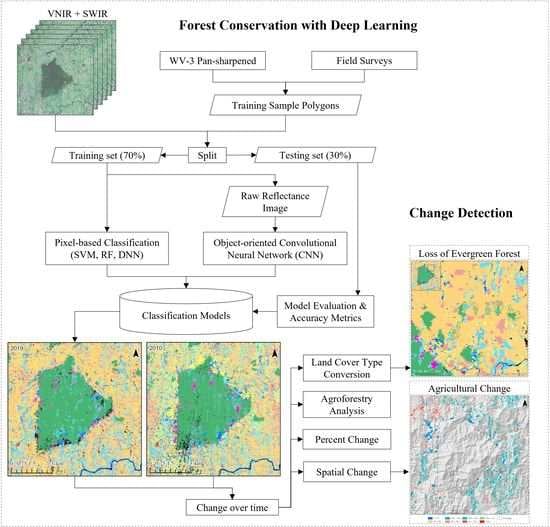
1. Introduction
The REDD+ (Reducing Emissions from Deforestation and Forest Degradation) program identifies halting and reversing forest loss and degradation, which is essential for mitigating climate change effects [
1,
2]. To implement REDD+ objectives at the national level, it is imperative to develop methodologies to accurately estimate forest types, forest cover area, forest degradation, and change as well as all forest restoration due to conservation efforts supported by the REDD+ program using satellite remote sensing. Efforts on documenting the impact of REDD+ payments on forest recovery and carbon sequestration in a fully automated manner is of special interest as ever-increasing stocks of very high-resolution satellite imagery with a global coverage present unprecedented challenges for big data analytics.
Due to its species richness and a loss of more than 70% of its original primary vegetation, Madagascar is considered to be one of eight biodiversity hotspots across the globe [
3]. It is estimated that 44% of Madagascar’s natural forest has been lost since 1953 due to anthropogenic activity [
4], which may be attributed to the increasing population, practice of slash-and-burn agriculture, grazing, fuelwood gathering, logging, economic development projects, cattle ranching, and mining [
5,
6]. The increasing deforestation results in an increasingly fragmented forest where at forest edges, the differing air temperature, moisture, and sunlight conditions impact both the abundance and distribution of species [
7]. Deforestation is a major threat to native species since 90% of species endemic to Madagascar are forest-dependent [
3]. Therefore, the study area provides a living laboratory for the studies of human–forest interactions, which has significant impact on our understanding of tropical forests and biodiversity at the global scale for better implementation of REDD+.
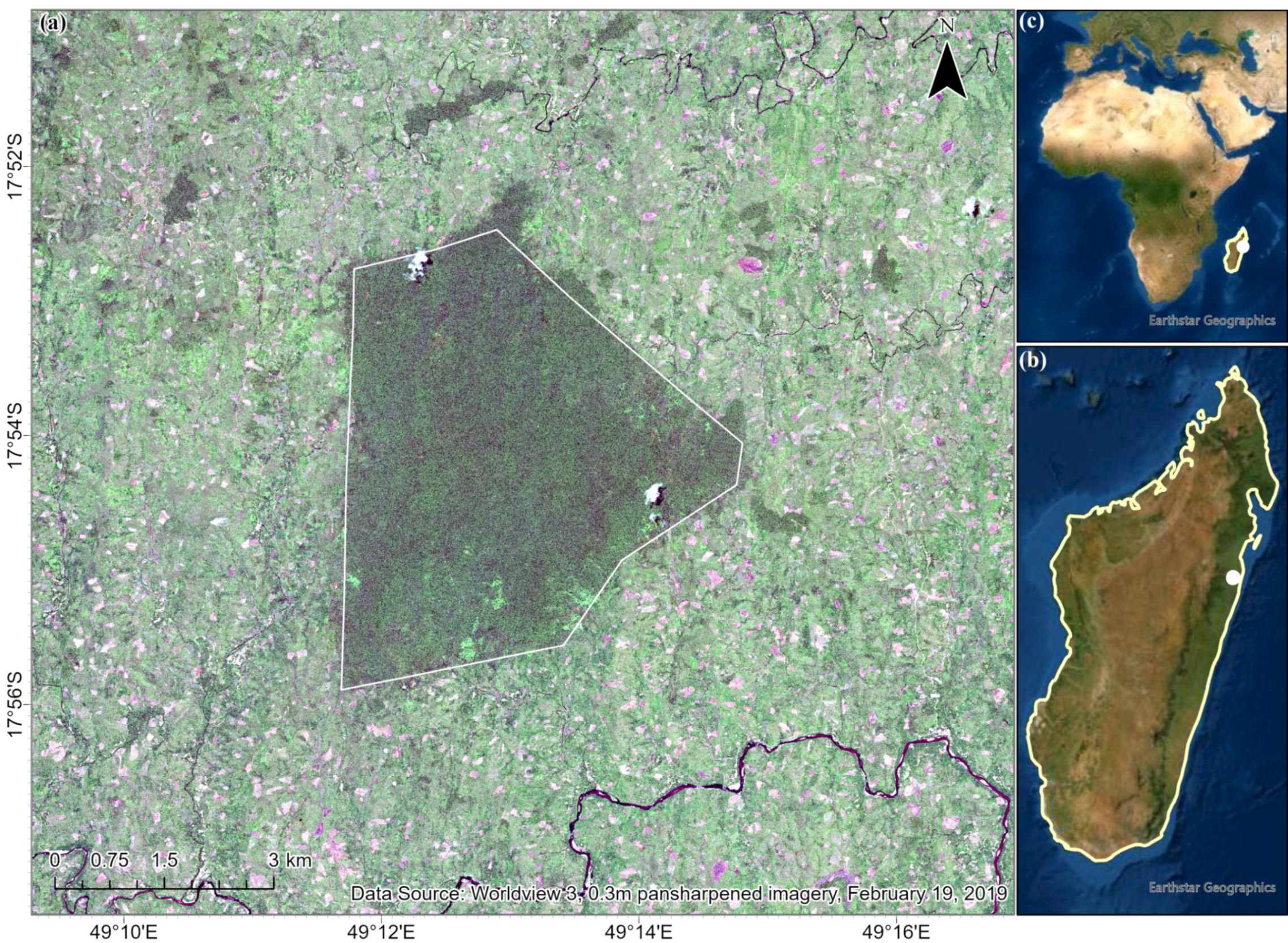
The Betampona Nature Reserve (BNR) is situated on the central–eastern coast of Madagascar. It was once contiguous with the Zahamena forest; however, extensive deforestation has resulted in it being one of the few remaining tracts of primary rainforest on the eastern coast [
6]. Although small and isolated, it has an extremely high plant species diversity per hectare when compared to other rainforests [
6]. As a result of its isolated nature and small extent, fauna endemic to the BNR is believed to be at higher risk [
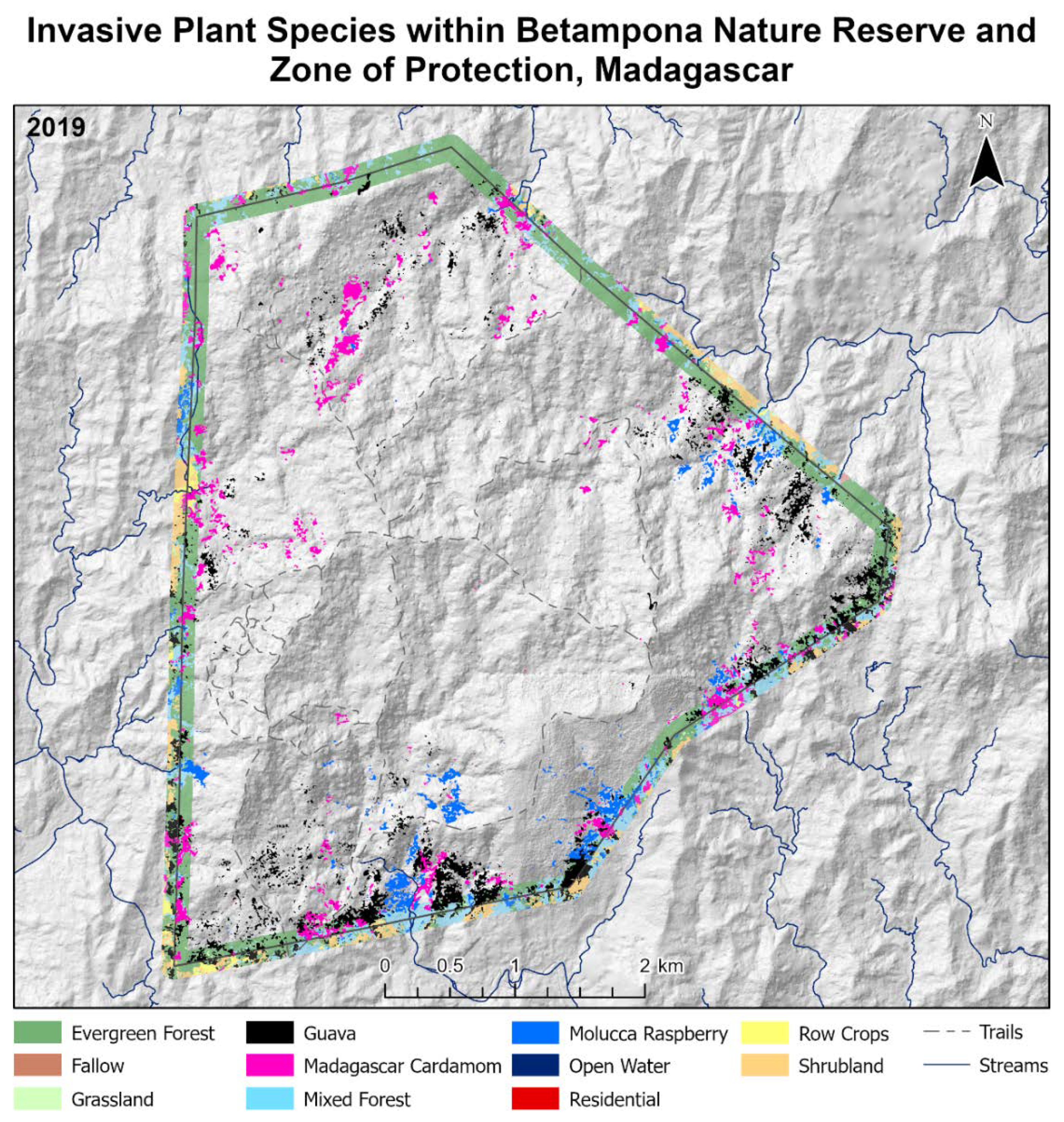
8]. Additionally, non-native invasive plant or animal species also threaten the biodiversity within the BNR [
9]. Three habitat-altering invasive plant species seen prominently within the BNR are Molucca Raspberry (
Rubus moluccanus), Madagascar Cardamom (
Afromomum angustifolium), and Strawberry Guava (
Psidium cattleianum) [
10]. Although Madagascar Cardamom is native to Madagascar, its weedlike characteristics in degraded areas within and surrounding the BNR make it invasive in the region [
11].
