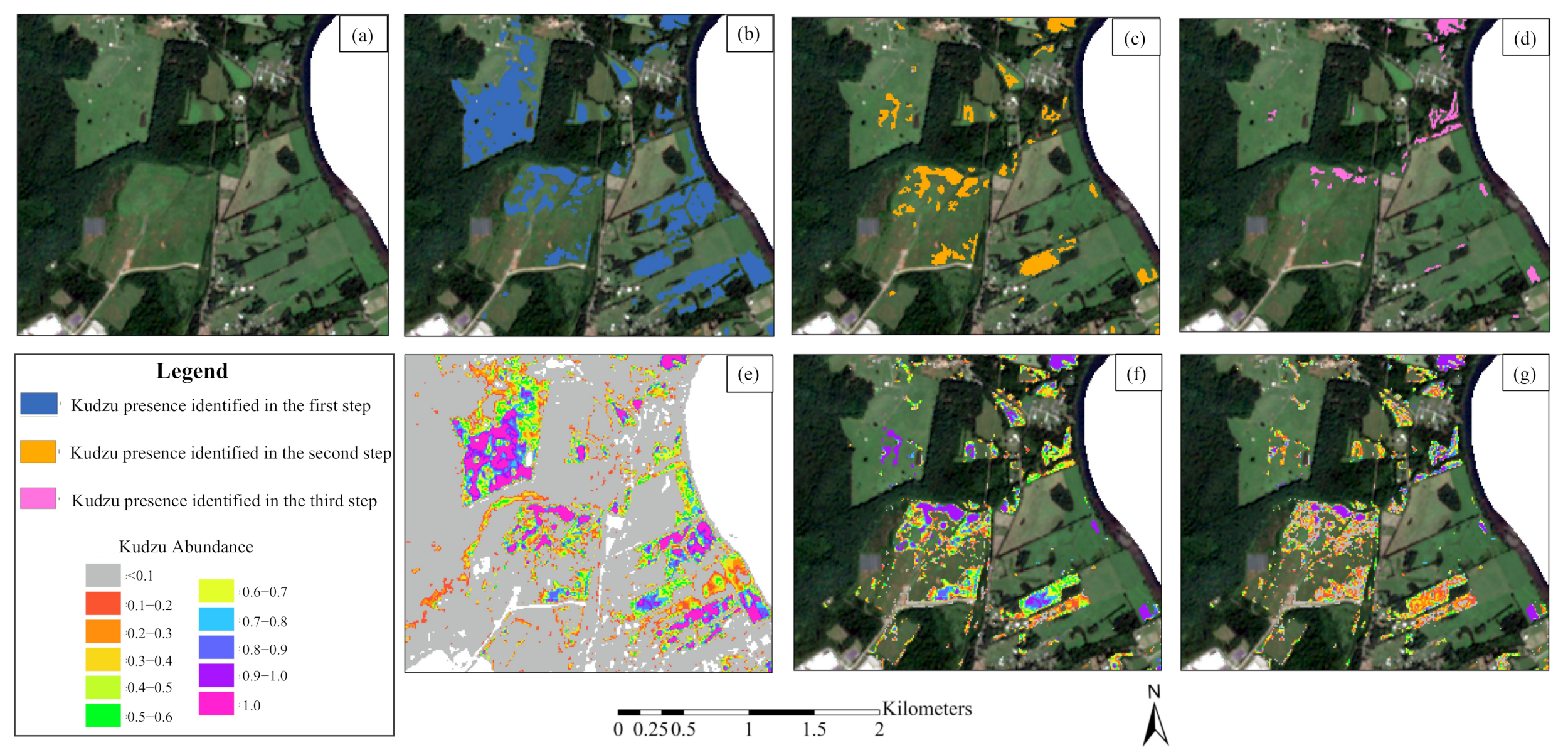
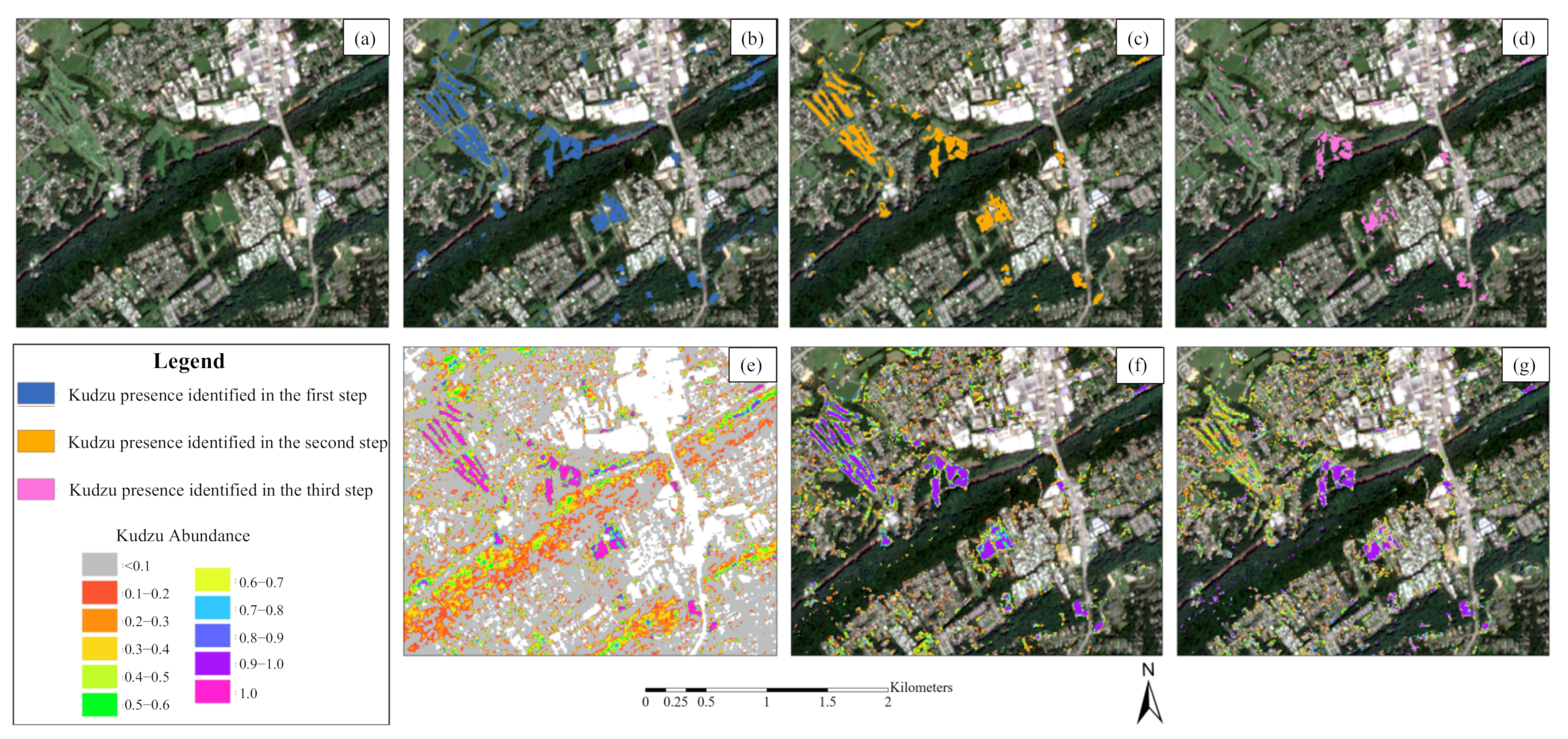
3.4. Kudzu Presence in Knox County
4. Discussion
4.1. Misclassification with the Surrounding Vegetation
4.2. Spectral Unmixing Model Selection for Kudzu Mapping
4.3. Future Improvements of the Proposed Kudzu Classification Approach
5. Conclusions
- The spectral unmixing approach is appropriate for kudzu mapping at the county scale using Sentinel-2 images and allows for continuous monitoring of large areas;
- Linear unmixing provides high producer’s accuracy but low user’s accuracy due to the misclassification of grasslands as kudzu;
- A phenology-based mask can be created based on the differences of kudzu abundance estimated from linear spectral unmixing and NDVI derived from the Sentinel-2 images. The use of this phenology-based mask improves the kudzu classification accuracy and decreases the computing expense for nonlinear spectral unmixing;
- The nonlinear unmixing analysis can refine the kudzu abundance estimation and presence classification, although an appropriate nonlinear model should be selected based on the performance assessment on the datasets and the physical interpretation of the spectral mixing scenarios;
- The refined kudzu presence map for Knox County gives user’s accuracy, producer’s accuracy, Jaccard index, and Kappa index values of 0.858, 0.907, 0.789, and 0.725, respectively, based on an optimal abundance reclassification threshold of 0.6;
- Kudzu plants are scattered in small patches along forest edges, roads, and vegetation tops near houses and infrastructure, especially in the northwestern and southeastern parts of Knox County.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frazier, A.; Wang, L. Characterizing spatial patterns of invasive species using sub–pixel classifications. Remote Sens. Environ. 2011, 115, 1997–2007. [Google Scholar] [CrossRef]
- Gavier-Pizarro, G.I.; Kuemmerle, T.; Hoyos, L.E.; Stewart, S.I.; Huebner, C.D.; Keuler, N.S.; Radeloff, V.C. Monitoring the invasion of an exotic tree (Ligustrum lucidum) from 1983 to 2006 with Landsat TM/ETM+ satellite data and Support Vector Machines in Córdoba, Argentina. Remote Sens. Environ. 2012, 122, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Hawthorne, T.; Elmore, V.; Strong, A.; Bennett-Martin, P.; Finnie, J.; Parkman, J.; Harris, T.; Singh, J.; Edwards, L.; Reed, J. Mapping non–native invasive species and accessibility in an urban forest: A case study of participatory mapping and citizen science in Atlanta, Georgia. Appl. Geogr. 2015, 56, 187–198. [Google Scholar] [CrossRef]
