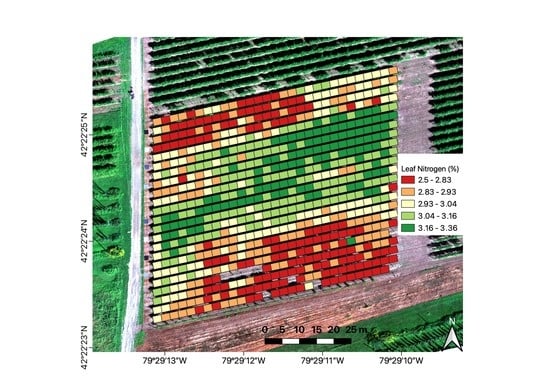
Assessing Grapevine Nutrient Status from Unmanned Aerial System (UAS) Hyperspectral Imagery
Abstract
:1. Introduction
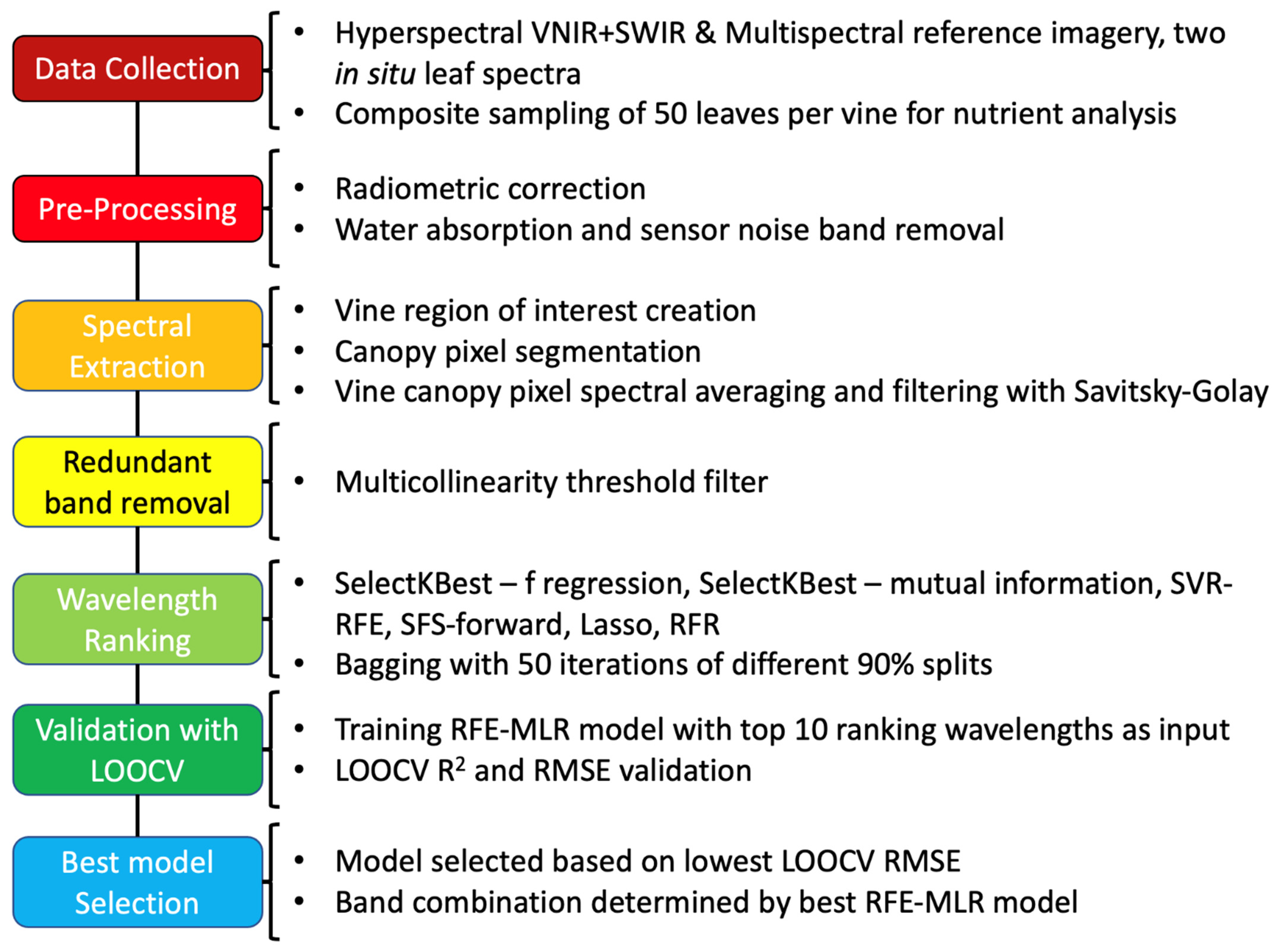
2. Materials and Methods
2.1. Study Area
2.2. Aerial Imaging Systems
2.3. Imaging Campaign
2.4. Sampling Plan
2.5. Data Preprocessing
2.5.1. Radiometric Correction
2.5.2. Vine Image Extraction
2.5.3. Canopy Segmentation and Spectral Extraction
2.6. Data Analysis
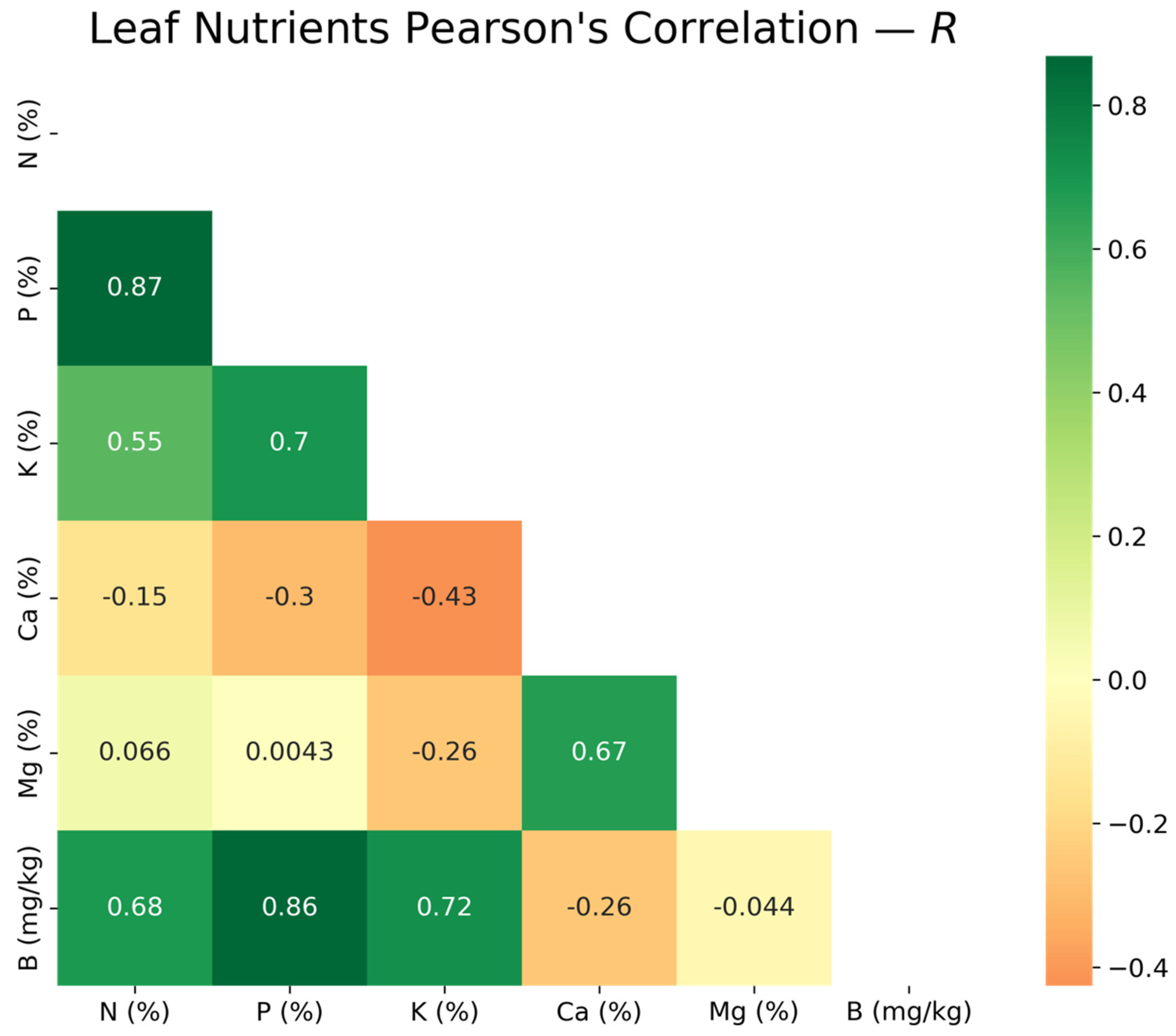
2.6.1. Multicollinearity Assessment
2.6.2. Ensemble Feature Selection Rankers
2.6.3. Ensemble Feature Selection Method
2.6.4. PLSR Comparison
3. Results
4. Discussion
4.1. Methodological Considerations
4.2. Regression Model Performance
4.3. Nitrogen Wavelength Selections vs. Spectral Features
4.4. Overall Wavelength Selection Spectral Regions
4.5. Future Research and Potential Improvments
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
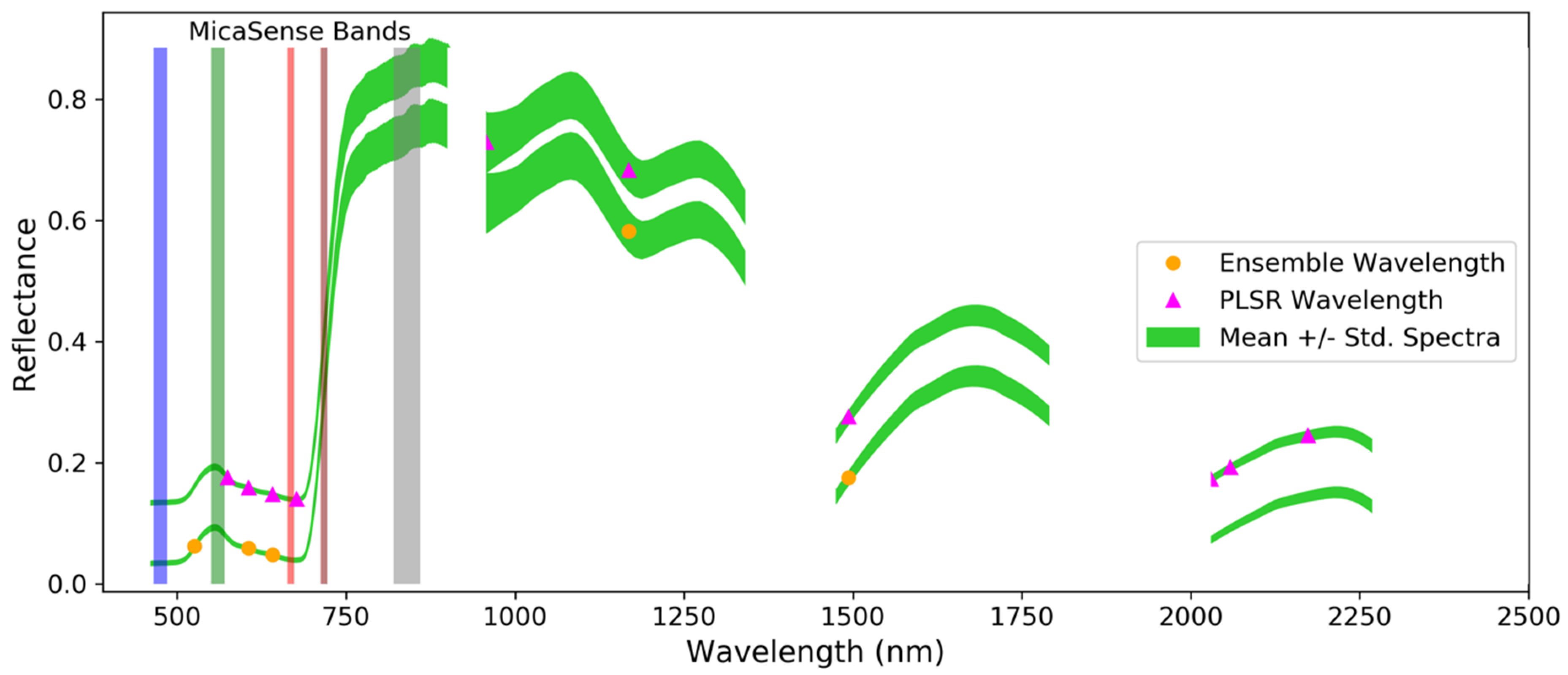
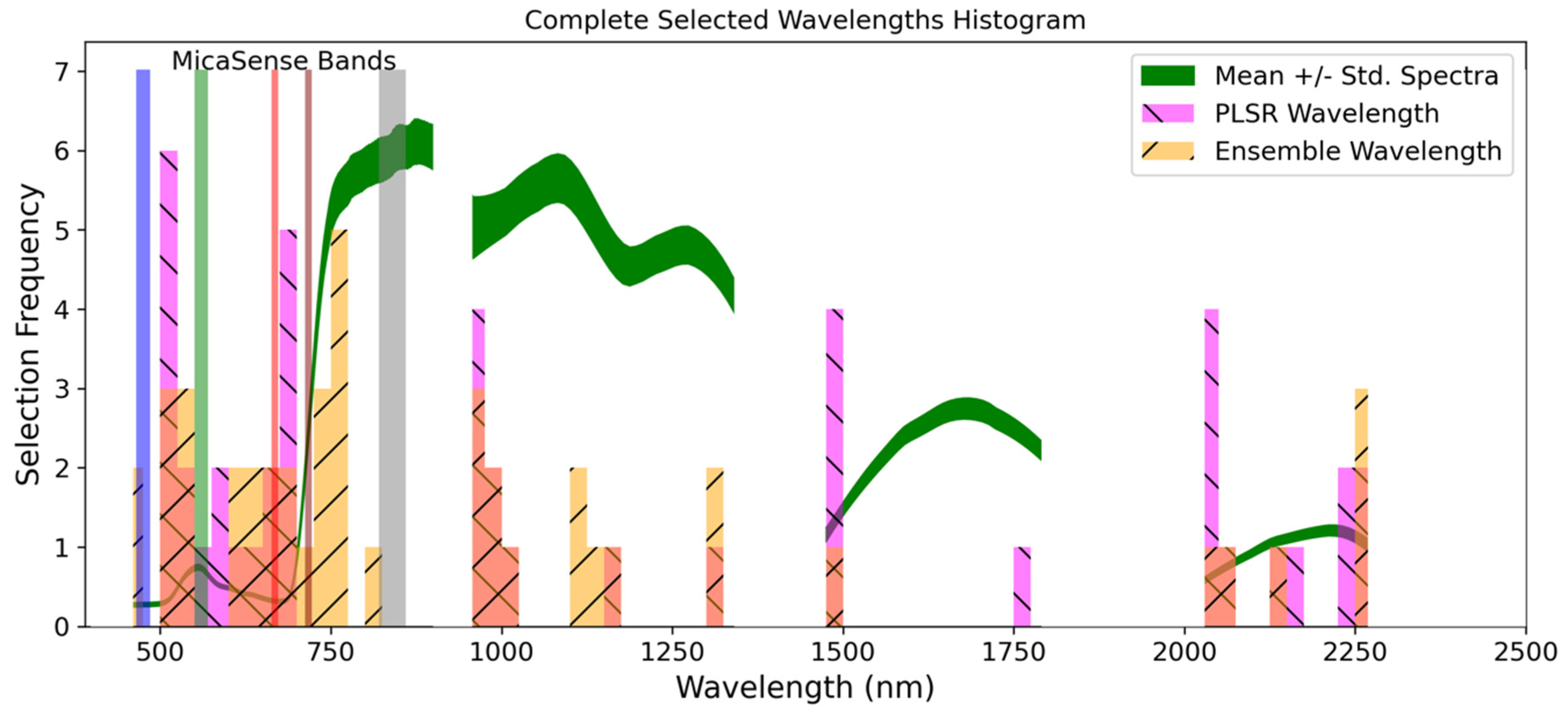
| Nutrient | #W a | Ensemble b Wavelengths (nm) | PLSR b Wavelengths (nm) |
|---|---|---|---|
| N | 5 (10) | 526, 606, 641, 1168, 1494 | 574, 606, 641, 677, 958, 1168, 1494, 2030, 2058, 2173 |
| P | 7, (9) | 677, 695, 730, 750, 879, 967, 2269 | 517, 548, 677, 967, 1302, 1494, 2030, 2231, 2269 |
| K | 5, (10) | 730, 967, 1120, 1312, 2058 | 519, 528, 552, 590, 672, 695, 967, 1513, 1762, 2058 |
| Ca | 9, (7) | 514, 661, 757, 772, 967, 1005, 1302, 2049, 2269 | 514, 523, 661, 1005, 1494, 2049, 2269 |
| Mg | 7, (10) | 514, 603, 643, 668, 692, 996, 2125 | 514, 523, 697, 704, 958, 996, 1790, 2058, 2125, 2269 |
| B | 8, (10) | 548, 719, 730, 764, 822, 977, 2231, 2269 | 474, 519, 548, 695, 719, 730, 764, 822, 977, 2144 |
References
- National Agricultural Statistics Service. Noncitrus Fruits and Nuts 2020 Summary; United States Department of Agriculture: Washington, DC, USA, 2021. [Google Scholar]
- Muñoz-Huerta, R.F.; Guevara-Gonzalez, R.G.; Contreras-Medina, L.M.; Torres-Pacheco, I.; Prado-Olivarez, J.; Ocampo-Velazquez, R.V. A Review of Methods for Sensing the Nitrogen Status in Plants: Advantages, Disadvantages and Recent Advances. Sensors 2013, 13, 10823–10843. [Google Scholar] [CrossRef]
- Schreiner, R.P.; Lee, J.; Skinkis, P.A. N, P, and K Supply to Pinot Noir Grapevines: Impact on Vine Nutrient Status, Growth, Physiology, and Yield. Am. J. Enol. Vitic. 2013, 64, 26–38. [Google Scholar] [CrossRef]