To effectively conserve biodiversity, an understanding of the human–environmental interactions is needed, since basic human needs have a direct impact on various conservation efforts through unsustainable agricultural practices, human encroachment, and bushmeat consumption [
4,
5]. Agriculture is the primary source of income for the locals around the BNR, with ‘tavy’, a cultural slash-and-burn system, being practiced by 80–85% of farmers [
5,
12]. To ensure a source of income, locals around the BNR amplify and expand agricultural production even if they are unable to sell their products to local markets [
12]. The poorest households around the BNR spend two-thirds of their income on food [
13] with many households experiencing chronic or temporary food insecurity, which is intensified by climate change-related events such as increased frequency of cyclones, causing flooding, landslides, and damage from powerful winds, accidents, illness, deaths, and animal and crop disease [
12].
These human–environmental interactions are often studied by mapping the landscape with different land cover and land uses using Remote Sensing. In recent studies, remote sensing has been used to study the BNR and surrounding ecosystems [
10,
14] and the entire island of Madagascar [
4], producing detailed Land Use or Land Cover maps through various methods [
15]. Freely available imagery data from Landsat missions is a major data source for both large- and small-scale classification maps [
15]. However, coarse resolution (10–100 m) images are not spatially detailed enough for forest conservation mapping projects [
16] or the identification of individual tree species [
17]. WorldView-3 (WV-3) has a high spatial resolution with a resolution of 1.2 m for visible bands (VNIR), 3.7 m for shortwave infrared (SWIR) bands, and 0.3 m in the panchromatic band. With the increased spatial resolution, pixel-based models that use only spectral information within each pixel for classification are not as effective as the within-class variation is much greater than the between-class variation. Classification accuracy is reduced, and maps derived from pixel-based methods show a salt-and-pepper effect when created based on spectral information alone using high-resolution imagery.
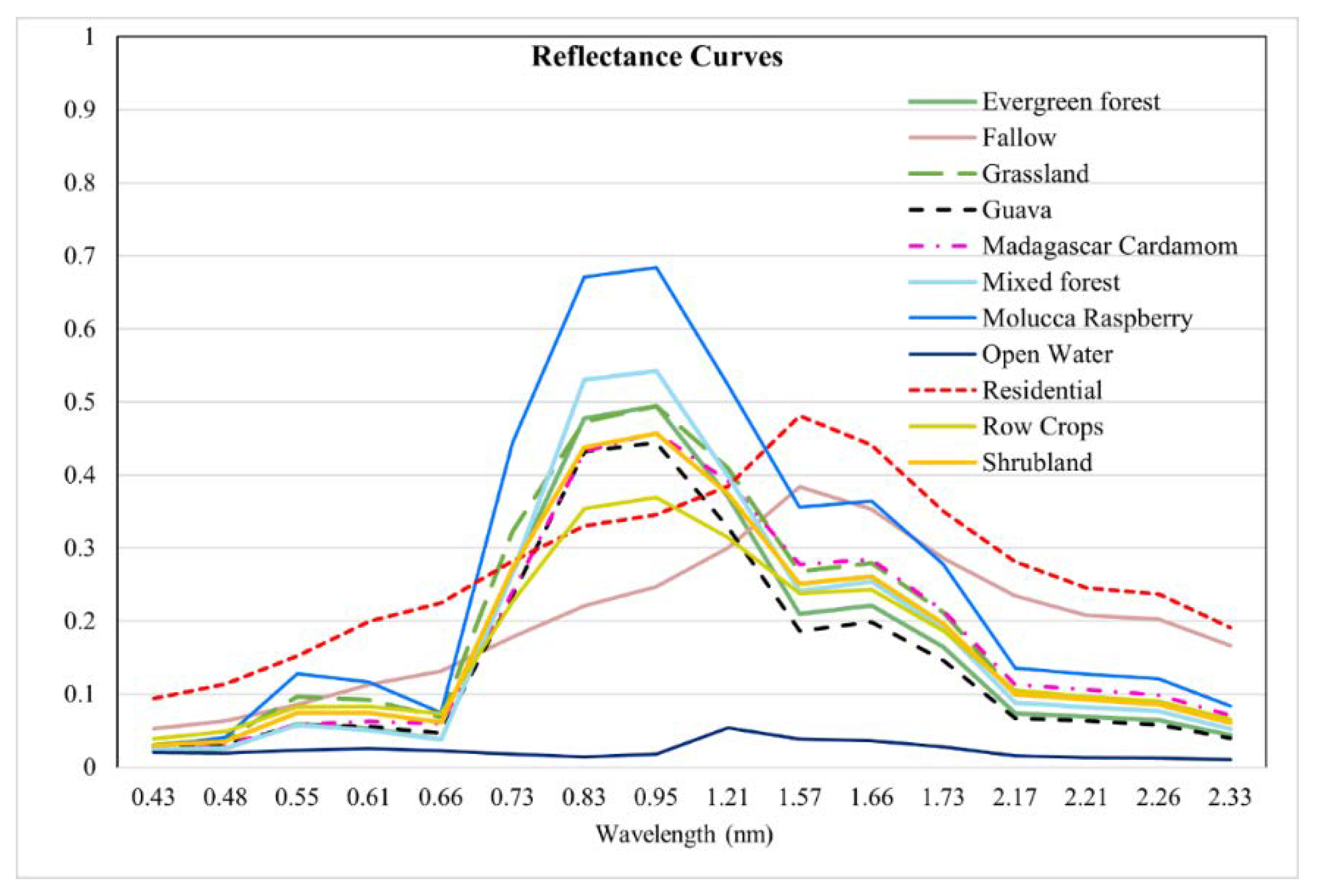
While the spatial and texture dimension of the remotely sensed imagery provides information regarding feature extent, the spectral data provide information that can be used to differentiate between classes [
15]. The shortwave infrared (SWIR) bands within the WV-3 satellite provide added values for improved vegetation mapping. The SWIR bands can detect non-pigment biochemical constituents of plants including water, cellulose, and lignin [
18]. SWIR-4 (1729.5 nm), SWIR-5 (2163.7 nm), and SWIR-8 (2329.2 nm) can detect characteristic absorption features of nitrogen, cellulose, and lignin, respectively [
18]. The inclusion of SWIR bands for tree-species classification has been shown to increase the accuracy to varying degrees [
18,
19].
