Abstract
Sodic soils adversely affect crop production over extensive areas of rain-fed cropping worldwide, with particularly large areas in Australia. Crop phenotyping may assist in identifying cultivars tolerant to soil sodicity. However, studies to identify the most appropriate traits and reliable tools to assist crop phenotyping on sodic soil are limited. Hence, this study evaluated the ability of multispectral, hyperspectral, 3D point cloud, and machine learning techniques to improve estimation of biomass and grain yield of wheat genotypes grown on a moderately sodic (MS) and highly sodic (HS) soil sites in northeastern Australia. While a number of studies have reported using different remote sensing approaches and crop traits to quantify crop growth, stress, and yield variation, studies are limited using the combination of these techniques including machine learning to improve estimation of genotypic biomass and yield, especially in constrained sodic soil environments. At close to flowering, unmanned aerial vehicle (UAV) and ground-based proximal sensing was used to obtain remote and/or proximal sensing data, while biomass yield and crop heights were also manually measured in the field. Grain yield was machine-harvested at maturity. UAV remote and/or proximal sensing-derived spectral vegetation indices (VIs), such as normalized difference vegetation index, optimized soil adjusted vegetation index, and enhanced vegetation index and crop height were closely corresponded to wheat genotypic biomass and grain yields. UAV multispectral VIs more closely associated with biomass and grain yields compared to proximal sensing data. The red-green-blue (RGB) 3D point cloud technique was effective in determining crop height, which was slightly better correlated with genotypic biomass and grain yield than ground-measured crop height data. These remote sensing-derived crop traits (VIs and crop height) and wheat biomass and grain yields were further simulated using machine learning algorithms (multitarget linear regression, support vector machine regression, Gaussian process regression, and artificial neural network) with different kernels to improve estimation of biomass and grain yield. The artificial neural network predicted biomass yield (R2 = 0.89; RMSE = 34.8 g/m2 for the MS and R2 = 0.82; RMSE = 26.4 g/m2 for the HS site) and grain yield (R2 = 0.88; RMSE = 11.8 g/m2 for the MS and R2 = 0.74; RMSE = 16.1 g/m2 for the HS site) with slightly less error than the others. Wheat genotypes Mitch, Corack, Mace, Trojan, Lancer, and Bremer were identified as more tolerant to sodic soil constraints than Emu Rock, Janz, Flanker, and Gladius. The study improves our ability to select appropriate traits and techniques in accurate estimation of wheat genotypic biomass and grain yields on sodic soils. This will also assist farmers in identifying cultivars tolerant to sodic soil constraints.
1. Introduction
Sodic soils occupy 581 million hectares globally, representing one of the significant constraints to agricultural production [1,2]. Worldwide, Australia has the widest cover of sodic soils (340 million hectares) [2]. Wheat is an important rain-fed crop grown in northeastern Australia, and sodicity poses a major threat to its production as it limits plant available water in the root zone [3,4,5]. To increase farm productivity and profitability, one important strategy is to identify wheat genotypes with better tolerance to sodic soil conditions. To do this, it is of importance to be able to evaluate and compare the performance of genotypes in terms of their growth and yield response in sodic soil [6].
Crop phenotyping, i.e., the evaluation and quantitative measurement of complex traits based on different characteristics over different growth stages [7], is widely used to identify adaptive traits and quantify plant-soil-environment interactions [8,9]. Phenotyping has traditionally relied on the collection of information about crop growth (e.g., biomass, leaf appearance, crop height, crop nutrient content) over the growing season using manual field sampling methods. However, manual methods are often time-consuming, labor-intensive, and potentially inaccurate [10,11]. The use of unmanned aerial vehicles fitted with a variety of powerful and high-resolution sensors has provided an alternative method of data collection that is inexpensive, as well as time- and labor-efficient, and which can provide accurate soil-crop information in real time at a field to regional scale [12,13,14]. However, limited studies have been reported using these approaches for crop phenotyping in constrained environments, particularly on sodic soils. These sensor-based approaches may be useful for phenotyping crops/cultivars grown on sodic soils where spatial variability and incomplete canopy cover can be major challenges to representative sampling. Although a recent study reported the potential of a high-resolution UAV-thermal imaging sensor to evaluate physiological performance, water status, and growth of wheat cultivars on sodic soils [15], greater research is required using different cropping traits and multiple sensor-based approaches for phenotyping on sodic soils that can improve estimation of crop growth and yield for adaptation of wheat genotypes in sodic soil environments.