- Padilla, F.M.; Gallardo, M.; Peña-Fleitas, M.T.; De Souza, R.; Thompson, R.B. Proximal Optical Sensors for Nitrogen Management of Vegetable Crops: A Review. Sensors 2018, 18, 2083. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, Z.; Fairbairn, D.; Li, N.; Xu, B.; Feng, H.; Yang, G. Multi-LUTs Method for Canopy Nitrogen Density Estimation in Winter Wheat by Field and UAV Hyperspectral. Comput. Electron. Agric. 2019, 162, 174–182. [Google Scholar] [CrossRef]
- Lee, H.; Wang, J.; Leblon, B. Intra-Field Canopy Nitrogen Retrieval from Unmanned Aerial Vehicle Imagery for Wheat and Corn Fields. Can. J. Remote Sens. 2020, 46, 454–472. [Google Scholar] [CrossRef]
- Liu, S.; Li, L.; Gao, W.; Zhang, Y.; Liu, Y.; Wang, S.; Lu, J. Diagnosis of Nitrogen Status in Winter Oilseed Rape (Brassica Napus L.) Using in-Situ Hyperspectral Data and Unmanned Aerial Vehicle (UAV) Multispectral Images. Comput. Electron. Agric. 2018, 151, 185–195. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, T.; Yang, C.; Song, H.; Jiang, Z.; Zhou, G.; Zhang, D.; Feng, H.; Xie, J. Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection. Remote Sens. 2020, 12, 1403. [Google Scholar] [CrossRef]
- Näsi, R.; Viljanen, N.; Kaivosoja, J.; Alhonoja, K.; Hakala, T.; Markelin, L.; Honkavaara, E. Estimating Biomass and Nitrogen Amount of Barley and Grass Using UAV and Aircraft Based Spectral and Photogrammetric 3D Features. Remote Sens. 2018, 10, 1082. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Cheng, T.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Combining Unmanned Aerial Vehicle (UAV)-Based Multispectral Imagery and Ground-Based Hyperspectral Data for Plant Nitrogen Concentration Estimation in Rice. Front. Plant Sci. 2018, 9, 936. [Google Scholar] [CrossRef]
- Zha, H.; Miao, Y.; Wang, T.; Li, Y.; Zhang, J.; Sun, W.; Feng, Z.; Kusnierek, K. Improving Unmanned Aerial Vehicle Remote Sensing-Based Rice Nitrogen Nutrition Index Prediction with Machine Learning. Remote Sens. 2020, 12, 215. [Google Scholar] [CrossRef] [Green Version]
- Wen, D.; Tongyu, X.; Fenghua, Y.; Chunling, C.; Wen, D.; Tongyu, X.; Fenghua, Y.; Chunling, C. Measurement of Nitrogen Content in Rice by Inversion of Hyperspectral Reflectance Data from an Unmanned Aerial Vehicle. Ciência Rural 2018, 48, e20180008. [Google Scholar] [CrossRef]
