Patch-Wise Semantic Segmentation for Hyperspectral Images via a Cubic Capsule Network with EMAP Features
Abstract
:1. Introduction
- In theory, the neural network can extract any feature, as long as the network architecture is good enough. However, it is very complicated and time-consuming to design a neural network that can extract a specific geometric structure. Therefore, in this paper, EMAP features are used as the input of the network, which has the advantage of being able to extract rich spatial geometric features well.
- The cubic convolutional network can extract the spatial–spectral features of the hyperspectral image from the three dimensions, which is conducive to making full use of the existing information and improving the classification accuracy.
- The capsule network can further extract more discriminative deep features, such as spectra with the properties of heterogeneity and homogeneity, to better distinguish pixels at the class boundary.
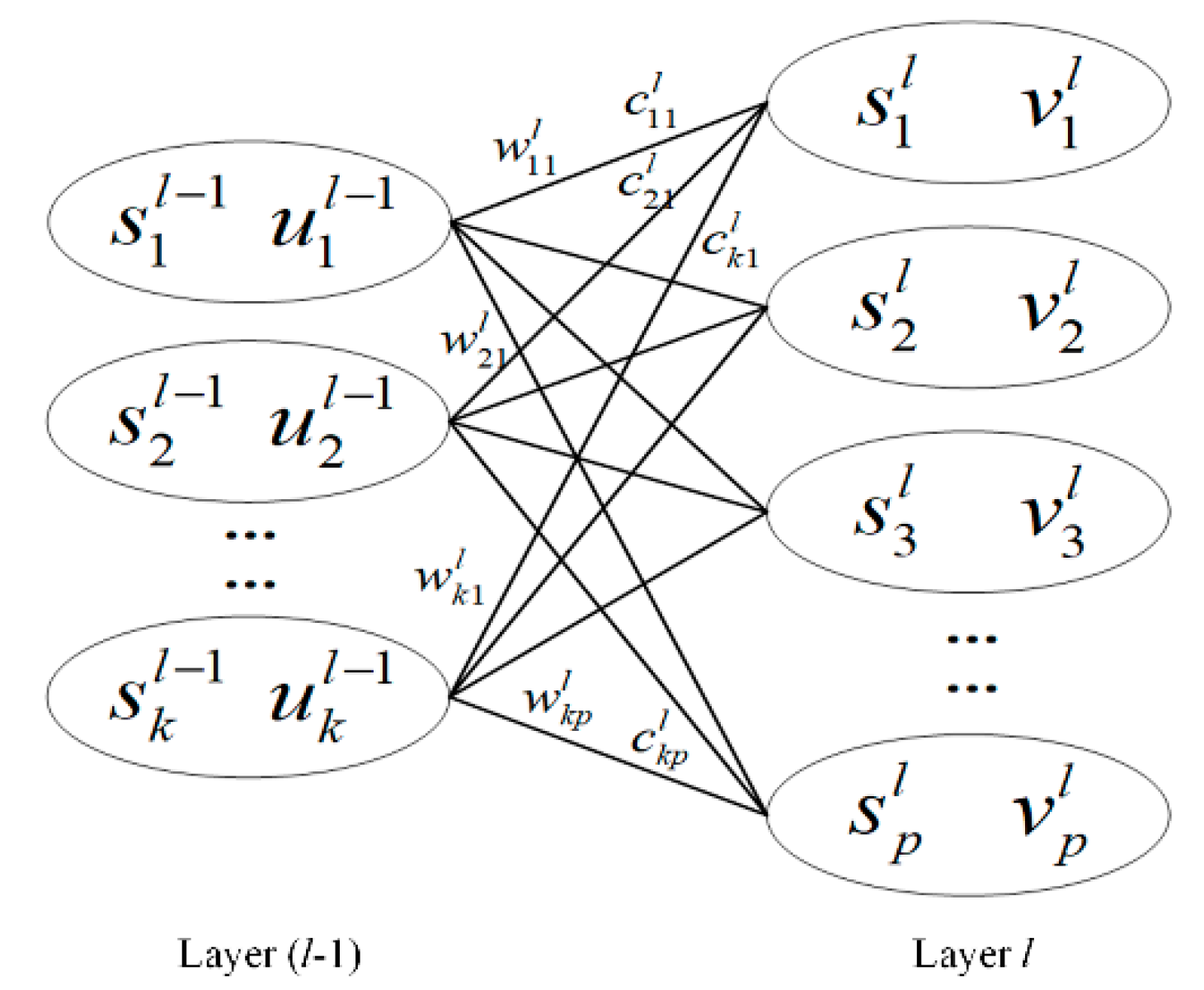
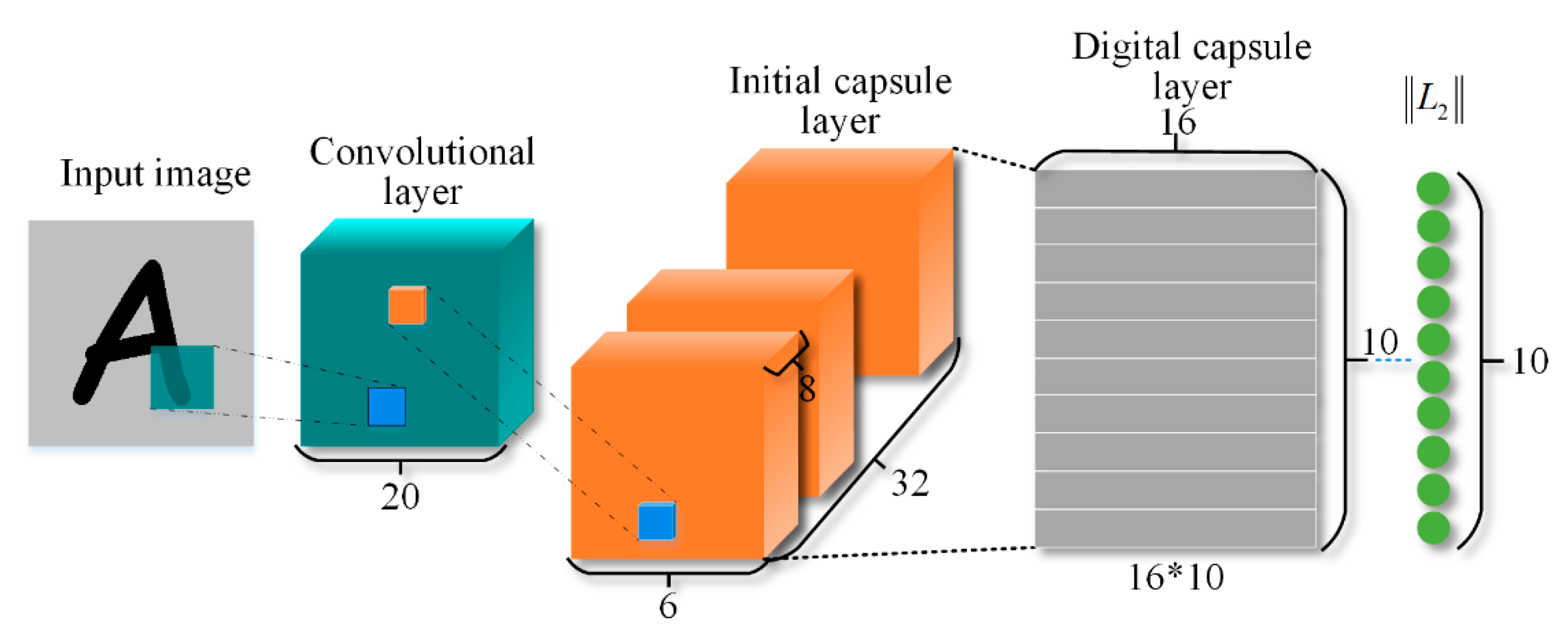
2. Deep Capsule Network
| Algorithm 1. Pseudo code of dynamic routing algorithm |
| 1. Initialization: the number of iterations , parameter , total number of iterations . 2. While , 3. Update the coupling coefficient . 4. Update the input vector . 5. Update the output vector . 6. Update the parameter . 7. Update the number of iterations . 8. End 9. Return the output vector . |
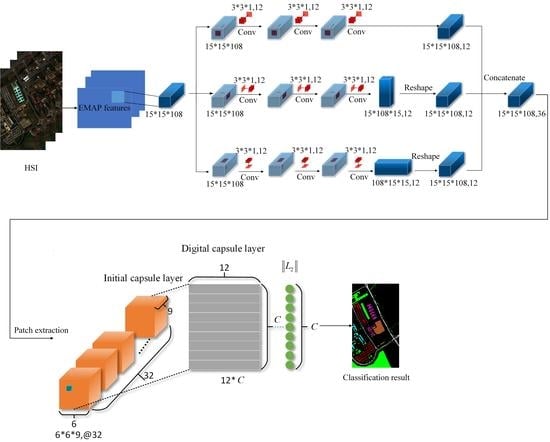
3. The Proposed Method
3.1. Extended Morphological Attribute Profile
3.2. Cubic Capsule Network with EMAP Features
| Algorithm 2. The pseudo code of EMAP–Cubic Caps network |
| 1. Input: Hyperspectral data X and corresponding label Y, the number of iterations 0, 0, total number of iterations = 100, = 3, learning rate = 0.0003. 2. Obtain hyperspectral data after EMAP feature extraction. 3. Divide into training set, verification set and test set, and input the training set and verification set into the cubic convolutional network. The sampling rate is shown in Table 1, Table 2 and Table 3. 4. While <, 5. Perform cubic convolution network. 6. + 1, algorithm 7. End 8. Input the feature map into the initial capsule layer. 9. Connect the initialized capsule to the digital capsule layer and use dynamic routing to update the parameters; see Algorithm 1 for specific steps. 10. Use the trained model to predict the test set. 11. Calculate OA, AA and Kappa. |
4. Experiment and Analysis
4.1. Experimental Data Set
4.1.1. Indian Pines
4.1.2. University of Pavia
4.1.3. Salinas
4.2. Experimental Setup
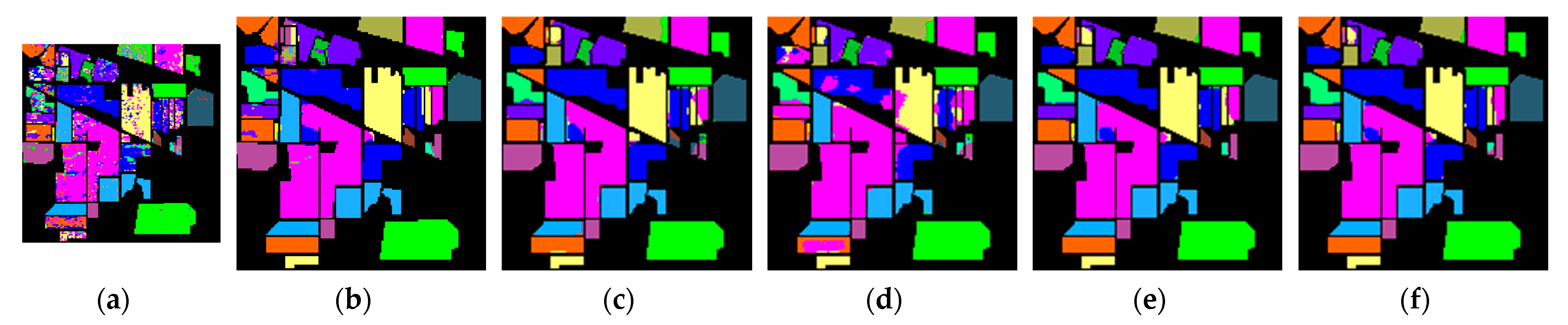
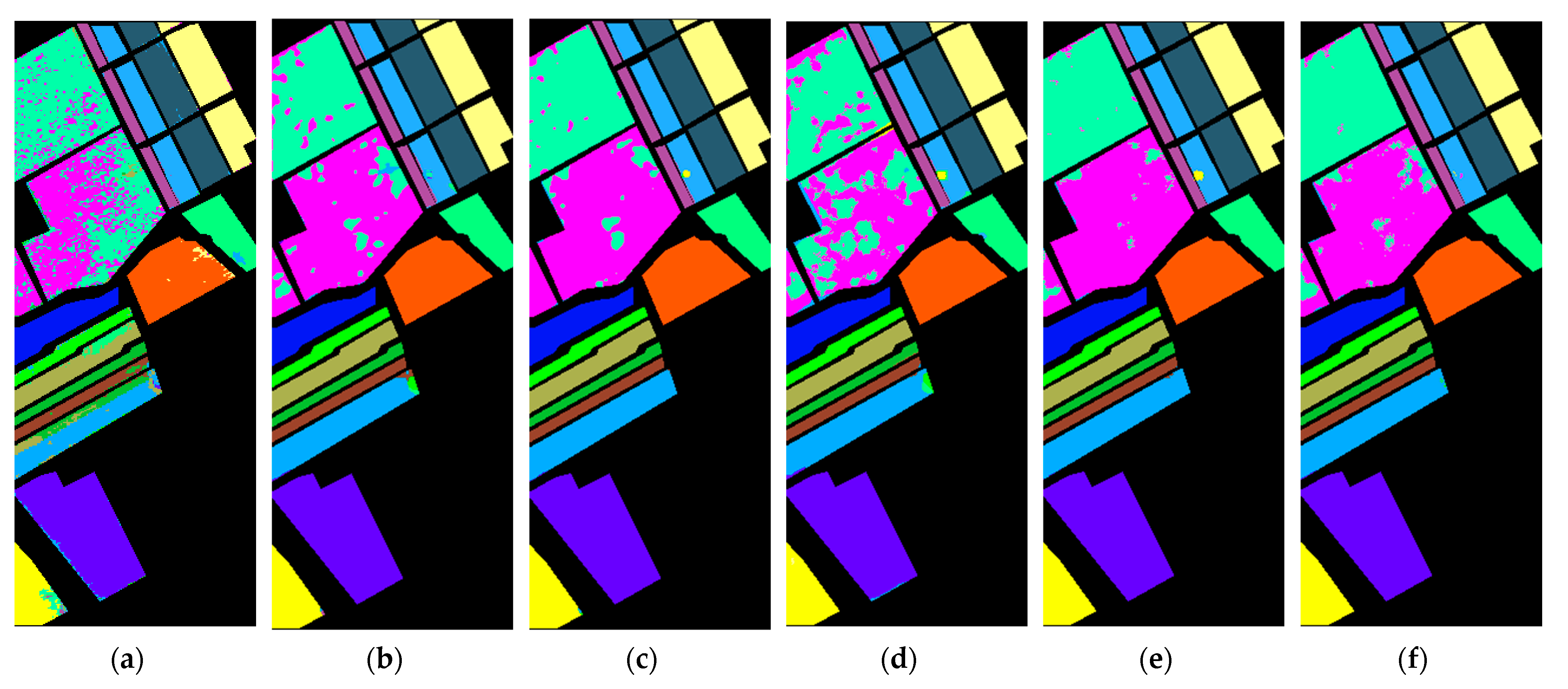
4.3. Experiment and Analysis