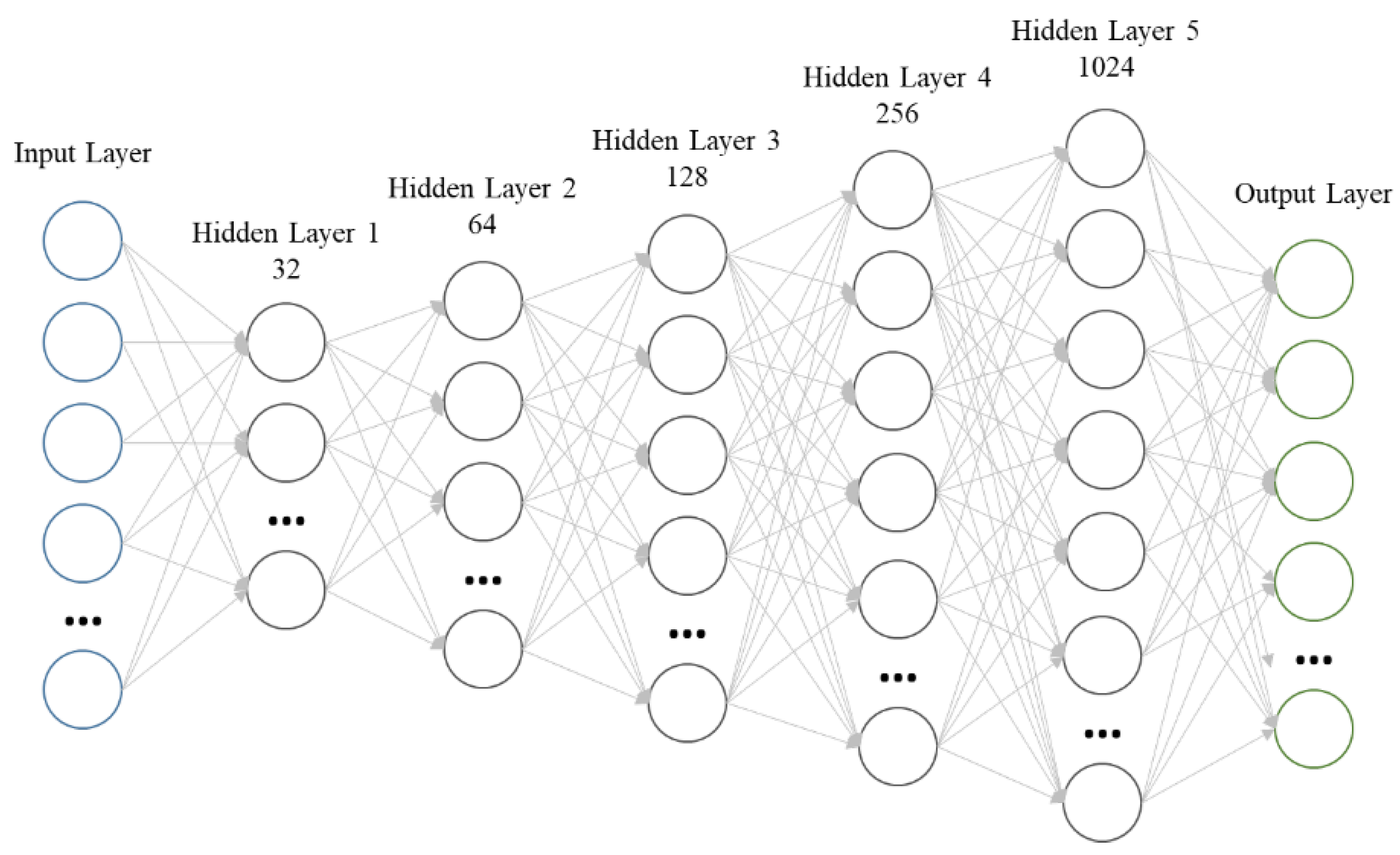
Support Vector Machines (SVM), Decision Trees (DT), Random Forests (RF), and Deep Neural Networks (DNN) have been used for image classification [
15]. However, these methods are dependent on handcrafted features and require domain expertise [
20]. Therefore, a shift is seen toward the use of deep learning algorithms for the creation of classification maps with Convolutional Neural Networks (CNNs) dominating because of their ability to extract relevant information automatically from imagery [
20]. CNNs are a type of Deep Neural Network model architecture that can extract various features automatically and out-perform pixel-based machine learning models [
19,
20,
21]. “Automated learning” here is the process in which a computer program is trained to learn and apply translationally invariant and spatially hierarchical patterns within the image through convolutional and pooling layers [
20]. Well-known deep CNN architectures used for image classification problems include AlexNet [
22], VGG Networks [
23], ResNet [
24], DenseNet [
25], among others. However, the fully connected layers at the end of any CNN architecture do not preserve the 2D input image and predict a single class for each input image. For mapping purposes, a per-pixel prediction of the 2D input image is required. Thus, new methods were developed, such as Fully Convolutional Networks (FCNNs) [
26] for image classification through pixel-wise semantic segmentation. Ref [
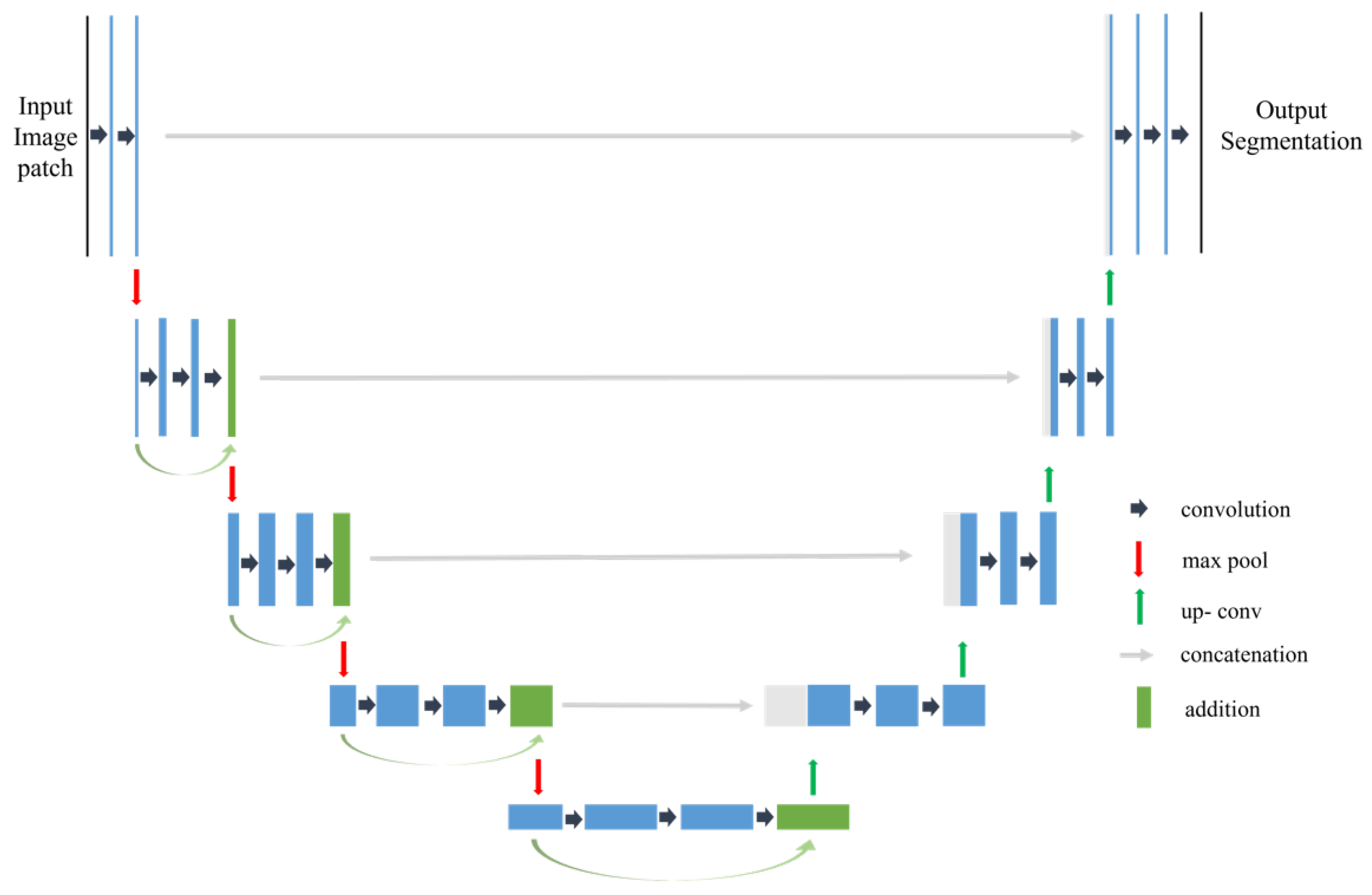
26] defined an architecture that replaces the dense fully connected layers with a deconvolutional layer. This FCNN architecture can create pixel-based classifications or semantic segmentation of the input image. Segmentation and classification of the input image occurs through the FCNN architecture, resulting in an end-to-end classification map. The input to the model is a 2D image, which results in a classification based on both spectral and textural features. SegNet and U-Net are types of FCNN for per-pixel semantic segmentation of input images [
27,
28].
