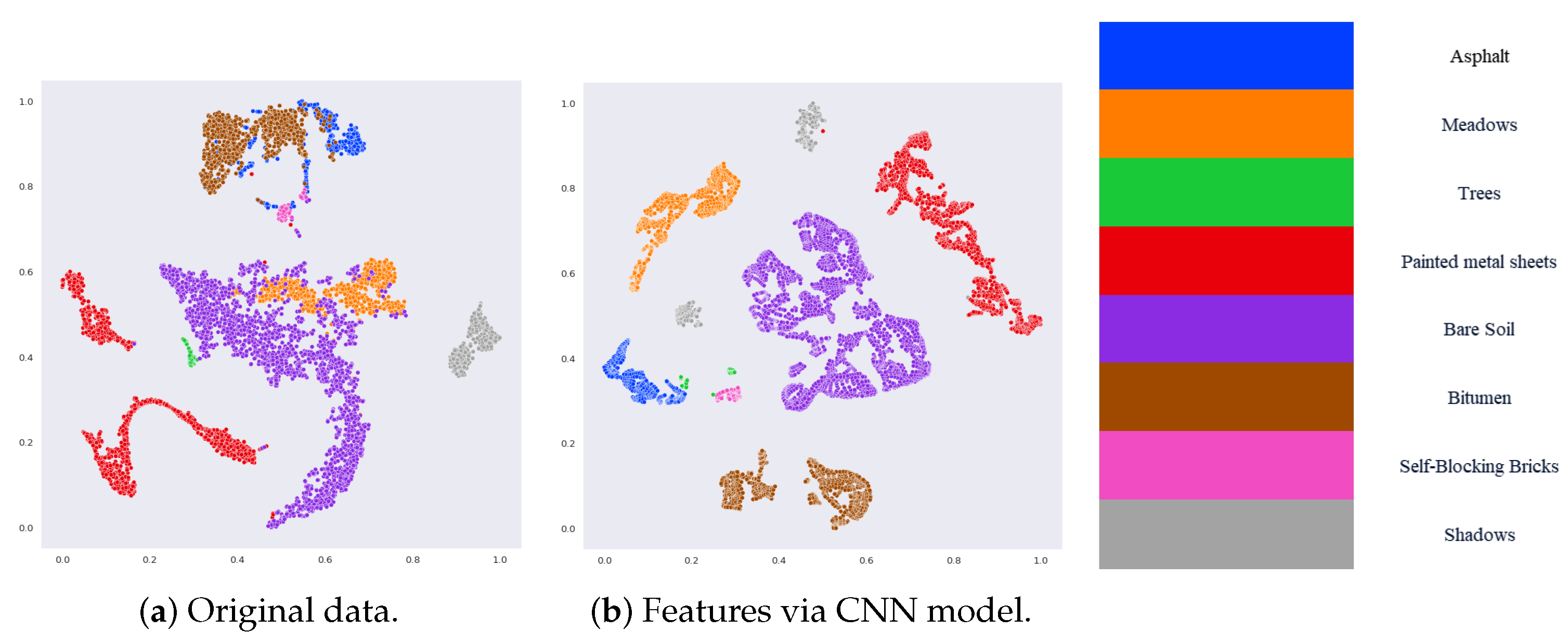
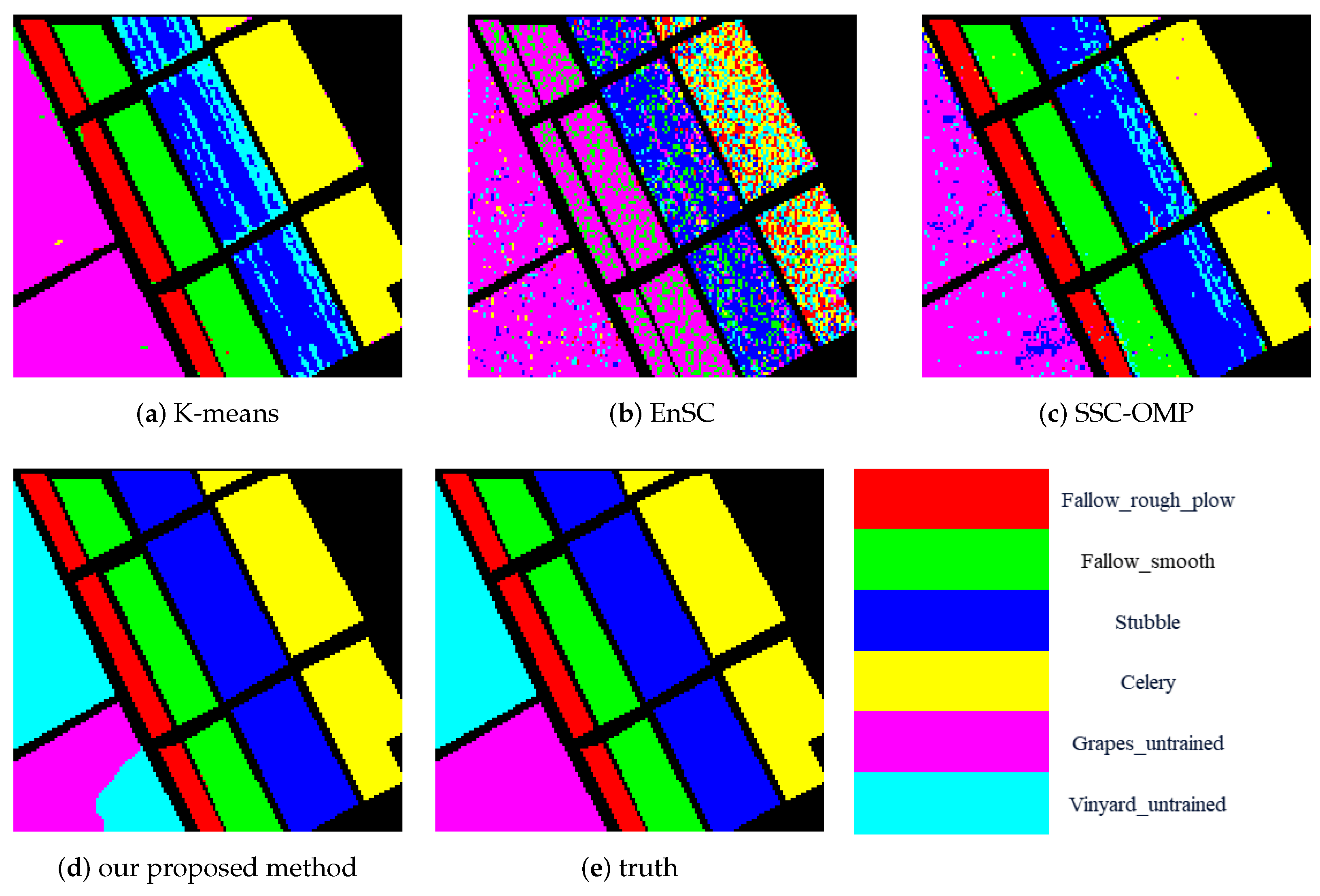
1. Introduction
Hyperspectral remote sensing has been widely used in many different fields [
1,
2,
3]. Hyperspectral image (HSI) classification is a fundamental issue and a hot topic in hyperspectral remote sensing. HSIs can provide rich spectral and spatial information, which improves the utility of HSIs in various applications. However, the abundant spectral information also causes a low classification accuracy, which is called the Hughes phenomenon. Moreover, the limited number of labeled hyperspectral samples also causes difficulties in hyperspectral image classification. In the real world, more and more hyperspectral data are becoming available with the development of information acquisition technology. However, most of these data are unlabeled, and labeling the data is an extremely laborious and time-consuming process. Nevertheless, HSI clustering focus on achieving a good classification performance without training labels. Thus, HSI clustering has attracted increasing levels of attention in recent years.
Some traditional methods used for natural images have been applied in the study of HSI clustering [
4,
5,
6,
7,
8]. The complex characteristics of HSIs strongly reduce their accuracy. Subsequently, more and more HSI clustering methods have been proposed. These methods can be divided into two main groups: spectral-only methods and spatial–spectral methods. Spectral-only methods ignore the spatial information of HSIs, which limits the performance of these methods. To improve accuracy, some spatial–spectral clustering methods have been proposed [
9,
10,
11,
12].