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, Q.; Zhang, L.; Zhang, B.; Tong, X.; Chanussot, J. Foreword to the special issue on hyperspectral remote sensing: Theory, methods, and applications. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Gevaert, C.M.; Suomalainen, J.; Tang, J.; Kooistra, L. Generation of Spectral-Temporal Response Surfaces by Combining Multispectral Satellite and Hyperspectral UAV Imagery for Precision Agriculture Applications. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3140–3146. [Google Scholar] [CrossRef]
- Booysen, R.; Gloaguen, R.; Lorenz, S.; Zimmermann, R.; Nex, P. The Potential of Multi-Sensor Remote Sensing Mineral Exploration: Examples from Southern Africa. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 6027–6030. [Google Scholar]
- Moghaddam, T.M.; Razavi, S.; Taghizadeh, M. Applications of Hyperspectral Imaging in Grains and Nuts Quality and Safety Assessment: A Review. J. Food Meas. Charact. 2013, 7, 129–140. [Google Scholar] [CrossRef]
- Ardouin, J.P.; Levesque, J.; Rea, T.A. A Demonstration of Hyperspectral Image Exploitation for Military Applications. In Proceedings of the 10th International Conference on Information Fusion, Quebec City, QC, Canada, 9–12 July 2007; pp. 1–8. [Google Scholar]
- He, C.; Sun, L.; Huang, W.; Zhang, J.; Zheng, Y.; Jeon, B. TSLRLN: Tensor subspace low-rank learning with non-local prior for hyperspectral image mixed denoising. Signal Process. 2021, 184, 108060. [Google Scholar] [CrossRef]
- Sun, L.; He, C.; Zheng, Y.; Tang, S. SLRL4D: Joint restoration of subspace low-rank learning and non-local 4-D transform filtering for hyperspectral image. Remote Sens. 2020, 12, 2979. [Google Scholar] [CrossRef]
- Ye, Q.; Zhao, H.; Li, Z.; Yang, X.; Gao, S.; Yin, T.; Ye, N. L1-Norm Distance Minimization-Based Fast Robust Twin Support Vector kk -Plane Clustering. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 4494–4503. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Li, Z.; Ye, Q.; Yin, H.; Liu, Q.; Chen, X.; Fan, X.; Yang, W.; Yang, G. Learning Robust Discriminant Subspace Based on Joint L2, p- and L2, s-Norm Distance Metrics. IEEE Trans. Neural Netw. Learn. Syst. 2020. Available online: https://pubmed.ncbi.nlm.nih.gov/33180734/ (accessed on 15 July 2021). [CrossRef] [PubMed]
- Ye, Q.; Li, Z.; Fu, L.; Zhang, Z.; Yang, W.; Yang, G. Nonpeaked Discriminant Analysis for Data Representation. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3818–3832. [Google Scholar] [CrossRef]
- Ye, Q.; Yang, J.; Liu, F.; Zhao, C.; Ye, N.; Yin, T. L1-Norm Distance Linear Discriminant Analysis Based on an Effective Iterative Algorithm. IEEE Trans. Circuits Syst. Video Technol. 2018, 28, 114–129. [Google Scholar] [CrossRef]