- Mackay, A. Climate change 2007: Impacts, adaptation and vulnerability. Contribution of Working Group II to the fourth assessment report of the Intergovernmental Panel on Climate Change. J. Environ. Qual. 2008, 37, 2407. [Google Scholar] [CrossRef]
- Beck, K.G.; Zimmerman, K.; Schardt, J.D.; Stone, J.; Lukens, R.R.; Reichard, S.; Randall, J.; Cangelosi, A.A.; Cooper, D.; Thompson, J.P. Invasive species defined in a policy context: Recommendations from the Federal Invasive Species Advisory Committee. Invasive Plant Sci. Manag. 2008, 1, 414–421. [Google Scholar] [CrossRef]
- Norambuena, H.; Escobar, S. Control biologico del espinillo en Chiloe. Tierra Adentro 2007, 77, 50–52. [Google Scholar]
- Tamura, M.; Tharayil, N. Plant litter chemistry and microbial priming regulate the accrual, composition and stability of soil carbon in invaded ecosystems. New Phytol. 2014, 203, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.R.; Prinz, T.; Ziller, S.R.; Thiele, J.; Heringer, G.; Meira-Neto, J.A.; Buttschardt, T.K. Open–source processing and analysis of aerial imagery acquired with a low–cost unmanned aerial system to support invasive plant management. Front. Environ. Sci. 2017, 5, 44. [Google Scholar] [CrossRef] [Green Version]
- Müllerová, J.; Brůna, J.; Bartaloš, T.; Dvořák, P.; Vítková, M.; Pyšek, P. Timing is important: Unmanned aircraft vs. satellite imagery in plant invasion monitoring. Front. Plant Sci. 2017, 8, 887. [Google Scholar] [CrossRef] [Green Version]
- Bradley, B.A. Remote detection of invasive plants: A review of spectral, textural and phenological approaches. Biol. Invasions 2014, 16, 1411–1425. [Google Scholar] [CrossRef]
- Xun, L.; Wang, L. An object–based SVM method incorporating optimal segmentation scale estimation using Bhattacharyya Distance for mapping salt cedar (Tamarisk spp.) with QuickBird imagery. GISci. Remote Sens. 2015, 52, 257–273. [Google Scholar] [CrossRef]