Additionally, to solve problems relating to high dimensionality, some methods based on sparse subspace clustering (SSC) [
13] have been proposed. Those methods rely on clustering HSI data in the low-dimensional subspace. However, the subspace that HSI data exists in is usually non-linear. This limits the performance of these clustering methods.
Recently, deep learning has achieved great success in the computer vision field [
14,
15,
16,
17]. To handle the challenge of nonlinearity, many deep learning-based methods have been proposed. Zhong et al. [
18] proposed a spectral–spatial residual network (SSRN) based on ResNet [
19]. Inspired by DenseNet [
20], Wang et al. [
21] designed a fast dense spectral–spatial convolution network (FDSSC). Ma et al. [
22] adopted a two-branch architecture and proposed a double-branch multi-attention mechanism network (DBMA). Li et al. [
23] introduced the self-attention mechanism to their double-branch dual-attention mechanism network (DBDA).
For HSI clustering, most of the existing deep-learning-based clustering methods can be divided into two steps: feature extraction via deep learning models and traditional clustering. Auto-encoders are used in deep clustering as feature extractors under unsupervised conditions. By encoding images into features and reconstructing images from the features, the model can extract features from HSIs without labels. Based on these features, traditional clustering methods or classification layers can be used to obtain the clustering result. For example, Zeng et al. [
24] proposed a Laplacian regularized deep subspace clustering method (LRDSC) for HSI clustering. In this method, a 3D auto-encoder network with skip connections is used to extract spatial–spectral features. Lei et al. [
25] designed a multi-scale auto-encoder to obtain spatial–spectral information for HSI clustering. Inputs at different scales can provide different types of information, but can increase the computation significantly.
However, the auto-encoder used for HSI processing requires an inordinate amount of computational resources due to the need to reconstruct the input data. Recently, contrastive learning was proposed as a means to extract features under unsupervised conditions. Unlike autoencoders, contrastive learning models operate on different augmented views of the same input image. Since these methods do not require image reconstruction, they require fewer computational resources. Li et al. [
26] proposed a clustering method based on contrastive learning.
To the best of our knowledge, there has been little research on contrastive learning methods for HSI processing. The contrastive learning methods used for typical RGB images can not be applied directly to HSI processing because some typical RGB image augmentation methods are not available for HSIs. For example, color distortion for typical RGB images will destroy spectral information when used on HSIs. We explore HSI augmentation by removing the spectral information of some non-central pixels. Different methods of selecting pixels to remove spectral information can be considered as different HSI augmentation methods.
In this paper, we propose a clustering method for HSIs based on contrastive learning. Firstly, we use contrastive learning methods to train a CNN model to extract features from HSIs. Then, we apply a spectral clustering algorithm to these features. The main contributions of our study are summarized as follows.
Inspired by DBMA and DBDA, we designed a double-branch dense spectral–spatial network for HSI clustering. These two branches can extract spectral and spatial features separately, avoiding the huge computation caused by multi-scale inputs. To reduce the computational load further, we remove the attention blocks in DBDA and DBMA.
We use contrastive learning to explore spatial–spectral information. We augment the image by removing the spectral information of some non-central pixels. Different methods of selecting pixels to remove spectral information can provide different augmented views of the HSI block.