- Mohanapriya, N.; Kalaavathi, B. Adaptive Image Enhancement using Hybrid Particle Swarm Optimization and Watershed Segmentation. Intell. Autom. Soft Comput. 2018, 25, 1–11. [Google Scholar] [CrossRef]
- Imani, M.; Ghassemian, H. An Overview on Spectral and Spatial Information Fusion for Hyperspectral Image Classification: Current Trends and Challenges. Inf. Fusion 2020, 59, 59–83. [Google Scholar] [CrossRef]
- Sun, L.; Wu, F.; He, C.; Zhan, T.; Liu, W.; Zhang, D. Weighted Collaborative Sparse and L1/2 Low-Rank Regularizations with Superpixel Segmentation for Hyperspectral Unmixing. IEEE Geosci. Remote Sens. Lett. 2020. Available online: https://ieeexplore.ieee.org/document/9186305 (accessed on 15 July 2021). [CrossRef]
- Chen, S.; Ouyang, Y.; Lin, C.; Chang, C.I. Iterative Support Vector Machine for Hyperspectral Image Classification. In Proceedings of the 2011 IEEE International Geoscience and Remote Sensing Symposium, Vancouver, BC, Canada, 24–29 July 2011; pp. 1712–1715. [Google Scholar]
- Tan, K.; Du, P. Classification of hyperspectral image based on morphological profiles and multi-kernel SVM. In Proceedings of the 2010 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Spectral–Spatial Hyperspectral Image Segmentation Using Subspace Multinomial Logistic Regression and Markov Random Fields. IEEE Trans. Geosci. Remote Sens. 2012, 50, 809–823. [Google Scholar] [CrossRef]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spectral–Spatial Hyperspectral Image Classification via Multiscale Adaptive Sparse Representation. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7738–7749. [Google Scholar] [CrossRef]
- Yi, C.; Nasrabadi, N.M.; Tran, T.D. Hyperspectral Image Classification Using Dictionary-Based Sparse Representation. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3973–3985. [Google Scholar]
- Gu, Y.; Liu, T.; Jia, X.; Benediktsson, J.A.; Chanussot, J. Nonlinear Multiple Kernel Learning with Multiple-Structure-Element Extended Morphological Profiles for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3235–3247. [Google Scholar] [CrossRef]
- Plaza, A.; Martinez, P.; Plaza, J.; Perez, R. Dimensionality reduction and classification of hyperspectral image data using sequences of extended morphological transformations. IEEE Trans. Geosci. Remote Sens. 2005, 43, 466–479. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Ma, C.; Chen, Y.; Zheng, Y.; Shim, H.; Wu, Z.; Jeon, B. Low Rank Component Induced Spatial-Spectral Kernel Method for Hyperspectral Image Classification. IEEE Trans. Circuits Syst. Video Technol. 2020, 30, 3829–3842. [Google Scholar] [CrossRef]