- Hunt, E.R.; Horneck, D.A.; Spinelli, C.B.; Turner, R.W.; Bruce, A.E.; Gadler, D.J.; Brungardt, J.J.; Hamm, P.B. Monitoring Nitrogen Status of Potatoes Using Small Unmanned Aerial Vehicles. Precis. Agric. 2018, 19, 314–333. [Google Scholar] [CrossRef]
- Agüera, F.; Carvajal, F.; Pérez, M. Measuring Sunflower Nitrogen Status from an Unmanned Aerial Vehicle-Based System and an on the Ground Device. In Proceedings of the The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Göttingen, Germany, 6 September 2012; Copernicus GmbH: Göttingen, Germany, 2012; Volume XXXVIII-1-C22, pp. 33–37. [Google Scholar]
- Atzberger, C. Advances in Remote Sensing of Agriculture: Context Description, Existing Operational Monitoring Systems and Major Information Needs. Remote Sens. 2013, 5, 949–981. [Google Scholar] [CrossRef] [Green Version]
- Kokaly, R.F.; Clark, R.N. Spectroscopic Determination of Leaf Biochemistry Using Band-Depth Analysis of Absorption Features and Stepwise Multiple Linear Regression. Remote Sens. Environ. 1999, 67, 267–287. [Google Scholar] [CrossRef]
- Miphokasap, P.; Wannasiri, W. Estimations of Nitrogen Concentration in Sugarcane Using Hyperspectral Imagery. Sustainability 2018, 10, 1266. [Google Scholar] [CrossRef] [Green Version]
- Zhou, K.; Cheng, T.; Zhu, Y.; Cao, W.; Ustin, S.L.; Zheng, H.; Yao, X.; Tian, Y. Assessing the Impact of Spatial Resolution on the Estimation of Leaf Nitrogen Concentration Over the Full Season of Paddy Rice Using Near-Surface Imaging Spectroscopy Data. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Kokaly, R.F. Investigating a Physical Basis for Spectroscopic Estimates of Leaf Nitrogen Concentration. Remote Sens. Environ. 2001, 75, 153–161. [Google Scholar] [CrossRef]
- Pullanagari, R.R.; Kereszturi, G.; Yule, I.J. Mapping of Macro and Micro Nutrients of Mixed Pastures Using Airborne AisaFENIX Hyperspectral Imagery. ISPRS J. Photogramm. Remote Sens. 2016, 117, 1–10. [Google Scholar] [CrossRef]
- Yao, X.; Huang, Y.; Shang, G.; Zhou, C.; Cheng, T.; Tian, Y.; Cao, W.; Zhu, Y. Evaluation of Six Algorithms to Monitor Wheat Leaf Nitrogen Concentration. Remote Sens. 2015, 7, 14939–14966. [Google Scholar] [CrossRef] [Green Version]