- Ji, W.; Wang, L. Phenology–guided saltcedar (Tamarix spp.) mapping using Landsat TM images in western US. Remote Sens. Environ. 2016, 173, 29–38. [Google Scholar] [CrossRef]
- Alderman, D.H. Channing Cope and the making of a miracle vine. Geogr. Rev. 2004, 94, 157–177. [Google Scholar] [CrossRef]
- Gerald, E.W.; Brendon, M.H.; Kudzu Vine, L. Pueraria montana, Adventive in Southern Ontario. Can. Field-Naturalist 2012, 126, 31–33. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Abidi, M.; Carrasco, L.; McNelis, J.; Tran, L.; Li, Y.; Grant, J. Mapping vegetation at species level with high–resolution multispectral and lidar data over a large spatial area: A case study with Kudzu. Remote Sens. 2020, 12, 609. [Google Scholar] [CrossRef] [Green Version]
- Britt, K.E. An Ecological Study of the Kudzu Bug in East Tennessee: Life History, Seasonality, and Phenology. Master’s Thesis, University of Tennessee, Knoxville, TN, USA, 2016. [Google Scholar]
- Jensen, T.; Seerup Hass, F.; Seam Akbar, M.; Holm Petersen, P.; Jokar Arsanjani, J. Employing machine learning for detection of invasive species using Sentinel-2 and AVIRIS data: The case of Kudzu in the United States. Sustainability 2020, 12, 3544. [Google Scholar] [CrossRef]
- Cheng, Y.-B.; Tom, E.; Ustin, S.L. Mapping an invasive species, kudzu (Pueraria montana), using hyperspectral imagery in western Georgia. J. Appl. Remote Sens. 2007, 1, 013514. [Google Scholar] [CrossRef]
- Yu, J.; Chen, D.; Lin, Y.; Ye, S. Comparison of linear and nonlinear spectral unmixing approaches: A case study with multispectral TM imagery. Int. J. Remote Sens. 2017, 38, 773–795. [Google Scholar] [CrossRef]
- Loope, L.L. An overview of problems with introduced plant species in national parks and biosphere reserves of the United States. Alien Plant Invasions Nativ. Ecosyst. Hawaii Manag. Res. 1992, 3, 28. [Google Scholar]
- The Census Bureau’s Population Estimates Program (PEP). Population and Housing Unit Estimates, 17 June 2021; The United States Census Bureau: Houtland, MD, USA, 2020. Available online: https://www.census.gov/programs–surveys/popest.html (accessed on 26 June 2021).
- Yamazaki, D.; Tawatari, R.; Yamaguchi, T.; O’Loughlin, F.; Neal, J.C.; Sampson, C.C.; Kanae, S.; Bates, P.D. Knoxville Topographic Map, Elevation, Relief. Available online: https://en–gb.topographic–map.com (accessed on 4 October 2021).