- Sun, L.; Ma, C.; Chen, Y.; Shim, H.; Wu, Z.; Jeon, B. Adjacent Superpixel-Based Multiscale Spatial-Spectral Kernel for Hyperspectral Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 1905–1919. [Google Scholar] [CrossRef]
- Wei, L.; Hung, C.; Han, Y. Modified PSO Algorithm on Recurrent Fuzzy Neural Network for System Identification. Intell. Autom. Soft Comput. 2019, 25, 329–341. [Google Scholar]
- Wu, H.; Liu, Q.; Liu, X. A Review on Deep Learning Approaches to Image Classification and Object Segmentation. Comput. Mater. Contin. 2019, 60, 575–597. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lu, W.; Li, F.; Peng, X.; Zhang, R. Deep Feature Fusion Model for Sentence Semantic Matching. Comput. Mater. Contin. 2019, 61, 601–616. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, X.; Xiao, B.; Cheng, X.; Wu, Y.; Chen, S. Multi-Task Convolution Operators with Object Detection for Visual Tracking. IEEE Trans. Circuits Syst. Video Technol. 2021. Available online: https://ieeexplore.ieee.org/document/9395485 (accessed on 15 July 2021). [CrossRef]
- Yue, J.; Zhao, W.; Mao, S.; Liu, H. Spectral–spatial Classification of Hyperspectral Images Using Deep Convolutional Neural Networks. Remote Sens. Lett. 2015, 6, 468–477. [Google Scholar] [CrossRef]
- Zhen, X.; Yu, H.Y.; Zheng, K.; Gao, L.R.; Song, M. A Novel Classification Framework for Hyperspectral Image Classification Based on Multiscale Spectral-Spatial Convolutional Network. In Proceedings of the 2021 11th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 March 2021; pp. 1–5. [Google Scholar]
- Yu, C.; Zhao, M.; Song, M.; Wang, Y.; Li, F.; Han, R.; Chang, C.I. Hyperspectral Image Classification Method Based on CNN Architecture Embedding with Hashing Semantic Feature. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 1866–1881. [Google Scholar] [CrossRef]
- Nyan, L.; Alexander, G.; Naing, M.; Do, M.; Htet, A. Hyperspectral Remote Sensing Images Classification Using Fully Convolutional Neural Network. In Proceedings of the 2021 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (ElConRus), Moscow, Russia, 26–29 January 2021; pp. 2166–2170. [Google Scholar]
- Mou, L.; Zhu, X. Learning to Pay Attention on Spectral Domain: A Spectral Attention Module-Based Convolutional Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 99, 1–13. [Google Scholar] [CrossRef]
- Gao, L.; Gu, D.; Zhuang, L.; Ren, J.; Yang, D.; Zhang, B. Combining T-distributed Stochastic Neighbor Embedding with Convolutional Neural Networks for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2020, 8, 1368–1372. [Google Scholar] [CrossRef]
- Zhang, M.; Li, W.; Du, Q. Diverse Region-Based CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. A Publ. IEEE Signal Process. Soc. 2018, 27, 2623–2624. [Google Scholar] [CrossRef] [PubMed]