Remote sensing spectral information and traits obtained from visible and near-infrared (VNIR) wavelengths can be used to determine crop health, vigor, moisture content, and growth [16]. With recent advances in the airborne LIDAR and/or UAV red-green-blue (RGB) sensor-based 3D point cloud techniques in high-throughput phenotyping (HTP), it is also possible to obtain high-resolution crop architectural traits (height, volume, and canopy cover) [17,18]. These architectural traits have been reported as important in influencing aboveground biomass and/or grain yield variation [19,20,21]. In addition, a wide range of proximal and handheld instruments, such as EMI (Geonics EM38®, Genonics LTD, Canada) [22,23], GreenSeeker® handheld sensor [24], and ASD FieldSpec® range [25,26], are now available to provide sound, detailed information on soil and crop characteristics at close distances. Hyperspectral sensors with hundreds of spectral bands have demonstrated potential for assessing and predicting crop performance and yield [26]. Researchers have suggested that using combined trait information derived from a number of sensors can be advantageous to quantify complex crop information on non-sodic soil [27,28]. However, the effectiveness of these remote sensing techniques has only been reported in the absence of soil constraints. These combined traits information may, thus, also be useful for assessing the performance of crops growing in sodic soils. However, there is currently little published information available to identify appropriate physiological traits that can be used to evaluate and forecast the relative performance of crops and/or cultivars using these proximal and handheld instruments with that of UAVs on sodic soils. Hence, the identification of robust techniques is required for phenotyping on sodic soils.
A wide range of vegetation indices (VIs), such as normalized difference vegetation index (NDVI), enhanced vegetation index (EVI), and optimized soil adjusted vegetation index (OSAVI), can be computed from multispectral and/or hyperspectral data collected using UAVs or proximal sensing. These VIs provide information about crop health, greenness, and vigor [29,30,31]. The NDVI is one of the most well-known VIs for monitoring crop growth and predicting grain yield [32,33,34,35,36]. However, sparse vegetation or soils that generate high reflectance (especially in dryland agriculture) can adversely affect the reliability of NDVI. Rondeaux et al. [37] reported that OSAVI was an improved model of NDVI that could measure canopy reflectance by using 0.16 as an optimal value for reducing the variation of canopy reflectance in the presence of soil background conditions. In contrast, Gao et al. [38] reported that EVI can be more responsive to canopy cover, structural variation, and architecture in comparison to NDVI, especially at a regional scale [39]. Huete et al. [40] also suggested that EVI decouples the background effects from the canopy and is less sensitive to atmospheric influence. Furthermore, NDVI and EVI together can be used to detect temporal vegetation cover and provide insights into canopy biophysical information. While a number of different VIs exist to predict crop growth and yield, it is often difficult to determine which is the most useful for a specific study, and the relative performance of VIs can vary depending on the site-specific soil and environmental conditions. There is also limited information on the usefulness of VIs in crop growth and yield estimation, particularly on sodic soils. Hence, a comprehensive assessment of these VIs derived from a different remote and/or proximal sensors is required to select appropriate traits for reliable crop growth and yield forecasting on sodic soils.
An accurate and reliable seasonal crop growth and yield forecast is essential for agriculture risk assessment and improving productivity. A number of multivariate predictive machine learning (ML) algorithms can be used in agricultural yield forecasting and modeling [41,42,43,44]. ML has been proven to improve decision making and provide reliable outcomes that are consistent and robust [45,46,47]. Although a number of studies have reported the usefulness of various ML algorithms, including support vector machine (SVM), artificial neural network (ANN), classification and regression tree (CRT), Gaussian process regression (GPR), and multilinear regression (MLR), for agricultural yield estimation at various spatial extensions (Table 1), the authors are currently unaware of any published assessment on the usefulness of these integrated ML and optical remote sensing-based approaches for crop growth and yield estimations, especially in constrained sodic soils. Hence, a thorough evaluation of these ML approaches is required to determine its utility for crop growth and yield prediction on sodic soils.