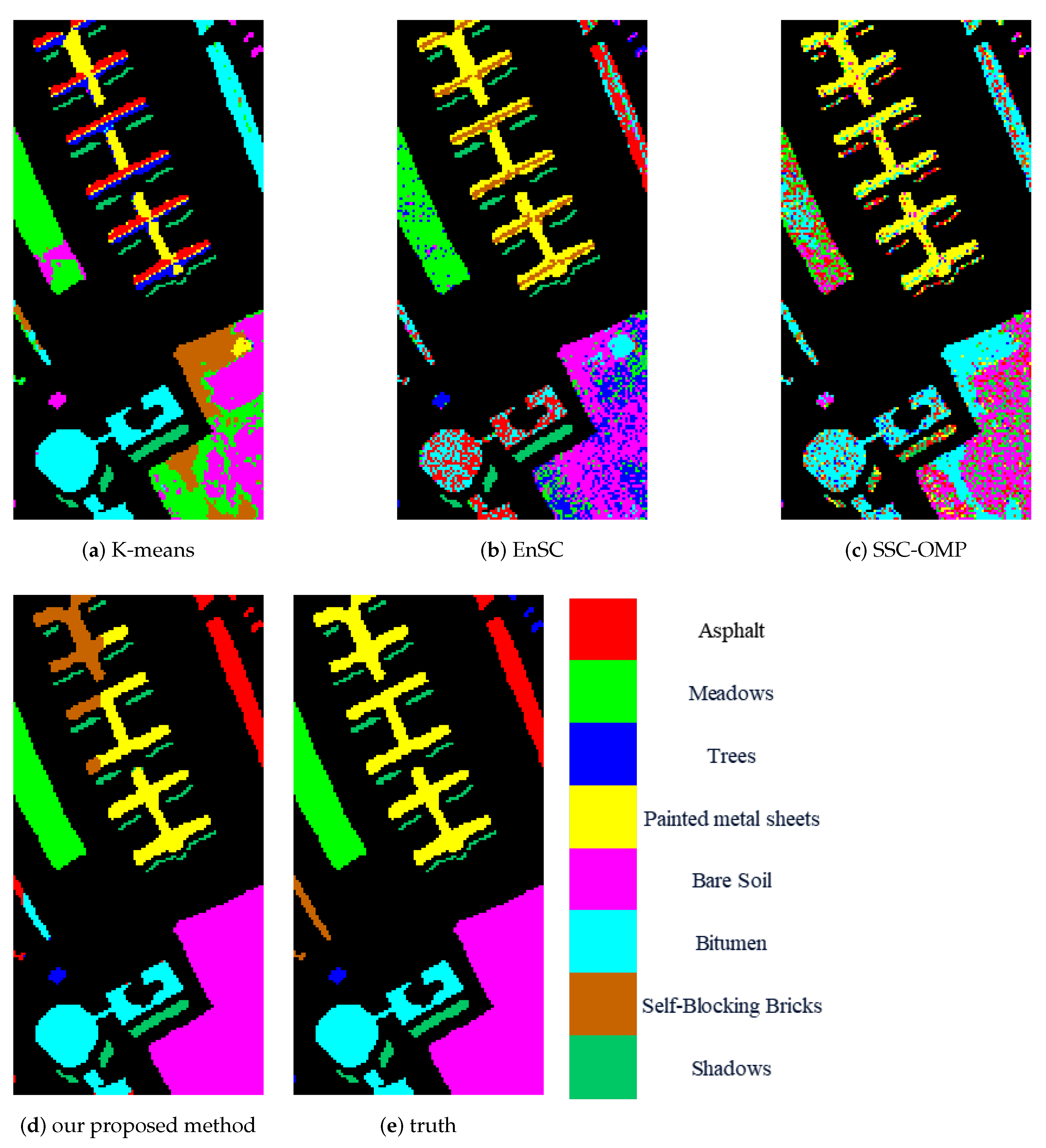
The experimental results obtained over three publicly available HSI datasets demonstrate the superiority of our proposed method compared to other state-of-the-art methods.
The rest of this paper is organized as follows. A brief overview of related work is presented in
Section 2. Our proposed method is described in
Section 3.
Section 4 and
Section 5 provide an analysis of the results and a discussion.
3. Method
Our proposed method consists of two stages: training and testing. Firstly, we used two augmented versions of HSI to train our CNN model. After training, we used the CNN model to obtain the features. Finally, we applied the spectral clustering algorithm based on the features to obtain the clustering result.