- Murali, K.; Sarma, T.; Bindu, C. A 3d-Deep CNN Based Feature Extraction and Hyperspectral Image Classification. In Proceedings of the 2020 IEEE India Geoscience and Remote Sensing Symposium (InGARSS), Ahmedabad, India, 1–4 December 2020; pp. 229–232. [Google Scholar]
- Roy, S.; Krishna, G.; Dubey, S.; Chaudhuri, B. HybridSN: Exploring 3-D–2-D CNN Feature Hierarchy for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2020, 17, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Ge, Z.; Cao, G.; Li, X.; Fu, P. Hyperspectral Image Classification Method Based on 2D–3D CNN and Multibranch Feature Fusion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 5776–5788. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Jiang, Y.; Wang, P.; Shen, Q.; Shen, C. Hyperspectral Classification Based on Lightweight 3-D-CNN With Transfer Learning. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5813–5828. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zheng, X.; Lu, X. A Supervised Segmentation Network for Hyperspectral Image Classification. IEEE Trans. Image Process. 2021, 30, 2810–2825. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, J.; Luo, Z.; Michael, C. Spectral–Spatial Residual Network for Hyperspectral Image Classification: A 3-D Deep Learning Framework. IEEE Trans. Geosci. Remote Sens. 2017, 56, 847–858. [Google Scholar] [CrossRef]
- Yu, C.; Han, R.; Song, M.; Liu, C.; Chang, C.I. A Simplified 2D-3D CNN Architecture for Hyperspectral Image Classification Based on Spatial–Spectral Fusion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 2485–2501. [Google Scholar] [CrossRef]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic Routing Between Capsules. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- Zhu, K.; Chen, Y.; Ghamisi, P.; Jia, X.; Benediktsson, J.A. Deep Convolutional Capsule Network for Hyperspectral Image Spectral and Spectral-spatial Classification. Remote Sens. 2019, 11, 223. [Google Scholar] [CrossRef] [Green Version]
- Paoletti, M.E.; Haut, J.M.; Fernandez-Beltran, R.; Plaza, J.; Plaza, A.; Li, J. Capsule Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 4, 2145–2160. [Google Scholar] [CrossRef]
- Jia, S.; Liao, J.; Xu, M.; Li, Y.; Zhu, J.; Jia, X.; Li, Q. 3-D Gabor Convolutional Neural Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021. Available online: https://ieeexplore.ieee.org/document/9460777 (accessed on 15 July 2021). [CrossRef]
- Hinton, G.E.; Sabour, S.; Frosst, N. Matrix Capsules with EM Routing. In Proceedings of the 6th International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018; pp. 1–15. [Google Scholar]