FCNNs and their variants have been used for end-to-end per-pixel image classification of high-resolution satellite images [
29,
30,
31,
32,
33]. The number of classification classes varies with a maximum of 13 classes seen in [
30]. For studies with more than 10 classes, high-resolution GaoFen-1, GaoFen-2, IKONOS, WV-2, and Quickbird satellite imagery were classified [
29,
30,
32]. However, these studies that use fully convolutional networks focus on the classification of land covers for an urban landscape [
29,
32] and agricultural landscapes [
30] with a few focusing on ecological landscapes [
34].The importance of high spatial resolution (3.7 m) SWIR bands for image classification, especially forest cover classification, has been unknown because 3.7 m SWIR bands have not been previously available for commercial or academic research. To the authors’ best knowledge, FCNNs, that incorporate spatial features, have not been utilized for tropical forest cover mapping using raw WV-3 satellite imagery.
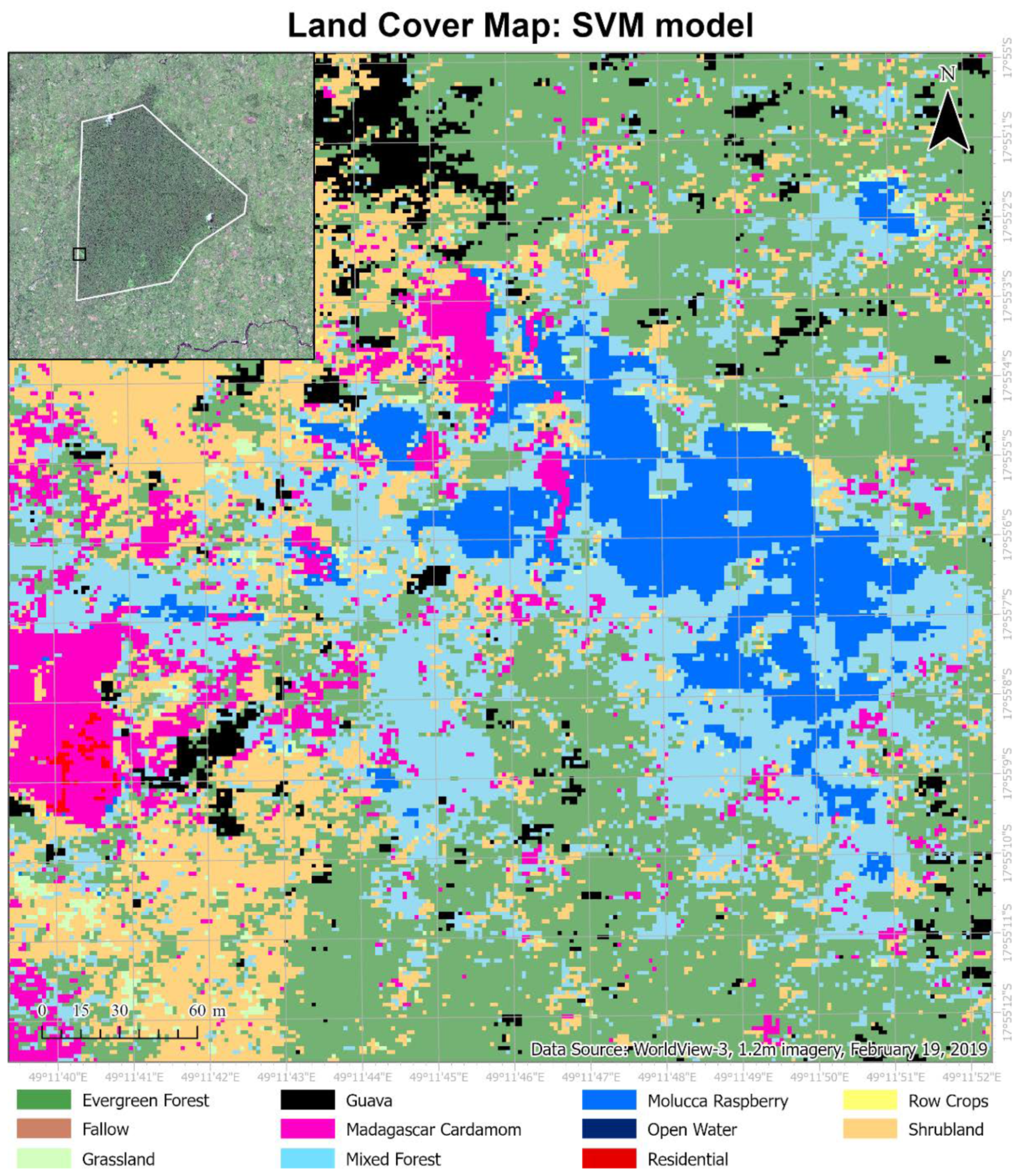
While considering the size of the BNR and more importantly, the rugged terrain, remotely sensed data is a top contender to map forest cover. It is hypothesized that (1) the end-to-end FCNN model (U-Net) will outperform other pixel-based methods (SVM, RF, DNN) due to the inclusion of textural features and end-to-end mapping capability, (2) the exclusion of SWIR bands would reduce the overall accuracy for all classification methods, with a greater reduction for SVM and RF, and (3) based on trends seen by [
14], a negative human influence on the BNR and surrounding habitat is expected. The objectives are to (1) to develop an end-to-end automated deep learning model for forest cover mapping using WV-3 imagery, (2) to evaluate the influence of WV-3 SWIR bands for tropical forest cover mapping, and to evaluate the impact of conservation efforts in the region by international organizations by analyzing land cover change from 2010 to 2019.