3.1. Augmentation in Our Experimental Method
We use two different composite methods to augment the HSI image. The augmentation methods are based on two steps. First, we use horizontal flip or vertical flip as the preliminary augmentation method. Then, we select some non-central pixels in the input blocks to remove spectral information. The different ways in which these pixels are selected can result in different augmentation methods, as illustrated in Algorithms 1 and 2, and
Figure 1. The size of the rectangular area in Algorithm 1 is not fixed.
| Algorithm 1 Selecting Random Rectangular Area to Remove Spectral Information. |
| 1: | Input: input image I; image size . |
| 2: | Output: augmented image . |
| 3: | Generate a matrix of the size () using 1 |
| 4: | Select a random submatrix in this matrix and change the elements inside to 0 |
| 5: | if the center point of the matrix is in the submatrix then |
| 6: | change the element of that point to 1 |
| 7: | end if |
| 8: | for i = 1 to c do |
| 9: | multiply the image in the ith channel by this matrix to obtain the augmented image |
| 10: | end for |
| 11: | Return the augmented image |
| Algorithm 2 Selecting Discrete Points to Remove Spectral Information. |
| 1: | Input: input image I; image size |
| 2: | Output: augmented image |
| 3: | Use 0 and 1 with the same probability to generate a random matrix of the size () |
| 4: | if the center point of the matrix is 0 then |
| 5: | change the element of that point to 1 |
| 6: | end if |
| 7: | for i = 1 to c do |
| 8: | multiply the image in the ith channel by this matrix to obtain the augmented image |
| 9: | end for |
| 10: | Return the augmented image |
3.2. Architectures of Our Experimental Models
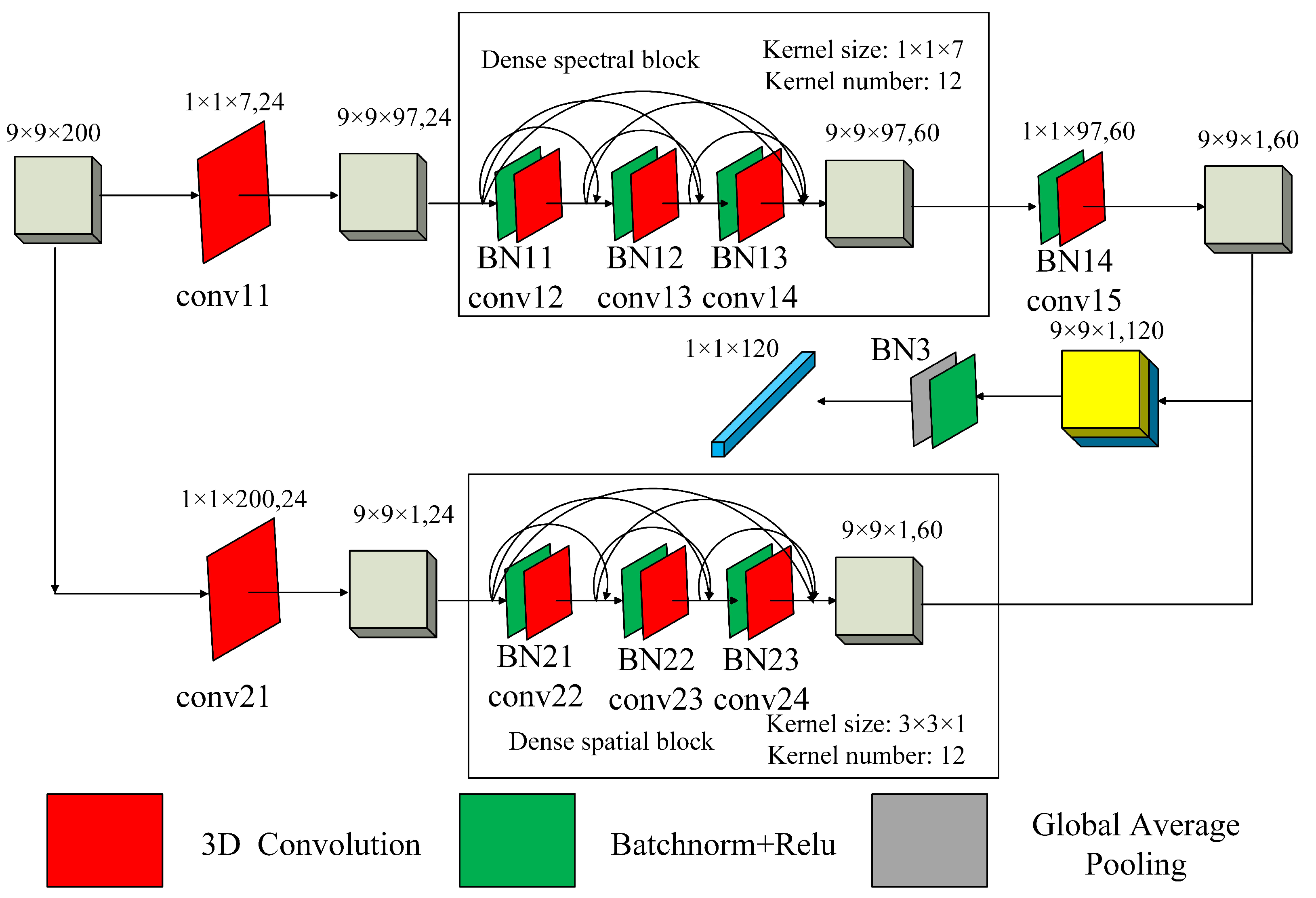
Our proposed method is illustrated in
Figure 2. We use a two-branch CNN model as the backbone model. The double-branch architecture can reduce the interference between spectral and spatial features. The backbone of the CNN model is shown in
Figure 3. To keep the network architecture the same for different hyperspectral images with different bands, we use the PCA method to reduce the dataset dimension to 100. The parameters of the 3D convolutions and batchnorms in our model are illustrated in
Table 1. A detailed introduction of these datasets is presented in
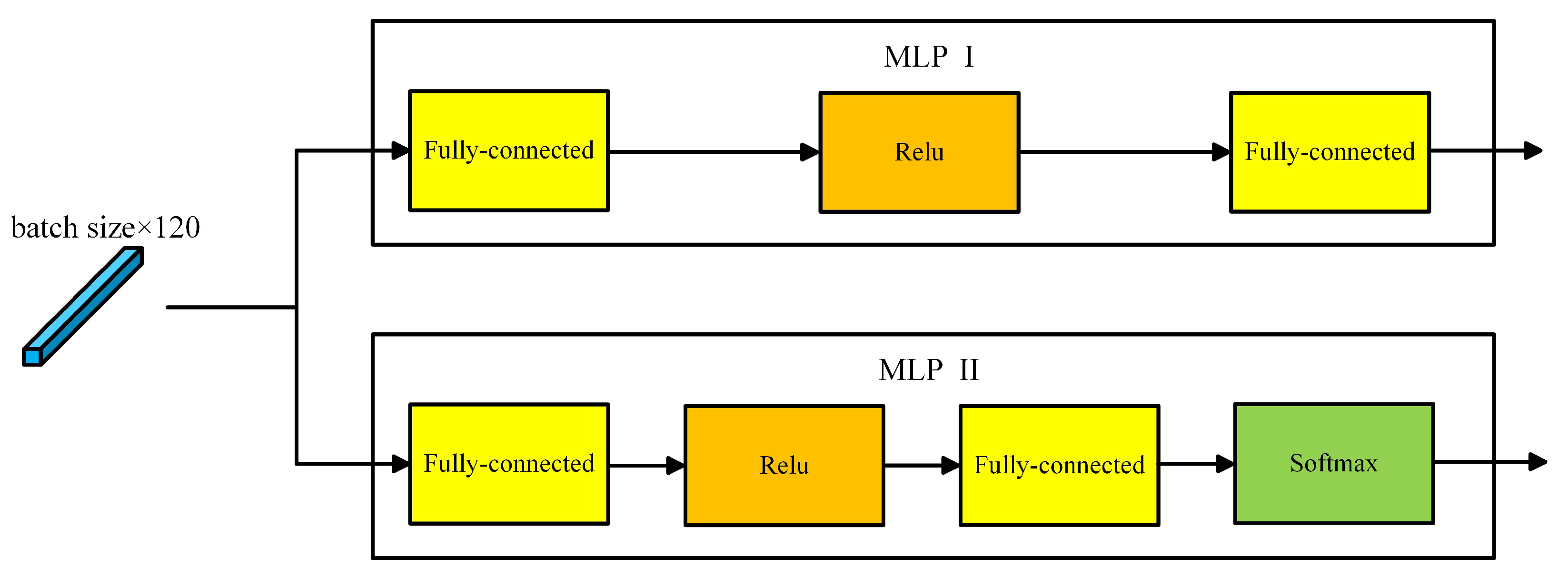
Section 4.1. The two MLPs in our method are shown in
Figure 4. The parameters of these MLPs can be seen in
Table 2. For MLP II, the final output dimension is equal to the cluster number.