- Wang, J.; Guo, S.; Huang, R.; Li, L.; Zhang, X.; Jiao, L. Dual-Channel Capsule Generation Adversarial Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021. Available online: https://ieeexplore.ieee.org/document/9328201 (accessed on 15 July 2021). [CrossRef]
- Xu, Q.; Wang, D.; Luo, B. Faster Multiscale Capsule Network with Octave Convolution for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2021, 2, 361–365. [Google Scholar] [CrossRef]
- Tuia, D.; Pacifici, F.; Kanevski, M.; Emery, W.J. Classification of Very High Spatial Resolution Imagery Using Mathematical Morphology and Support Vector Machines. IEEE Trans. Geosci. Remote Sens. 2009, 47, 3866–3879. [Google Scholar] [CrossRef]
- Mauro, D.M.; Benediktsson, J.A.; Björn, W.; Lorenzo, B. Morphological Attribute Profiles for the Analysis of Very High-Resolution Images. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3747–3762. [Google Scholar]
- Mura, M.; Villa, A.; Benediktsson, J.A.; Chanussot, J.; Bruzzone, L. Classification of Hyperspectral Images by Using Extended Morphological Attribute Profiles and Independent Component Analysis. IEEE Geosci. Remote Sens. Lett. 2011, 8, 542–546. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Mura, M.D.; Chanussot, J.; Du, P.; He, X. Random Subspace Ensembles for Hyperspectral Image Classification with Extended Morphological Attribute Profiles. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4768–4786. [Google Scholar] [CrossRef]





| Category | Class Name | Number of Training Samples | Number of Verification Samples | Number of Test Samples |
|---|---|---|---|---|
| 1 | Alfalfa | 2 | 1 | 43 |
| 2 | Corn-notill | 71 | 36 | 1321 |
| 3 | Corn-mintill | 41 | 21 | 768 |
| 4 | Corn | 11 | 6 | 220 |
| 5 | Grass-pasture | 24 | 12 | 447 |
| 6 | Grass-trees | 36 | 18 | 676 |
| 7 | Grass-pasture-mowed | 1 | 1 | 26 |
| 8 | Hay-windrowed | 23 | 12 | 443 |
| 9 | Oats | 1 | 1 | 18 |
| 10 | Soybean-notill | 48 | 24 | 900 |
| 11 | Soybean-mintill | 122 | 61 | 2272 |
| 12 | Soybean-clean | 29 | 15 | 549 |
| 13 | Wheat | 10 | 5 | 190 |
| 14 | Woods | 63 | 32 | 1170 |
| 15 | Buildings-grass-trees-drive | 19 | 10 | 357 |
| 16 | Store-steel-towers | 4 | 2 | 87 |
| total | 505 | 257 | 9487 |
| Category | Class Name | Number of Training Samples | Number of Verification Samples | Number of Test Samples |
|---|---|---|---|---|
| 1 | Asphalt | 30 | 15 | 6586 |
| 2 | Meadows | 30 | 15 | 18,604 |
| 3 | Gravel | 30 | 15 | 2054 |
| 4 | Trees | 30 | 15 | 3019 |
| 5 | Painted metal sheets | 30 | 15 | 1300 |
| 6 | Bare Soil | 30 | 15 | 4984 |
| 7 | Bitumen | 30 | 15 | 1285 |
| 8 | Self-Blocking Bricks | 30 | 15 | 3637 |
| 9 | Shadows | 30 | 15 | 902 |
| total | 270 | 135 | 42,371 |
| Category | Class Name | Number of Training Samples | Number of Verification Samples | Number of Test Samples |