- Berger, K.; Verrelst, J.; Féret, J.-B.; Wang, Z.; Wocher, M.; Strathmann, M.; Danner, M.; Mauser, W.; Hank, T. Crop Nitrogen Monitoring: Recent Progress and Principal Developments in the Context of Imaging Spectroscopy Missions. Remote Sens. Environ. 2020, 242, 111758. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Pu, R.; Li, Z.; Li, H.; Xu, X.; Song, X.; Yang, X.; Zhao, C. An Overview of Crop Nitrogen Status Assessment Using Hyperspectral Remote Sensing: Current Status and Perspectives. Eur. J. Agron. 2021, 124, 126241. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral Dimensionality Reduction for Biophysical Variable Statistical Retrieval. ISPRS J. Photogramm. Remote. Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Friedel, M.; Hendgen, M.; Stoll, M.; Löhnertz, O. Performance of Reflectance Indices and of a Handheld Device for Estimating In-Field the Nitrogen Status of Grapevine Leaves. Aust. J. Grape Wine Res. 2020, 26, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Omidi, R.; Moghimi, A.; Pourreza, A.; El-Hadedy, M.; Eddin, A.S. Ensemble Hyperspectral Band Selection for Detecting Nitrogen Status in Grape Leaves. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 286–293. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Marchetto, P.M. Ensemble Feature Selection for Plant Phenotyping: A Journey from Hyperspectral to Multispectral Imaging. IEEE Access 2018, 6, 56870–56884. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-Method Ensemble Selection of Spectral Bands Related to Leaf Biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E.; Schmidtlein, S. Brightness-Normalized Partial Least Squares Regression for Hyperspectral Data. J. Quant. Spectrosc. Radiat. Transf. 2010, 111, 1947–1957. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of Variability in Canopy Reflectance and the Convergent Properties of Plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing Canopy Biochemistry from Imaging Spectroscopy and Its Application to Ecosystem Studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Curran, P.J. Remote Sensing of Foliar Chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Schnug, E.; Haneklaus, S. Sulphur Deficiency Symptoms in Oilseed Rape (Brasica Napus L.)—The Aesthetics of Starvation. Phyton 2005, 45, 79. [Google Scholar]
- Rustioni, L.; Grossi, D.; Brancadoro, L.; Failla, O. Iron, Magnesium, Nitrogen and Potassium Deficiency Symptom Discrimination by Reflectance Spectroscopy in Grapevine Leaves. Sci. Hortic. 2018, 241, 152–159. [Google Scholar] [CrossRef]