3.3. Summary of Our Experimental Method
The overall architecture of our proposed method is shown in Algorithm 3 and
Figure 3. Firstly, we use different augmentations to generate different views of input. Then, we traine the CNN model. After training, we can obtain the features of input HSIs via the CNN model. Finally, we use the spectral clustering algorithm based on the features to obtain the clustering result.
| Algorithm 3 Our proposed clustering algorithm. |
| 1: | Input: dataset I; pixel block size ; training epochs E; batch size N. |
| 2: | Output: cluster assignments. |
| 3: | Sample pixel block of size from the dataset I |
| 4: | //training |
| 5: | for epoch = 1 to E do |
| 6: | compute instance-level contrastive loss |
| 7: | compute cluster-level contrastive loss |
| 8: | compute overall contrastive loss |
| 9: | update the network |
| 10: | end for |
| 11: | //test |
| 12: | Extract features using the CNN model |
| 13: | Use spectral clustering algorithm to obtain the clustering result |
We utilize overall contrastive loss to guide the training process. The overall contrastive loss consists of two parts: instance-level contrastive loss and cluster-level contrastive loss .
In this paper, the mini-batch size is
N. After two types of image augmentations on each input image
, our proposed method works based on
samples
. For a specific sample
, there are a positive pair
and
negative pairs between this sample with the augmented visions of other input images. We can obtain
using MLP I. The instance-level contrastive loss is calculated based on the cosine similarity of each pair. The similarity is computed by
where
and
. The cluster-level contrastive loss
is calculated using the following equations.
where
is the instance-level temperature parameter.
is the loss for the sample
and
is the loss for the sample
.
For cluster-level contrastive loss
, we use the MLP II output
,
.
are the two types of image augmentations,
N is the batch size, and
K is the cluster number.
is the
ith column of
, which is the representation of cluster
i under the data augmentation
a. There is one positive pair
and
negative pairs. The cluster-level contrastive loss is calculated based on the cosine similarity of each pair. The similarity is computed by
where
and
. The instance-level contrastive loss
is calculated using the following equations.
where
is the cluster-level temperature parameter.
is the loss for the sample
and
is the loss for the sample
.
H(
Y) prevents most instances from being assigned to the same cluster.
The overall contrastive loss
is calculated using the following equation:
After training, we can use the model to extract features. Then, we use the spectral clustering algorithm to obtain the final clustering result. To the best of our knowledge, we are the first to propose a contrastive learning-based HSI clustering method. Moreover, we explore the HSI augmentation method that we apply to our proposed clustering method.
6. Conclusions and Future Research
In this paper, we proposed a contrastive learning method for HSI clustering. The contrastive learning method extracts spatial–spectral information based on different augmented views of HSI. We removed the spectral information of some non-central pixels to augment the HSIs. Different methods of selecting the pixels to remove spectral information can be regarded as different augmentation methods. Based on the augmented views of samples, the CNN model was trained under supervision using instance-level and cluster-level contrastive loss. After training, the CNN model was used to extract features from input pixel blocks. Finally, according to the features, we conducted spectral clustering to obtain the clustering result. The experimental results achieved on three public datasets confirmed the superiority of our proposed method. However, our proposed method also has some disadvantages. Because spectral clustering has the computational complexity of O(n) and the space complexity of O(n), it is not suitable for use on large datasets.
In the future, we will focus on HSI data augmentation. More augmentation methods for use on HSIs will be studied, such as rotation, GAN-based augmentation, and so on. We will also try to find a more effective method for selecting non-central pixels to remove the corresponding spectral information. Moreover, we will try to study our proposed method under more challenging conditions, such as luminosity, atmospheric conditions, spatial data sparsity, and noisy spectral data.