|---|---|---|---|---|
| 1 | Brocoli_green_weeds_1 | 20 | 10 | 1979 |
| 2 | Brocoli_green_weeds_2 | 20 | 10 | 3696 |
| 3 | Fallow | 20 | 10 | 1946 |
| 4 | Fallow_rough_plow | 20 | 10 | 1364 |
| 5 | Fallow_smooth | 20 | 10 | 2648 |
| 6 | Stubble | 20 | 10 | 3929 |
| 7 | Celery | 20 | 10 | 3549 |
| 8 | Grapes_untrained | 20 | 10 | 11,241 |
| 9 | Soil_vineyard_develop | 20 | 10 | 6173 |
| 10 | Corn_senesced_green_ weeds | 20 | 10 | 3248 |
| 11 | Lettuce_romaine_4wk | 20 | 10 | 1038 |
| 12 | Lettuce_romaine_5wk | 20 | 10 | 1897 |
| 13 | Lettuce_romaine_6wk | 20 | 10 | 886 |
| 14 | Lettuce_romaine_7wk | 20 | 10 | 1040 |
| 15 | Vineyard_untrained | 20 | 10 | 7238 |
| 16 | Vineyard_vertical_trellis | 20 | 10 | 1777 |
| total | 320 | 160 | 53,649 |
| Category | EMAP–SVM | DR–CNN | SSRN | 3D–Caps | Proposed Methods | |
|---|---|---|---|---|---|---|
| Cubic-Caps | EMAP–Cubic-Caps | |||||
| 1 | 66.04 | 83.33 | 100 | 97.30 | 95.35 | 100 |
| 2 | 75.30 | 96.15 | 96.91 | 95.00 | 95.48 | 98.83 |
| 3 | 40.86 | 96.16 | 95.79 | 93.88 | 96.89 | 98.47 |
| 4 | 51.38 | 98.98 | 98.99 | 99.47 | 98.56 | 99.49 |
| 5 | 90.42 | 95.80 | 95.35 | 96.47 | 96.03 | 96.19 |
| 6 | 97.42 | 95.87 | 96.15 | 99.11 | 95.89 | 97.92 |
| 7 | 66.67 | 92.59 | 96.29 | 73.91 | 65.38 | 100 |
| 8 | 97.11 | 89.98 | 89.78 | 98.25 | 99.34 | 100 |
| 9 | 20.83 | 89.47 | 94.74 | 88.89 | 100 | 75.00 |
| 10 | 71.40 | 92.39 | 92.52 | 89.18 | 93.58 | 97.58 |
| 11 | 98.13 | 98.38 | 98.02 | 74.35 | 98.70 | 98.94 |
| 12 | 80.26 | 92.65 | 92.50 | 98.97 | 95.46 | 94.48 |
| 13 | 88.53 | 100 | 100 | 100 | 100 | 100 |
| 14 | 93.61 | 93.88 | 93.73 | 93.72 | 94.72 | 98.71 |
| 15 | 76.05 | 98.40 | 98.09 | 96.87 | 97.21 | 96.67 |
| 16 | 70.34 | 100 | 100 | 92.55 | 98.85 | 95.35 |
| OA (%) | 76.52 | 95.70 | 95.75 | 90.20 | 96.47 | 98.20 |
| AA (%) | 74.02 | 94.62 | 96.18 | 92.995 | 95.08 | 96.72 |
| Kappa | 0.7383 | 0.9436 | 0.9569 | 0.9015 | 0.9598 | 0.9795 |
| Category | EMAP–SVM | DR–CNN | SSRN | 3D–Caps | Proposed Methods | |
|---|---|---|---|---|---|---|
| Cubic-Caps | EMAP–Cubic-Caps | |||||
| 1 | 71.29 | 98.63 | 98.70 | 98.41 | 98.86 | 99.63 |
| 2 | 75.75 | 98.34 | 98.46 | 97.49 | 99.75 | 99.76 |
| 3 | 72.97 | 99.30 | 98.96 | 79.20 | 99.10 | 99.85 |
| 4 | 91.80 | 90.73 | 91.68 | 99.86 | 98.24 | 98.98 |
| 5 | 99.33 | 99.32 | 99.39 | 99.77 | 99.92 | 100 |
| 6 | 71.55 | 74.53 | 78.33 | 58.82 | 93.57 | 95.15 |
| 7 | 87.60 | 99.61 | 99.45 | 90.60 | 93.92 | 95.73 |
| 8 | 67.29 | 97.15 | 97.55 | 87.11 | 95.57 | 97.59 |
| 9 | 99.31 | 100 | 100 | 100 | 99.45 | 99.77 |
| OA (%) | 76.45 | 94.30 | 95.15 | 88.30 | 98.15 | 98.81 |
| AA (%) | 81.88 | 95.29 | 95.84 | 90.14 | 97.60 | 98.49 |