- Camino, C.; González-Dugo, V.; Hernández, P.; Sillero, J.C.; Zarco-Tejada, P.J. Improved Nitrogen Retrievals with Airborne-Derived Fluorescence and Plant Traits Quantified from VNIR-SWIR Hyperspectral Imagery in the Context of Precision Agriculture. Int. J. Appl. Earth Obs. 2018, 70, 105–117. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A Model of Leaf Optical Properties Spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Baret, F.; Jacquemoud, S.; Guyot, G.; Leprieur, C. Modeled Analysis of the Biophysical Nature of Spectral Shifts and Comparison with Information Content of Broad Bands. Remote Sens. Environ. 1992, 41, 133–142. [Google Scholar] [CrossRef]
- Moghimi, A.; Pourreza, A.; Zuniga-Ramirez, G.; Williams, L.E.; Fidelibus, M.W. A Novel Machine Learning Approach to Estimate Grapevine Leaf Nitrogen Concentration Using Aerial Multispectral Imagery. Remote Sens. 2020, 12, 3515. [Google Scholar] [CrossRef]
- Wolf, T. Wine Grape Production Guide for Eastern North America (NRAES 145); Natural Resource, Agriculture, and Engineering Service (NRAES): New York, NY, USA, 2008. [Google Scholar]
- Bates, T.R.; Dunst, R.M.; Joy, P. Seasonal Dry Matter, Starch, and Nutrient Distribution in “Concord” Grapevine Roots. HortScience 2002, 37, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.L.; Schulte, E.E. Digestion of Plant Tissue for Analysis by ICP Emission Spectroscopy. Commun. Soil Sci. Plant Anal. 1985, 16, 943–958. [Google Scholar] [CrossRef]
- L3Harris ENVI. Available online: https://www.l3harrisgeospatial.com/Software-Technology/ENVI (accessed on 11 March 2021).
- Using ENVI: Atmospheric Correction—Empirical Line Correction. Available online: https://www.l3harrisgeospatial.com/docs/atmosphericcorrection.html#empirical_line_calibration (accessed on 24 May 2021).
- Boggs, T. Spectral Python (SPy). Available online: http://www.spectralpython.net (accessed on 11 March 2021).