- Climate in Knoxville, Tennessee. Available online: https://www.bestplaces.net/climate/city/tennessee/knoxville (accessed on 4 October 2021).
- Interactive United States Köppen Climate Classification Map. Available online: www.plantmaps.com (accessed on 4 October 2021).
- All About the Humid Subtropical Climate. Available online: https://321boat.com/all–about–the–humid–subtropical–climate/ (accessed on 24 September 2021).
- Royimani, L.; Mutanga, O.; Odindi, J.; Dube, T.; Matongera, T.N. Advancements in satellite remote sensing for mapping and monitoring of alien invasive plant species (AIPs). Phys. Chem. Earth Parts A/B/C 2019, 112, 237–245. [Google Scholar] [CrossRef]
- SUHET. Sentinel-2 User Handbook; European Space Agency: Paris, France, 2015; Available online: https://sentinel.esa.int/web/sentinel/home (accessed on 24 July 2021).
- Clark, M.L.; Aide, T.M.; Grau, H.R.; Riner, G. A scalable approach to mapping annual land cover at 250 m using MODIS time series data: A case study in the Dry Chaco ecoregion of South America. Remote Sens. Environ. 2010, 114, 2816–2832. [Google Scholar] [CrossRef]
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Heinz, D.C. Fully constrained least squares linear spectral mixture analysis method for material quantification in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2001, 39, 529–545. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Roberts, D.A.; Stow, D.A.; An, L.; Hall, S.J.; Yabiku, S.T.; Kyriakidis, P.C. Mapping understory invasive plant species with field and remotely sensed data in Chitwan, Nepal. Remote Sens. Environ. 2020, 250, 112037. [Google Scholar] [CrossRef]
- Diao, C.; Wang, L. Development of an invasive species distribution model with fine–resolution remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2014, 30, 65–75. [Google Scholar] [CrossRef]
- Heylen, R.; Burazerovic, D.; Scheunders, P. Fully constrained least squares spectral unmixing by simplex projection. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4112–4122. [Google Scholar] [CrossRef]
- Steve, C. Invasive Plant Risk Assessment: Kudzu Pueraria Montana var. Lobata. Available online: https://www.daf.qld.gov.au/__data/assets/pdf_file/0004/74137/IPA-Kudzu-Risk-Assessment.pdf (accessed on 9 October 2021).