| Kappa | 0.6985 | 0.9254 | 0.9364 | 0.8493 | 0.9755 | 0.9842 |
| Category | EMAP–SVM | DR–CNN | SSRN | 3D–Caps | Proposed Methods | |
|---|---|---|---|---|---|---|
| Cubic-Caps | EMAP–Cubic-Caps | |||||
| 1 | 92.90 | 100 | 100 | 100 | 100 | 100 |
| 2 | 92.77 | 96.89 | 100 | 100 | 100 | 100 |
| 3 | 93.49 | 98.78 | 97.55 | 94.98 | 100 | 100 |
| 4 | 87.87 | 95.84 | 94.86 | 95.34 | 98.60 | 99.34 |
| 5 | 90.61 | 99.92 | 99.49 | 98.43 | 99.96 | 99.96 |
| 6 | 85.51 | 99.85 | 99.90 | 100 | 100 | 100 |
| 7 | 89.12 | 100 | 100 | 99.58 | 100 | 99.97 |
| 8 | 51.43 | 93.24 | 93.73 | 83.76 | 99.66 | 98.90 |
| 9 | 93.61 | 98.68 | 99.50 | 99.36 | 99.77 | 99.91 |
| 10 | 66.20 | 88.68 | 95.38 | 91.76 | 97.00 | 98.44 |
| 11 | 87.70 | 100 | 93.97 | 94.26 | 99.07 | 99.89 |
| 12 | 82.91 | 99.95 | 100 | 99.89 | 100 | 100 |
| 13 | 77.19 | 98.22 | 100 | 99.53 | 100 | 100 |
| 14 | 71.08 | 99.71 | 94.32 | 97.79 | 99.69 | 98.68 |
| 15 | 64.01 | 73.81 | 83.06 | 60.50 | 86.57 | 99.34 |
| 16 | 94.78 | 99.94 | 100 | 94.47 | 99.88 | 96.88 |
| OA (%) | 76.74 | 93.15 | 95.44 | 88.95 | 97.39 | 98.55 |
| AA (%) | 82.57 | 96.47 | 96.98 | 94.35 | 98.70 | 99.08 |
| Kappa | 0.7539 | 0.9239 | 0.9493 | 0.8774 | 0.9709 | 0.9838 |
| Competing Methods | IN | UP | Salinas | |
|---|---|---|---|---|
| DR–CNN [34] | Train. (min) | 7.86 | 7.62 | 7.74 |
| Test.(s) | 55.71 | 97.19 | 281.25 | |
| SSRN [40] | Train. (min) | 2.62 | 1.41 | 2.06 |
| Test.(s) | 4.49 | 12.24 | 25.91 | |
| 3D–Caps [43] | Train. (min) | 1.52 | 1.15 | 1.38 |
| Test.(s) | 4.44 | 11.46 | 17.56 | |
| Cubic-Caps | Train. (min) | 1.94 | 1.22 | 1.84 |
| Test. (s) | 5.43 | 14.36 | 27.64 | |
| EMAP–Cubic-Caps | Train. (min) | 1.76 | 1.29 | 1.50 |
| Test.(s) | 4.21 | 13.33 | 25.47 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Song, X.; Guo, H.; Zhao, G.; Wang, J. Patch-Wise Semantic Segmentation for Hyperspectral Images via a Cubic Capsule Network with EMAP Features. Remote Sens. 2021, 13, 3497. https://doi.org/10.3390/rs13173497
Sun L, Song X, Guo H, Zhao G, Wang J. Patch-Wise Semantic Segmentation for Hyperspectral Images via a Cubic Capsule Network with EMAP Features. Remote Sensing. 2021; 13(17):3497. https://doi.org/10.3390/rs13173497
Chicago/Turabian StyleSun, Le, Xiangbo Song, Huxiang Guo, Guangrui Zhao, and Jinwei Wang. 2021. "Patch-Wise Semantic Segmentation for Hyperspectral Images via a Cubic Capsule Network with EMAP Features" Remote Sensing 13, no. 17: 3497. https://doi.org/10.3390/rs13173497
APA StyleSun, L., Song, X., Guo, H., Zhao, G., & Wang, J. (2021). Patch-Wise Semantic Segmentation for Hyperspectral Images via a Cubic Capsule Network with EMAP Features. Remote Sensing, 13(17), 3497. https://doi.org/10.3390/rs13173497