Table 1.
Findings from the previous literatures.
Accurate crop growth and yield prediction can be reliant on the nature and importance of input cropping traits and data variability. It can also be specific to different environments and sites. Hence, a thorough evaluation of these approaches is necessary for adaptation in sodic soils. The present study proposes an integrated optical remote sensing and ML-based framework to quantify wheat genotypic traits to assist estimation of growth and yield on sodic soils. The research objectives are to (1) assess the utility of a variety of remote sensing techniques and traits to determine crop growth and yield of wheat genotypes on sodic soils, (2) examine the abilities of different ML algorithms coupled with remote sensing-derived biophysical traits to improve prediction of wheat biomass and grain yields, and (3) quantify the impacts of sodic soils on wheat crop growth and yield of to identify tolerant genotypes on sodic soils. The present study tests if integrated optical remote sensing and ML-based techniques can accurately forecast yield and help to distinguish tolerant genotypes on sodic soil that can assist agronomists/researchers to select appropriate techniques/traits for phenotyping on sodic soils by reducing the need for extensive, labor-intensive, and manual methods of phenotyping to examine the traits and crop growth. In addition, the study may also guide decisions for farmers to select genotypes tolerant to sodic soil constraints to maximize productivity in sodic soil environments.
2. Materials and Methods
2.1. Site Selection and Soil Sampling
The experiment was performed at two rain-fed sites, one moderately sodic (MS) (28.15°S and 150.22°E) and the other highly sodic (HS) (28.08°S and 150.15°E), near Goondiwindi in northeastern Australia. Both sites have well-structured gray vertisols with high clay content and are located at an average elevation of 268 m above mean sea level. During the normal crop growing season from May to October, air temperature varied between ~5 and ~35 °C for both sites with seasonal means of 14.7 °C for the MS and 14.9 °C for the HS site [15,54]. Mean relative humidity was 54.6% for the MS and 54.3% for the HS site, and total in-season crop rainfall was 86.0 mm for the MS and 85.6 mm for the HS site (Figure 1) [15].
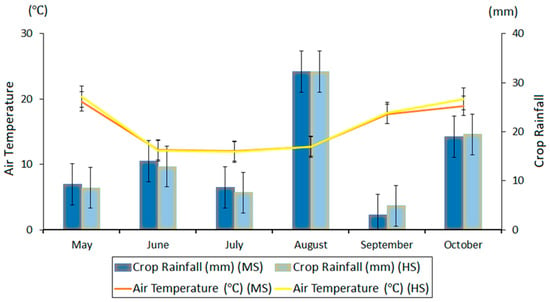
Figure 1.
In-season crop rainfall and air temperature (monthly mean) of the moderately sodic (MS) and highly sodic (HS) sites in the wheat-growing season of 2018 [15]. The error bars represent the standard error of the monthly mean values.
At each site, soil samples were collected from a minimum of eight points using a hydraulic sampling rig to take 50 mm diameter soil cores to a depth of 150 cm. The soil samples were dried at 40 °C and ground to <2 mm. In a 1:5 soil water suspension [55], pH, EC, and chloride (Cl) were measured; the EC of saturated extract (ECse) was then computed from EC 1:5, and the clay content was determined using the pipette method [56,57]. Exchangeable Na and the cation exchange capacity (CEC) were measured using a 1 M NH4Cl (pH 8.5) extraction solution [58], and ESP was calculated from exchangeable Na relative to CEC. The volumetric moisture content as a percentage was determined after drying samples at 105 °C to obtain the gravimetric soil moisture content before multiplying this by the soil bulk density (BD) [15]. The soil physicochemical properties of both sites are presented in Figure 2.
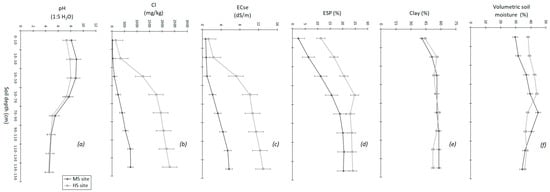
Figure 2.
Soil physicochemical properties including pH (a), Cl concentration (b), electrical conductivity (c), exchangeable sodium percentage (d), clay content (e), and initial volumetric soil moisture prior to sowing (f) for the moderately sodic and highly sodic sites at 0–150 cm soil depth. The error bars represent the standard error of the mean from eight sampling points at each site [15,59].