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef] [Green Version]
- Carlson, T.N.; Ripley, D.A. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sens. Environ. 1997, 62, 241–252. [Google Scholar] [CrossRef]
- Silván-Cárdenas, J.; Wang, L. Retrieval of subpixel Tamarix canopy cover from Landsat data along the Forgotten River using linear and nonlinear spectral mixture models. Remote Sens. Environ. 2010, 114, 1777–1790. [Google Scholar] [CrossRef]
- Fan, W.; Hu, B.; Miller, J.; Li, M. Comparative study between a new nonlinear model and common linear model for analysing laboratory simulated-forest hyperspectral data. Int. J. Remote Sens. 2009, 30, 2951–2962. [Google Scholar] [CrossRef]
- Altmann, Y.; Halimi, A.; Dobigeon, N.; Tourneret, J.-Y. Supervised nonlinear spectral unmixing using a postnonlinear mixing model for hyperspectral imagery. IEEE Trans. Image Process. 2012, 21, 3017–3025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heylen, R.; Parente, M.; Gader, P. A review of nonlinear hyperspectral unmixing methods. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1844–1868. [Google Scholar] [CrossRef]
- Tang, M.; Zhang, B.; Marinoni, A.; Gao, L.; Gamba, P. Multiharmonic postnonlinear mixing model for hyperspectral nonlinear unmixing. IEEE Geosci. Remote Sens. Lett. 2018, 15, 1765–1769. [Google Scholar] [CrossRef]
- Story, M.; Congalton, R.G. Accuracy assessment: A user’s perspective. Photogramm. Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Somodi, I.; Čarni, A.; Ribeiro, D.; Podobnikar, T. Recognition of the invasive species Robinia pseudacacia from combined remote sensing and GIS sources. Biol. Conserv. 2012, 150, 59–67. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Liu, C.; Frazier, P.; Kumar, L. Comparative assessment of the measures of thematic classification accuracy. Remote Sens. Environ. 2007, 107, 606–616. [Google Scholar] [CrossRef]
- Tung, F.; LeDrew, E. The determination of optimal threshold levels for change detection using various accuracy indexes. Photogramm. Eng. Remote Sens. 1988, 54, 1449–1454. [Google Scholar]
- Schnell, A. What Is Kappa and How Does It Measure Inter–Rater Reliability? Available online: https://www.theanalysisfactor.com/kappa–measures–inter–rater–reliability/#respond (accessed on 9 October 2021).
- Bioucas-Dias, J.M.; Plaza, A.; Dobigeon, N.; Parente, M.; Du, Q.; Gader, P.; Chanussot, J. Hyperspectral unmixing overview: Geometrical, statistical, and sparse regression–based approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 354–379. [Google Scholar] [CrossRef] [Green Version]
- Dobigeon, N.; Tourneret, J.-Y.; Richard, C.; Bermudez, J.C.M.; McLaughlin, S.; Hero, A.O. Nonlinear unmixing of hyperspectral images: Models and algorithms. IEEE Signal Process. Mag. 2013, 31, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Shipman, H.; Adams, J.B. Detectability of minerals on desert alluvial fans using reflectance spectra. J. Geophys. Res. Solid Earth 1987, 92, 10391–10402. [Google Scholar] [CrossRef]
- Pacheco, A.; McNairn, H. Evaluating multispectral remote sensing and spectral unmixing analysis for crop residue mapping. Remote Sens. Environ. 2010, 114, 2219–2228. [Google Scholar] [CrossRef]
- Horler, D.; Dockray, M.; Barber, J. The red edge of plant leaf reflectance. Int. J. Remote Sens. 1983, 4, 273–288. [Google Scholar] [CrossRef]