5. Conclusions
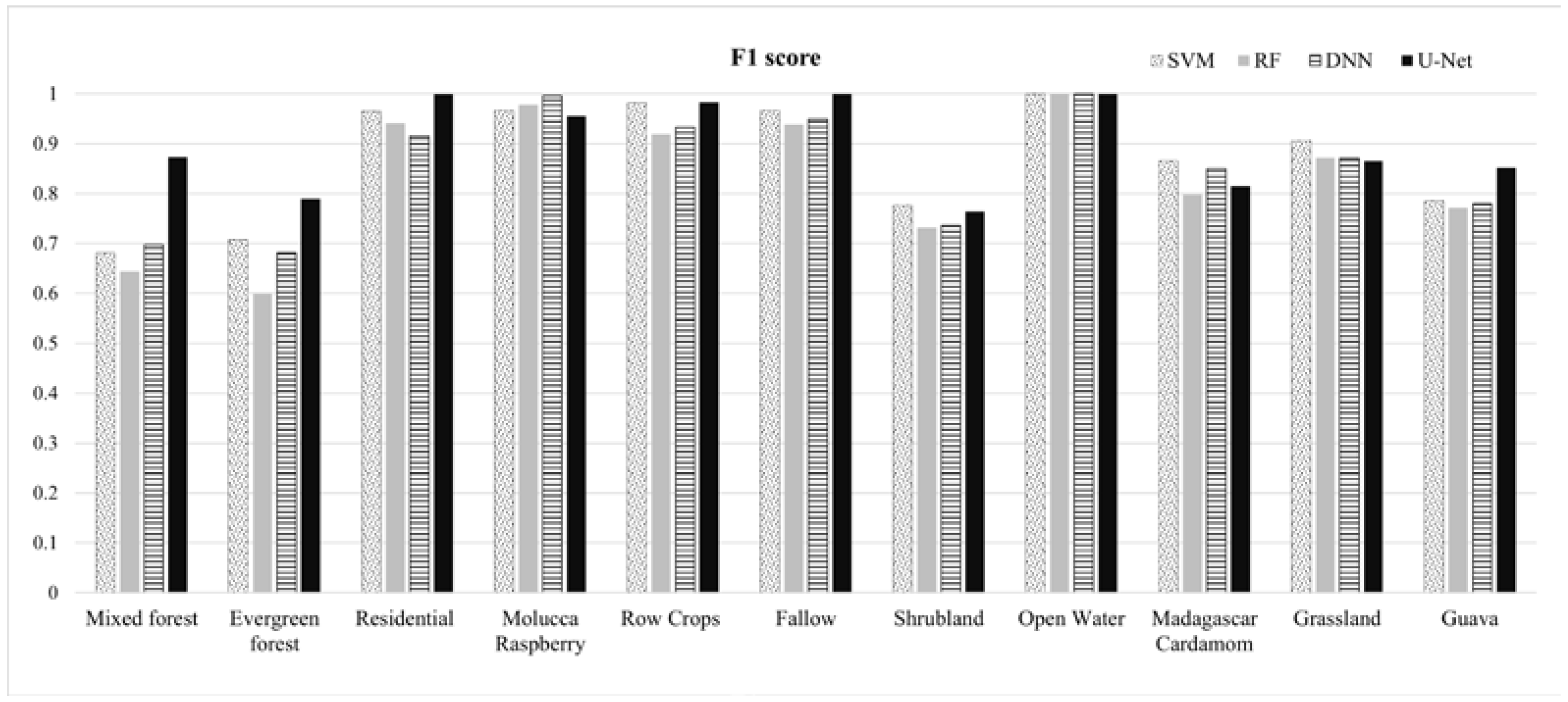
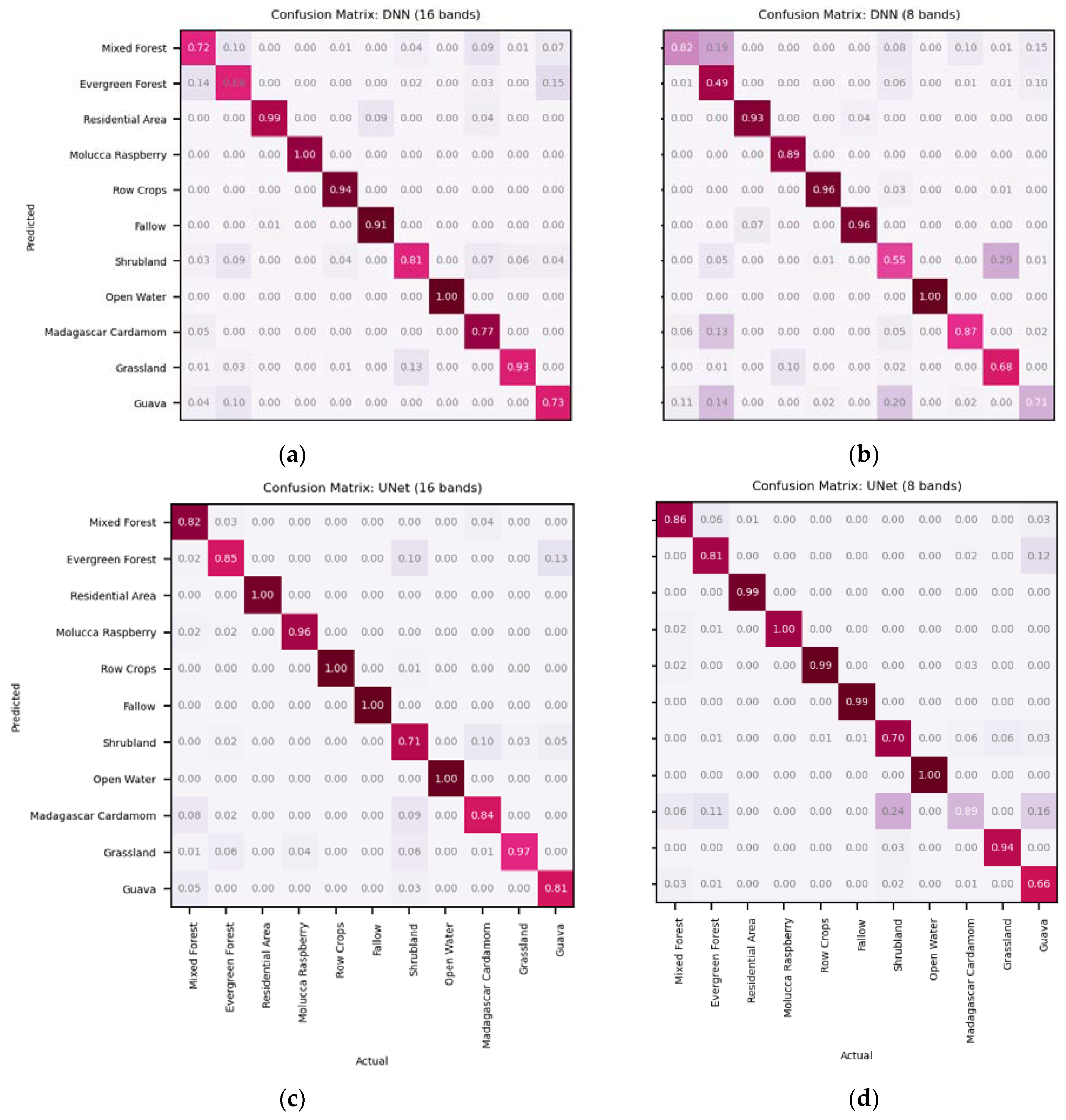
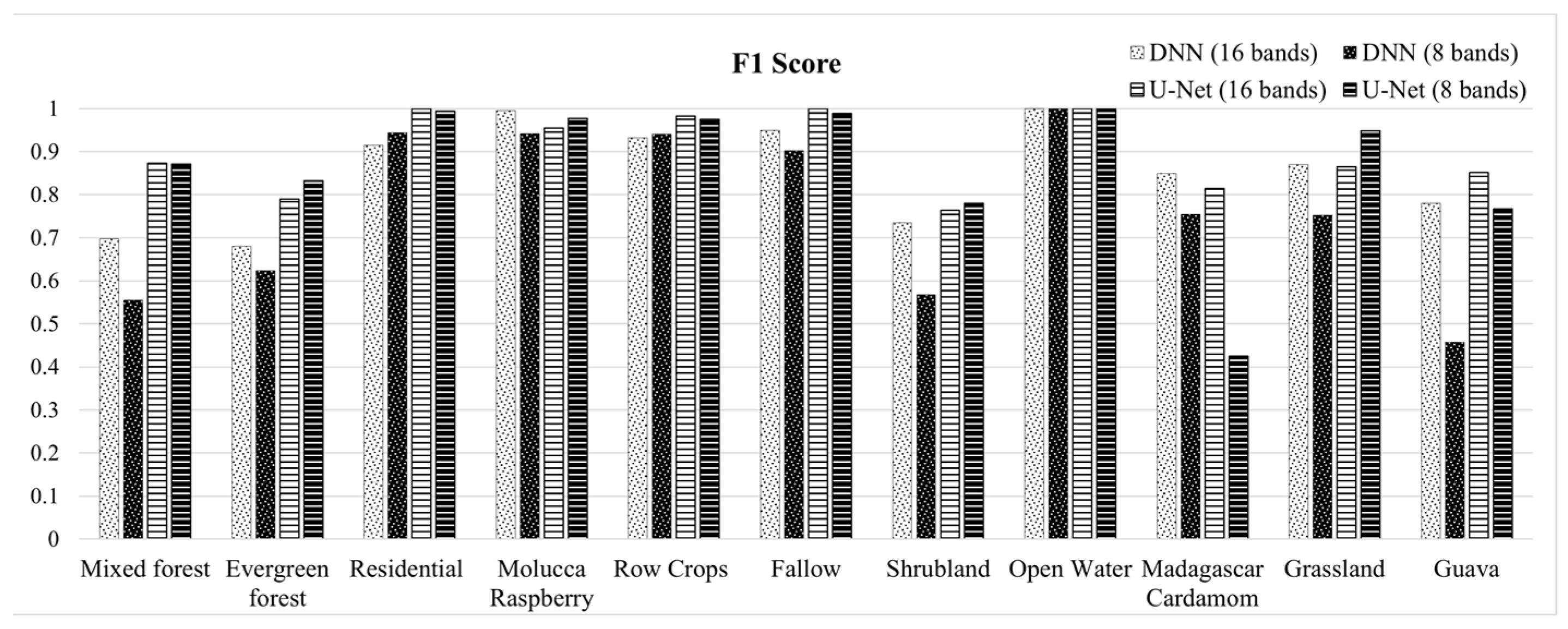
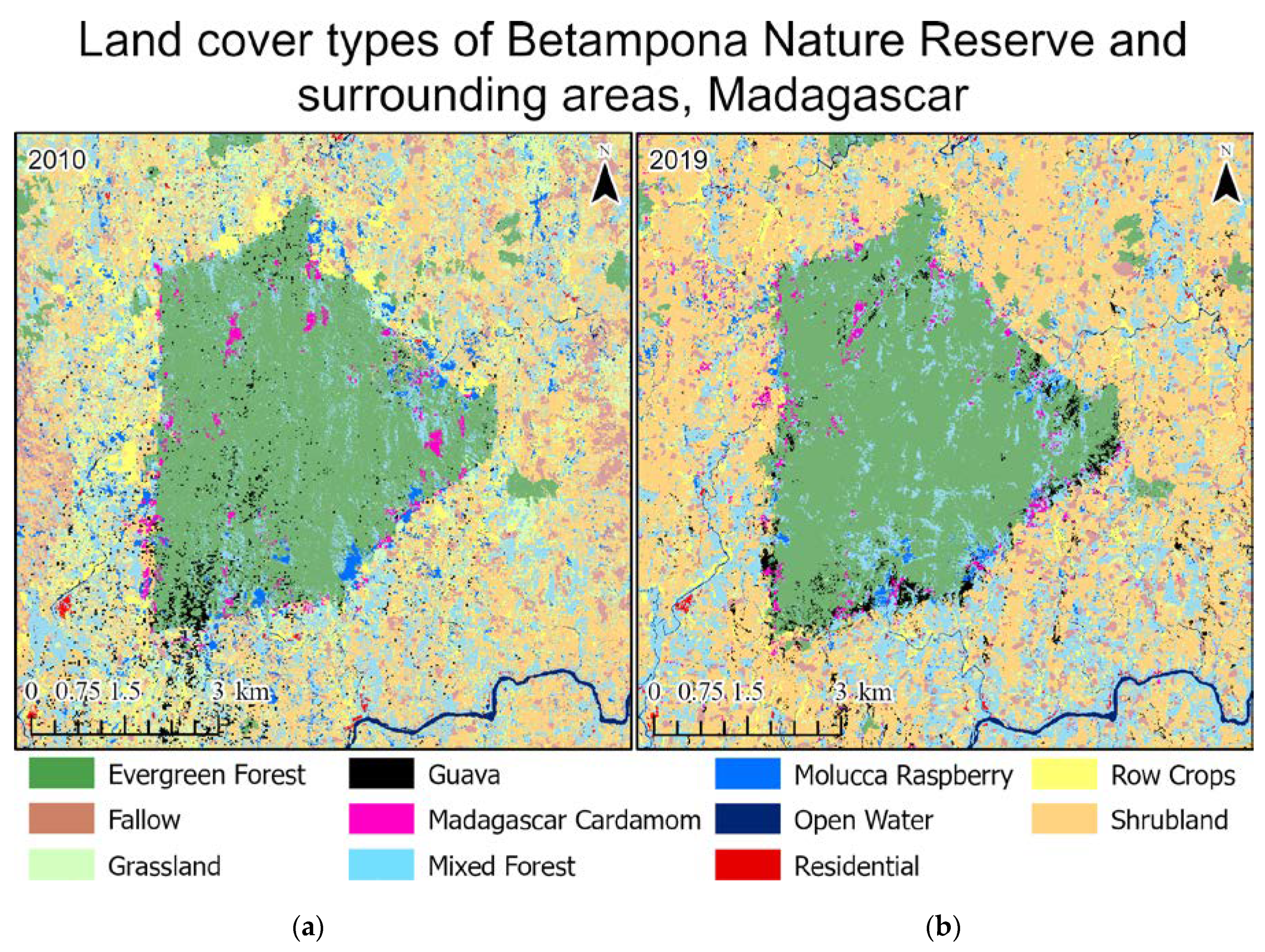
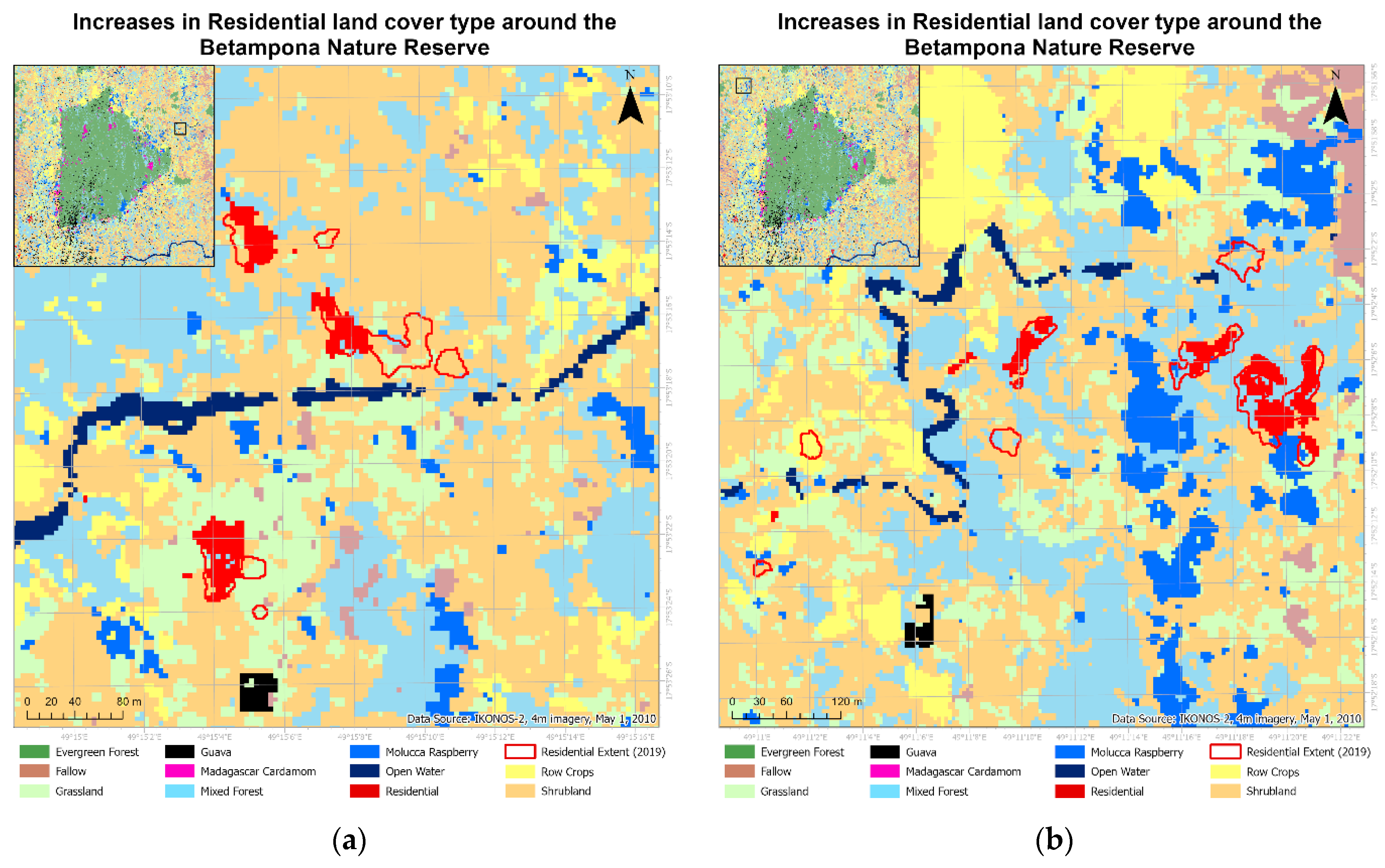
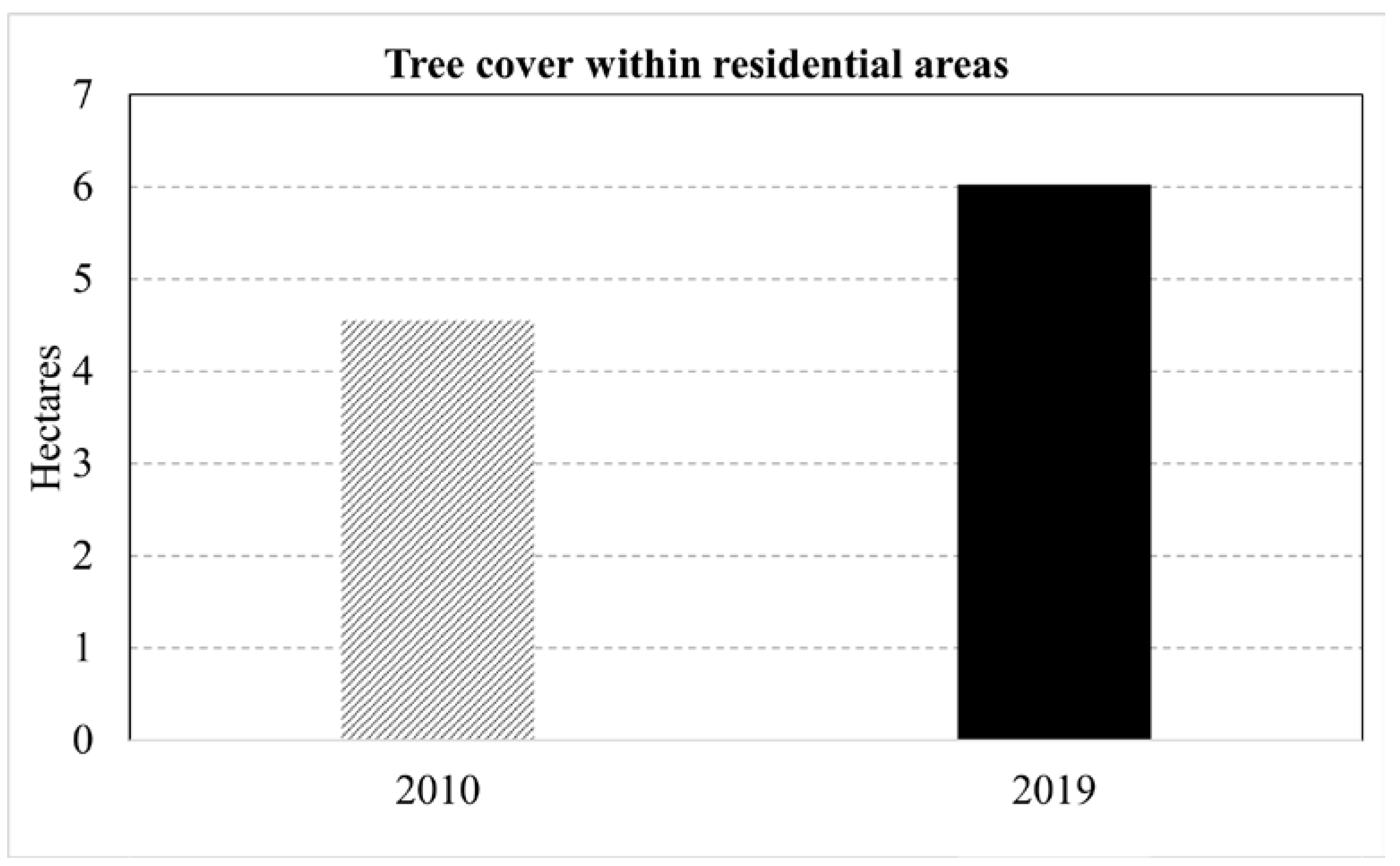
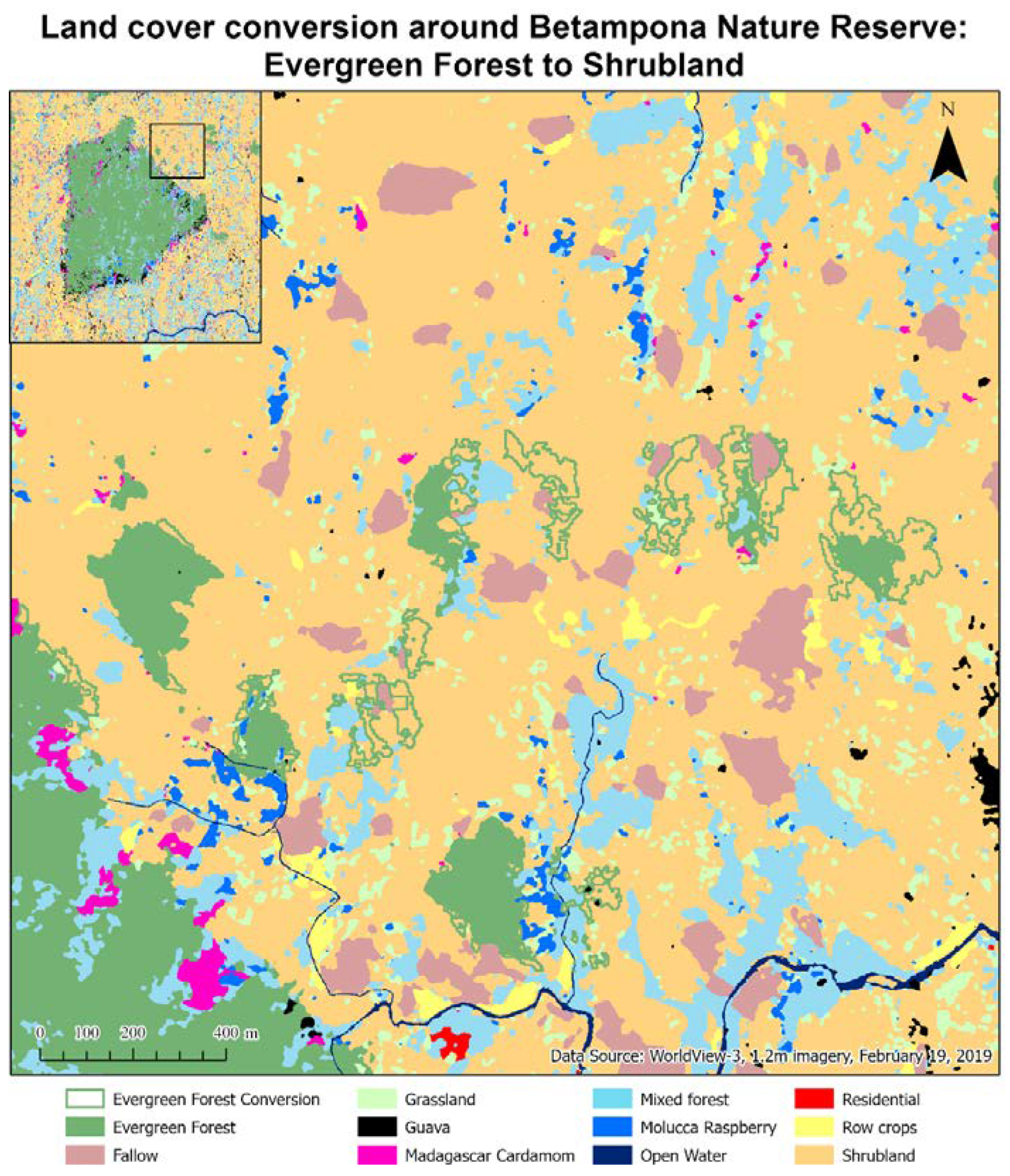
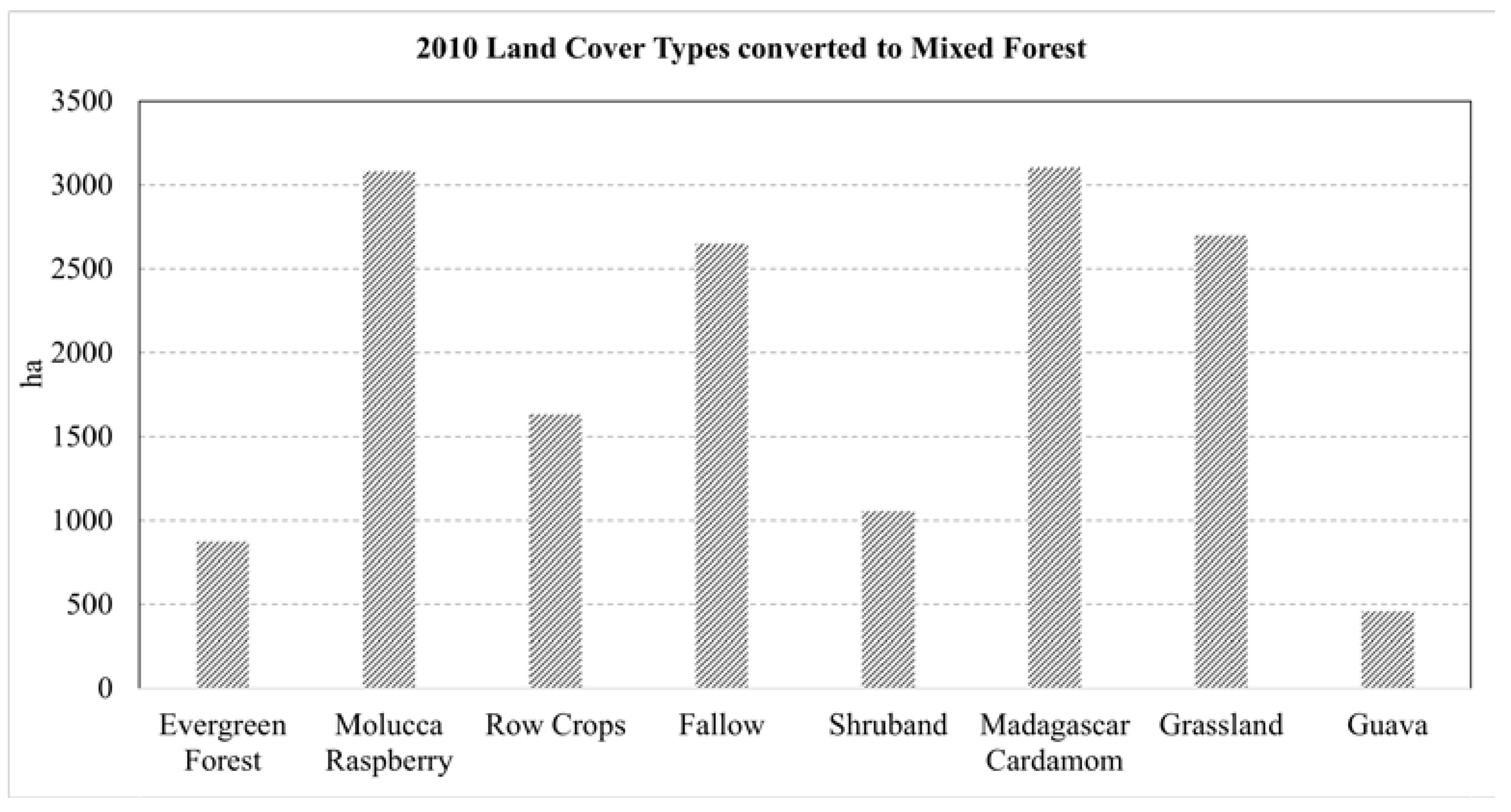
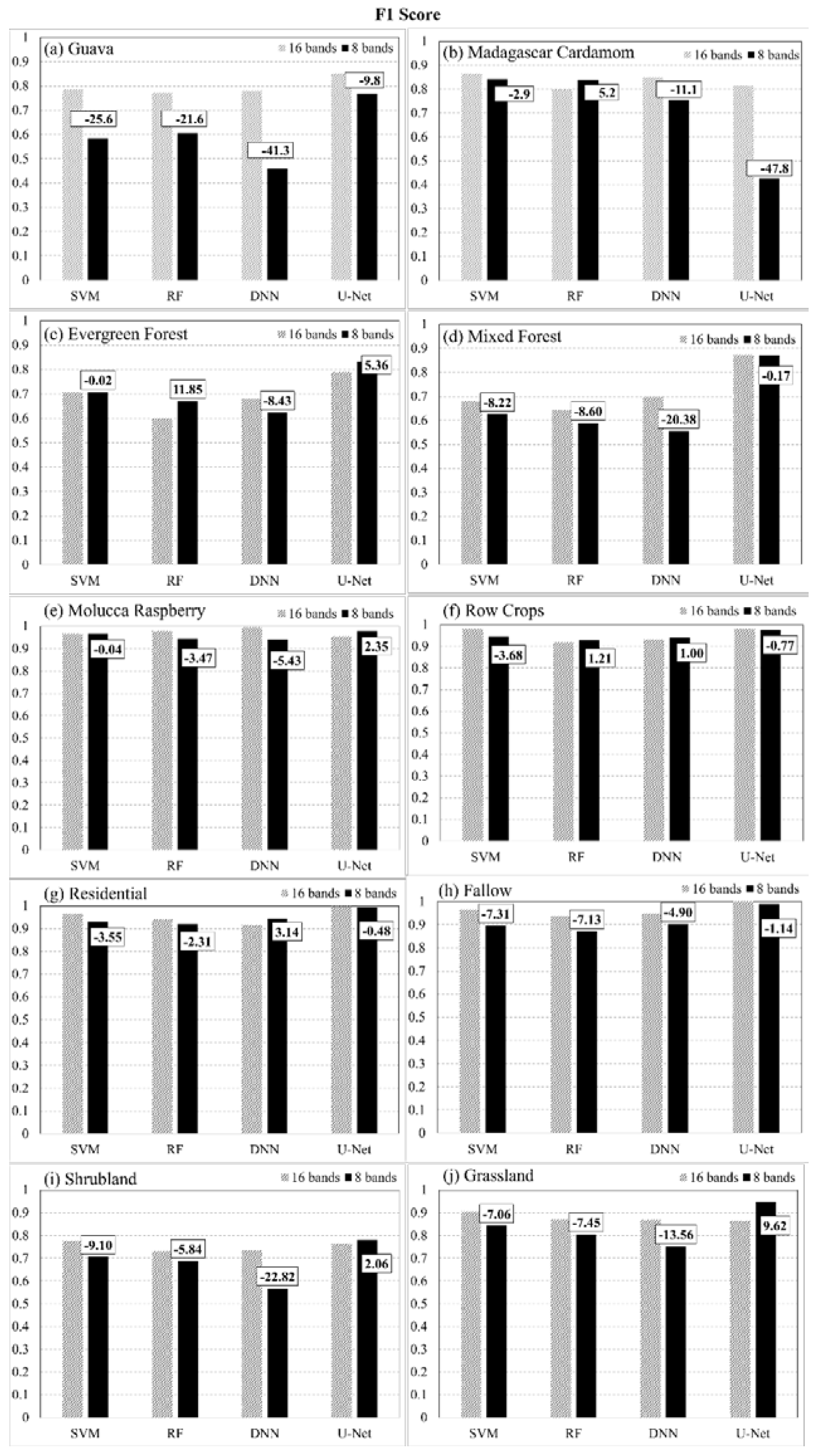
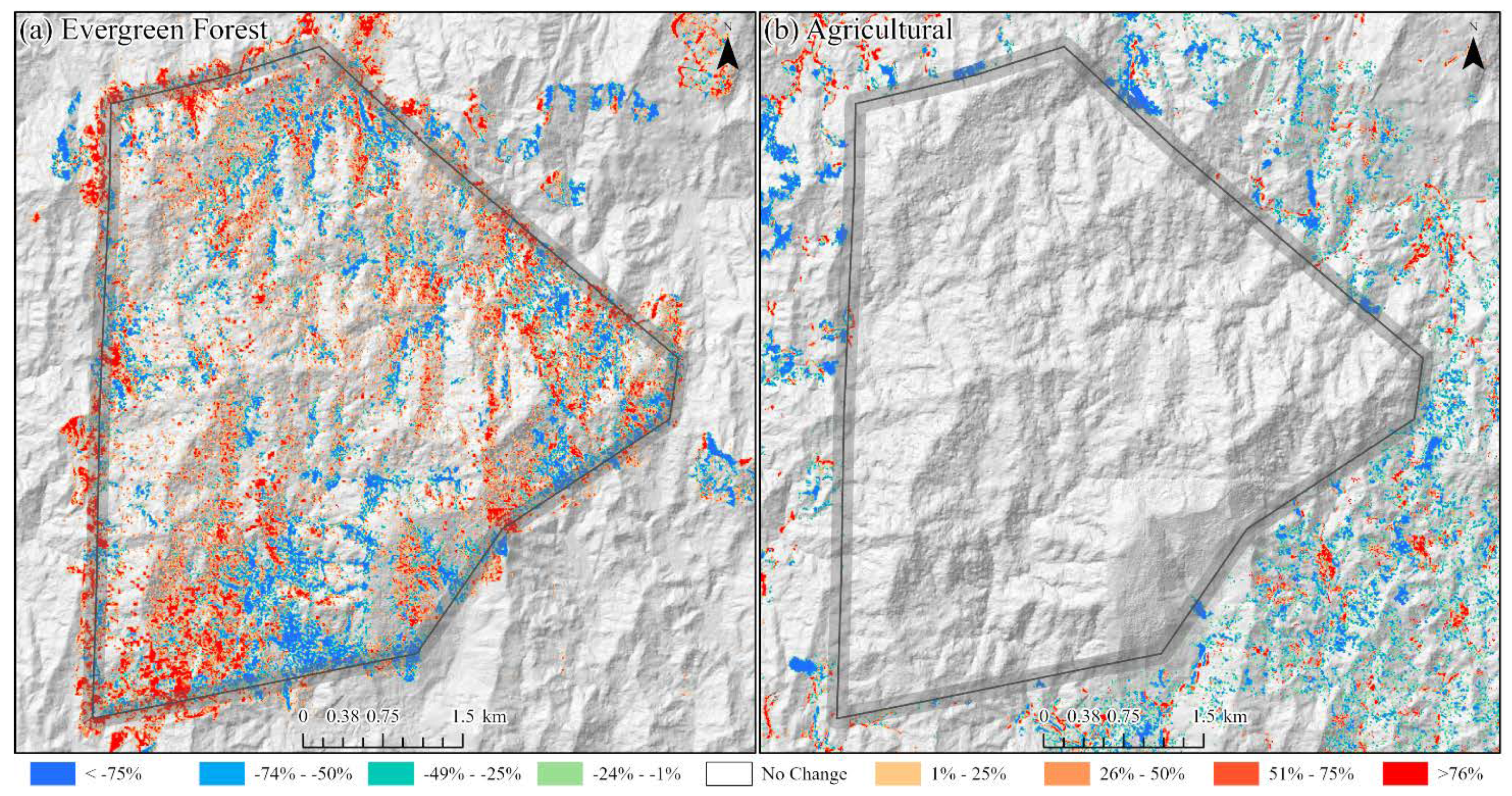
This paper demonstrated that FCNN models that utilize texture information in a fully automated, end-to-end mapping outperformed pixel-based conventional machine learning approaches in mapping tropical rainforest habitats. Our results showed that the FCNN model (U-Net) produced an overall accuracy of 90.9% compared to accuracies of 88.6%, 84.8%, and 86.6% for SVM, RF, and DNN models respectively. Moreover, the WV-3 SWIR bands increased the accuracy by 1.87% for the U-Net model. The decrease in accuracy without SWIR bands was