2.2. Experimental Design and Crop Biophysical Measurements
We used a randomized complete block design (RCBD) for the experiment with eight replications [15,54,59]. Four of the replications (72 plots) were used for destructive plant sampling, crop height, and biophysical measurements close to flowering stage, 110–112 days after sowing (DAS), and another four replications (72 plots) were used for grain yield measurements at maturity (152 DAS). Eighteen contrasting wheat genotypes were tested (Supplementary Table S1) [15,54,59], making a total of 144 plots (four columns and 36 rows) (Figure 3). Destructive sampling plots and grain harvest plots for each genotype in each replicate block were arranged in adjacent pairs. Each plot was 5 × 2 m at harvest and consisted of five planting rows with 30 cm spacing in between (Supplementary Figure S1) [15,54,59]. The crops were sown in late May and harvested in early November of 2018.
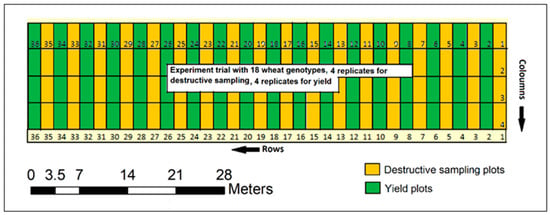
Figure 3.
Layout design of experimental trials. The destructive sampling plots (in odd numbers) marked by yellow color and yield plots (in even numbers) in green color show the alignment of each genotype side by side [15].
The aboveground biomass yield at both sites was harvested from a 3 × 0.5 m area from the middle three rows of each destructive sampling plots at 110–112 DAS (Supplementary Figure S1), stored in paper bags, and then dried at 70 °C for 72 h to determine plant dry weight (DW) [15,48]. In addition, crop heights were measured manually by placing a ruler vertically on the soil surface and measuring the height of plants from the middle three rows in each plot. Average plant height of a plot was then estimated using the mean of these measurements. Grain yield was measured from the yield plots by machine harvesting at crop maturity (152 DAS).
2.3. Remote Sensing Data Collection and Preprocessing
2.3.1. Proximal Sensing for Canopy Reflectance Measurements
We used a handheld GreenSeeker® 505 (Trimble Inc., Sunnyvale, CA, USA) active sensor to measure NDVI from tillering to crop maturity to acquire data on growth and greenness during these crop development stages. NDVI values range between 0 and +1. Higher values indicate greater canopy cover and/or greenness suggesting good canopy health. The sensor was conveyed at an average speed of 0.84 m/s and an elevation of 0.5 m from the top of the canopy from the middle three rows of each biomass sampling plot (Supplementary Figure S1). The measurements were taken continuously and at the same speed without stopping the sensor in between plots [15]. The field of view of the sensor was oval, and it changed with the elevation from the ground. Bare soil reflectance was measured separately from outside of the plot area and calibrated using a calibration panel to reduce the effect of soil reflectance on the canopy [15,59].
Canopy reflectance was also measured using a spectroradiometer (ASD FieldSpec® HandHeld 2, Malvern Panalytical Ltd., Malvern, UK) under cloud-free, wind-free, and sunny conditions between 11:00 a.m. and 3:00 p.m. Weekly in-situ monitoring of crop development was conducted, and a date near flowering (110–112 DAS) was selected for crop assessment. A date near flowering was chosen since this is the most crucial time before grain filling when the canopy is most fully developed and likely to modify the reflectance most efficiently [15]. The instrument was calibrated using a standard white spectralon calibration panel at each time of point shoot to reduce the effects of soil reflectance on canopy spectral measurements. The continuous spectra were recorded in Vis to NIR from 325 nm to 1075 nm with ±1 nm accuracy and a resolution of <3 nm for each spectrum. The field of view of the sensor was 25° and, in each plot, five spectral measurements of 0.2 m diameter area of the canopy were recorded from the middle three rows (Supplementary Figure S1) by locating the sensor in a vertical position 0.5 m above the canopy cover [26]. The measured canopy reflectance data were then exported into an ASCII file using ViewSpec Pro, an integrated package with RS3 spectral acquisition software (Analytical Spectral Devices Inc., Boulder, CO, USA) [60]. Subsequently, the raw data were exported into the R statistical software platform and converted into hyperspectral data by using the hyperSpec package [61]. The preprocessing (‘cleaning’) was carried out using a spreadsheet program. Accordingly, the bands introducing excessive variation (‘noise’) into the data (901–1050 nm) and very short wavelength bands (325–399 nm) were excluded.