- Wang, B.; Jia, K.; Liang, S.; Xie, X.; Wei, X.; Zhao, X.; Yao, Y.; Zhang, X. Assessment of Sentinel-2 MSI spectral band reflectances for estimating fractional vegetation cover. Remote Sens. 2018, 10, 1927. [Google Scholar] [CrossRef] [Green Version]


| Band Number | Central Wavelength (nm) | Bandwidth (nm) | Spatial Resolution (m) |
|---|---|---|---|
| 1—Coastal aerosol | 443 | 20 | 60 |
| 2—Blue | 490 | 65 | 10 |
| 3—Green | 560 | 35 | 10 |
| 4—Red | 665 | 30 | 10 |
| 5—Vegetation Red Edge | 705 | 15 | 20 |
| 6—Vegetation Red Edge | 740 | 15 | 20 |
| 7—Vegetation Red Edge | 783 | 20 | 20 |
| 8—Near-Infrared | 842 | 115 | 10 |
| 8b—Narrow Near-Infrared | 865 | 20 | 20 |
| 9—Water Vapor | 945 | 20 | 60 |
| 10—Cirrus | 1375 | 30 | 60 |
| 11—Short-wave Infrared | 1610 | 90 | 20 |
| 12—Short-wave Infrared | 2190 | 180 | 20 |
| Thresholds | 1 | 0.9 | 0.8 | 0.7 | 0.6 | 0.5 | 0.4 | 0.3 | 0.2 | 0.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| PA | 0.989 | 0.961 | 0.919 | 0.879 | 0.848 | 0.808 | 0.774 | 0.734 | 0.676 | 0.616 |
| UA | 0.511 | 0.475 | 0.432 | 0.396 | 0.367 | 0.333 | 0.289 | 0.245 | 0.195 | 0.152 |
| Jaccard | 0.508 | 0.466 | 0.416 | 0.376 | 0.344 | 0.309 | 0.266 | 0.225 | 0.179 | 0.138 |
| Kappa | 0.560 | 0.585 | 0.545 | 0.503 | 0.466 | 0.421 | 0.364 | 0.305 | 0.234 | 0.170 |
| Thresholds | 1 | 0.9 | 0.8 | 0.7 | 0.6 | 0.5 | 0.4 | 0.3 | 0.2 | 0.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| PA | 1.000 | 0.989 | 0.976 | 0.951 | 0.922 | 0.884 | 0.849 | 0.809 | 0.748 | 0.687 |
| UA | 0.802 | 0.801 | 0.785 | 0.764 | 0.738 | 0.689 | 0.632 | 0.572 | 0.530 | 0.506 |
| Jaccard | 0.802 | 0.794 | 0.771 | 0.734 | 0.695 | 0.632 | 0.568 | 0.504 | 0.450 | 0.411 |
| Kappa | 0.000 | 0.263 | 0.409 | 0.489 | 0.525 | 0.478 | 0.414 | 0.352 | 0.281 | 0.236 |
| Procedure Step | Computation Areas for Unmixing Models |
|---|---|
| Step 1 Linear unmixing | Knox County vegetated area, 1126.06 km2 |
| Step 3 Nonlinear unmixing | Phenology-based masked kudzu area, 134.19 km2 |
| Thresholds | 1 | 0.9 | 0.8 | 0.7 | 0.6 | 0.5 | 0.4 | 0.3 | 0.2 | 0.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| PA | 0.974 | 0.955 | 0.944 | 0.925 | 0.907 | 0.880 | 0.842 | 0.803 | 0.756 | 0.698 |
| UA | 0.875 | 0.872 | 0.867 | 0.864 | 0.858 | 0.852 | 0.830 | 0.790 | 0.715 | 0.615 |
| Jaccard | 0.855 | 0.837 | 0.824 | 0.807 | 0.789 | 0.763 | 0.718 | 0.662 | 0.581 | 0.486 |
| Kappa | 0.126 | 0.454 | 0.618 | 0.698 | 0.725 | 0.723 | 0.690 | 0.633 | 0.548 | 0.433 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, M.; Tang, M.; Li, Y. Phenology and Spectral Unmixing-Based Invasive Kudzu Mapping: A Case Study in Knox County, Tennessee. Remote Sens. 2021, 13, 4551. https://doi.org/10.3390/rs13224551
Shen M, Tang M, Li Y. Phenology and Spectral Unmixing-Based Invasive Kudzu Mapping: A Case Study in Knox County, Tennessee. Remote Sensing. 2021; 13(22):4551. https://doi.org/10.3390/rs13224551
Chicago/Turabian StyleShen, Ming, Maofeng Tang, and Yingkui Li. 2021. "Phenology and Spectral Unmixing-Based Invasive Kudzu Mapping: A Case Study in Knox County, Tennessee" Remote Sensing 13, no. 22: 4551. https://doi.org/10.3390/rs13224551
APA StyleShen, M., Tang, M., & Li, Y. (2021). Phenology and Spectral Unmixing-Based Invasive Kudzu Mapping: A Case Study in Knox County, Tennessee. Remote Sensing, 13(22), 4551. https://doi.org/10.3390/rs13224551