much greater for DNN (8.55%), SVM (5.42%) and RF (3.18%) models that use only spectral features for classification, demonstrating the potential of the FCNN approach without using expansive SWIR data collections. Comparing LCLU maps generated in 2010 and 2019, changes in classes were quantified to understand human interactions with the environment and quantify the impact of conservation efforts. A 44% increase in Residential is seen in the study area, and a decrease in Row Crops in the ZOP. Within the BNR, invasive plant species are seen to decrease, which is complemented by increases in Mixed (12%) and Evergreen Forest (0.7%). This increase in forest cover indicates a reversal of the trends described by Ghulam et al. (2014). In addition, a 32% increase is seen in the tree cover within residential areas either due to native tree maturing or successful agroforestry efforts by the MFG. Conservation efforts worldwide should consider the needs of the local human population. As such, future conservation efforts by the MFG include improving human health and recognizing the importance of people-oriented strategies in order to provide cash crops and a sustainable food source for the locals.
Machine/deep learning has been far advanced in some application domains; however, challenges exist for remote sensing applications especially for tropical forest monitoring due to in part the lack of sufficient training data representing various biomes and forest types globally that must be manually created and in part the challenges of managing and analyzing big satellite data at the global scale. We presented a fully automated deep learning model in this paper which can be applied to any part of the world. Taking advantage of cloud computing, the approach can be duplicated for national-scale forest cover mapping and change detection, which has significant impact on our understanding of tropical forests and biodiversity for better implementation of REDD+.