- Oshigami, S.; Yamaguchi, Y.; Uezato, T.; Momose, A.; Arvelyna, Y.; Kawakami, Y.; Yajima, T.; Miyatake, S.; Nguno, A. Mineralogical Mapping of Southern Namibia by Application of Continuum-Removal MSAM Method to the HyMap Data. Int. J. Remote Sens. 2013, 34, 5282–5295. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Miller, M.E.; Kianian, S.F.; Marchetto, P.M. A Novel Approach to Assess Salt Stress Tolerance in Wheat Using Hyperspectral Imaging. Front. Plant Sci. 2018, 9, 1182. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Jia, X.; Kuo, B.-C.; Crawford, M.M. Feature Mining for Hyperspectral Image Classification. Proc. IEEE 2013, 101, 676–697. [Google Scholar] [CrossRef]
- Hassanzadeh, A.; Murphy, S.P.; Pethybridge, S.J.; van Aardt, J. Growth Stage Classification and Harvest Scheduling of Snap Bean Using Hyperspectral Sensing: A Greenhouse Study. Remote Sens. 2020, 12, 3809. [Google Scholar] [CrossRef]
- Kraskov, A.; Stögbauer, H.; Grassberger, P. Estimating Mutual Information. Phys. Rev. E 2004, 69, 066138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, B.C. Mutual Information between Discrete and Continuous Data Sets. PLoS ONE 2014, 9, e87357. [Google Scholar] [CrossRef] [PubMed]
- Archibald, R.; Fann, G. Feature Selection and Classification of Hyperspectral Images With Support Vector Machines. IEEE Geosci. Remote Sens. Lett. 2007, 4, 674–677. [Google Scholar] [CrossRef]
- Rady, A.; Ekramirad, N.; Adedeji, A.A.; Li, M.; Alimardani, R. Hyperspectral Imaging for Detection of Codling Moth Infestation in GoldRush Apples. Postharvest Biol. Technol. 2017, 129, 37–44. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Selection and Shrinkage via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Chan, J.C.-W.; Paelinckx, D. Evaluation of Random Forest and Adaboost Tree-Based Ensemble Classification and Spectral Band Selection for Ecotope Mapping Using Airborne Hyperspectral Imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A Review of Variable Selection Methods in Partial Least Squares Regression. Chemom. Intell. Lab. Syst. 2012, 118, 62–69. [Google Scholar] [CrossRef]
- Giovos, R.; Tassopoulos, D.; Kalivas, D.; Lougkos, N.; Priovolou, A. Remote Sensing Vegetation Indices in Viticulture: A Critical Review. Agriculture 2021, 11, 457. [Google Scholar] [CrossRef]
- Gil-Pérez, B.; Zarco-Tejada, P.J.; Correa-Guimaraes, A.; Relea-Gangas, E.; Navas-Gracia, L.M.; Hernández-Navarro, S.; Sanz-Requena, J.F.; Berjón, A.; Martín-Gil, J. Remote Sensing Detection of Nutrient Uptake in Vineyards Using Narrow-Band Hyperspectral Imagery. Vitis 2010, 49, 167–173. [Google Scholar]
- Zarco-Tejada, P.J.; Berjón, A.; López-Lozano, R.; Miller, J.R.; Martín, P.; Cachorro, V.; González, M.R.; de Frutos, A. Assessing Vineyard Condition with Hyperspectral Indices: Leaf and Canopy Reflectance Simulation in a Row-Structured Discontinuous Canopy. Remote Sens. Environ. 2005, 99, 271–287. [Google Scholar] [CrossRef]
- Retzlaff, R.; Molitor, D.; Behr, M.; Bossung, C.; Rock, G.; Hoffmann, L.; Evers, D.; Udelhoven, T. UAS-Based Multi-Angular Remote Sensing of the Effects of Soil Management Strategies on Grapevine. OENO One 2015, 49, 85–102. [Google Scholar] [CrossRef] [Green Version]
- Romero, I.; García-Escudero, E.; Martín, I. Leaf Blade versus Petiole Analysis for Nutritional Diagnosis of Vitis Vinifera L. Cv. Tempranillo. Am. J. Enol. Vitic. 2013, 64, 50–64. [Google Scholar] [CrossRef]
- Schreiner, R.P.; Scagel, C.F. Leaf Blade versus Petiole Nutrient Tests as Predictors of Nitrogen, Phosphorus, and Potassium Status of ‘Pinot Noir’ Grapevines. HortScience 2017, 52, 174–184. [Google Scholar] [CrossRef]
- Filella, I.; Penuelas, J. The Red Edge Position and Shape as Indicators of Plant Chlorophyll Content, Biomass and Hydric Status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf Optical Properties in Higher Plants: Linking Spectral Characteristics to Stress and Chlorophyll Concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debnath, S.; Paul, M.; Rahaman, D.M.M.; Debnath, T.; Zheng, L.; Baby, T.; Schmidtke, L.M.; Rogiers, S.Y. Identifying Individual Nutrient Deficiencies of Grapevine Leaves Using Hyperspectral Imaging. Remote Sens. 2021, 13, 3317. [Google Scholar] [CrossRef]
- Ferwerda, J.G.; Skidmore, A.K.; Mutanga, O. Nitrogen Detection with Hyperspectral Normalized Ratio Indices across Multiple Plant Species. Int. J. Remote Sens. 2005, 26, 4083–4095. [Google Scholar] [CrossRef]
- Herrmann, I.; Karnieli, A.; Bonfil, D.J.; Cohen, Y.; Alchanatis, V. SWIR-Based Spectral Indices for Assessing Nitrogen Content in Potato Fields. Int. J. Remote Sens. 2010, 31, 5127–5143. [Google Scholar] [CrossRef]
- Kumar, L.; Schmidt, K.; Dury, S.; Skidmore, A. Imaging Spectrometry and Vegetation Science. In Imaging Spectrometry: Basic Principles and Prospective Applications; Meer, F.D., van der Jong, S.M.D., Eds.; Remote Sensing and Digital Image Processing; Springer: Dordrecht, The Netherlands, 2001; pp. 111–155. ISBN 978-0-306-47578-8. [Google Scholar]
- Asner, G.P. Biophysical and Biochemical Sources of Variability in Canopy Reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]


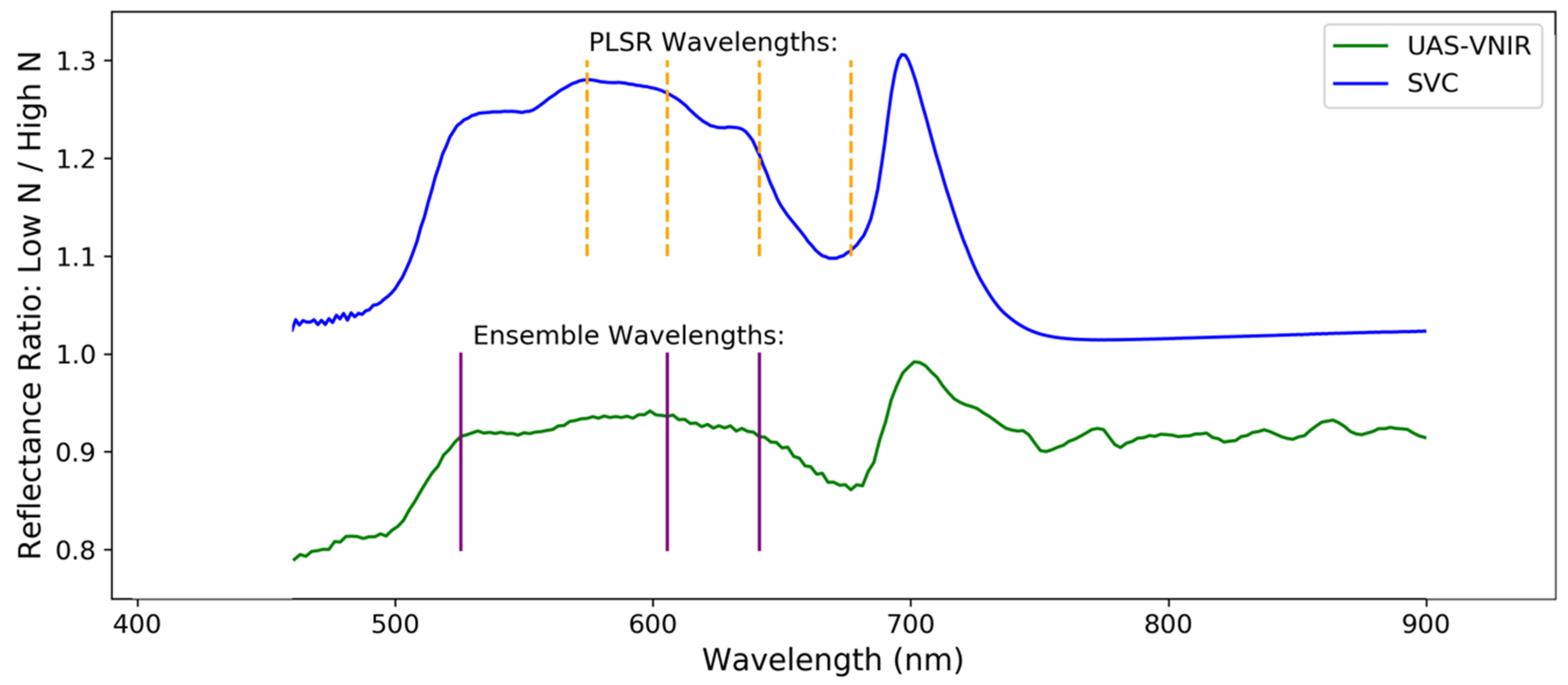
| Method: | Ensemble | PLSR | |||||
|---|---|---|---|---|---|---|---|
| Nutrient | IMT a | # W b | # W b | ||||
| N (%) | 27 | 5 | 0.44 | 0.17 | 10 | 0.45 | 0.17 |
| P (%) | 26 | 7 | 0.34 | 0.02 | 9 | 0.51 | 0.02 |
| K (%) | 24 | 5 | 0.26 | 0.13 | 10 | 0.42 | 0.12 |
| Ca (%) | 25 | 9 | 0.33 | 0.19 | 7 | 0.38 | 0.18 |
| Mg (%) | 26 | 7 | 0.23 | 0.05 | 10 | 0.27 | 0.04 |
| B (mg/kg) | 25 | 8 | 0.46 | 3.08 | 10 | 0.46 | 3.08 |
| Selected (nm) | Method | Absorption (nm) | Chemical | Behavior a |
|---|---|---|---|---|
| - | - | 430 | Chlorophyll a | Electron transition |
| - | - | 460 | Chlorophyll b | Electron transition |
| 526 | Ensemble | - | - | - |
| 574 | PLSR | - | - | - |
| 606 | Both | - | - | - |
| 641 | Both | 640 | Chlorophyll b | Electron transition |
| 677 | PLSR | 660 | Chlorophyll a | Electron transition |
| 958 | PLSR | 970 | Water | O-H bend, 1st overtone |
| 1168 | Both | - | - | - |
| 1494 | Both | 1510 | Protein, nitrogen | N-H stretch, 1st overtone |
| 2030 | PLSR | - | - | - |
| 2058 | PLSR | 2060 | Protein, nitrogen | N=H bend, 2nd overtone/N=H bend/N-H stretch |
| 2173 | PLSR | 2180 | Protein, nitrogen | N-H bend, 2nd overtone/C-H stretch/C-O stretch/C=O stretch/C-N stretch |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chancia, R.; Bates, T.; Vanden Heuvel, J.; van Aardt, J. Assessing Grapevine Nutrient Status from Unmanned Aerial System (UAS) Hyperspectral Imagery. Remote Sens. 2021, 13, 4489. https://doi.org/10.3390/rs13214489
Chancia R, Bates T, Vanden Heuvel J, van Aardt J. Assessing Grapevine Nutrient Status from Unmanned Aerial System (UAS) Hyperspectral Imagery. Remote Sensing. 2021; 13(21):4489. https://doi.org/10.3390/rs13214489
Chicago/Turabian StyleChancia, Robert, Terry Bates, Justine Vanden Heuvel, and Jan van Aardt. 2021. "Assessing Grapevine Nutrient Status from Unmanned Aerial System (UAS) Hyperspectral Imagery" Remote Sensing 13, no. 21: 4489. https://doi.org/10.3390/rs13214489
APA StyleChancia, R., Bates, T., Vanden Heuvel, J., & van Aardt, J. (2021). Assessing Grapevine Nutrient Status from Unmanned Aerial System (UAS) Hyperspectral Imagery. Remote Sensing, 13(21), 4489. https://doi.org/10.3390/rs13214489