2.3.2. UAV-Based Sensing
The multispectral data were obtained at both sites via UAV campaigns (Figure 4) at 110–112 DAS in sunny, cloud-free, and low-wind conditions during midday. The propeller aero points were positioned on the ground uniformly for recording GPS positioning information (Figure 4d) before starting the flight. The flight plan was set up in the Pix4D capture software platform (Pix4D S.A., Switzerland) using a 2D polygon mission tool with lower ground sample distance (GSD = 1.3 cm/px), 85% image overlapping, a field of view (FOV) of 85°, and 30 m flight height. A five-band multispectral RedEdge-M camera (MicaSense Inc., Seattle, DC, USA) (Figure 4c) with a 5.4 mm focal length was used to capture high-resolution (1280 × 960 pixels) multispectral imagery. The camera was mounted on a DJI Matrices 100 (SZ DJI Technology, Shenzhen, Guangdong, China) quadcopter. The imagery was acquired with bandwidth and the center wavelength of 20 nm at 475 nm (blue), 20 nm at 560 nm (green), 10 nm at 668 nm (red), 10 nm at 717 nm (red edge), and 40 nm at 840 nm (near infrared) [34,62]. The raw images were recorded in 16 bit raw GeoTiff files stored on a local digital card, with the images uploaded to ATLAS cloud (MicaSense Inc., Seattle, DC, USA) for visualization, further processing, and information extraction.
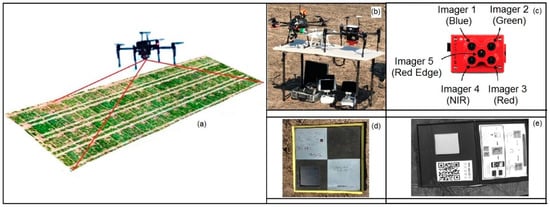
Figure 4.
(a) A representative UAV multispectral image of the site; (b) UAV platforms used at the experiment site; (c) RedEdge–M camera for the multispectral mission; (d) propeller aero point for recording GCPs for drone survey; (e) Calibrated Reflectance Panel.
Image preprocessing, including orientation correction, tie point matching, reflectance calibration, and orthomosaicing was performed in Agisoft Metashape 1.5.5 software platform (Agisoft LLC, St. Petersburg, Russia) [63]. A Calibrated Reflectance Panel (CRP) (MicaSense Inc., Seattle, DC, USA) was used before and after the individual flights for calibrating GeoTiff layers of five spectral bands for the conversion of digital number (DN) into reflectance (Figure 4e). Pixels of the canopy were segmented from background soil reflectance using Otsu’s thresholding method [64,65] in the MATLAB R2020a image processing platform (The Mathworks® Inc., Natick, MA, USA) (Supplementary Figure S2), which finds the optimal threshold by maximizing the weighted sum of between-class variances. Lastly, statistics of plot-wise mean canopy reflectance were extracted from the canopy pixels using the zonal statistics tool in Arc GIS 10.7.1 software platform (ESRI Ltd., Redlands, CA, USA).
In addition, a high-resolution RGB camera (DJI Technology, Shenzhen, Guangdong, China), fitted with a 24 mm lens with a field of view (FOV) of 25° × 20° and 5472 × 3648 pixel resolution, was used to acquire RGB images of the site at 110–112 DAS. The camera was mounted on a DJI Phantom 4 Professional UAV at 30 m flight height with a GSD of 1.3 cm/px, 85% image overlapping, and a FOV of 85°. The RGB mission planning and setup was similar to that used for the UAV multispectral mission. The images were then loaded and aligned in Agisoft Metashape 1.5.5 software platform (Agisoft LLC, St. Petersburg, Russia) for orthomosaicing and further preprocessing and point cloud extraction. Initially, a sparse point cloud was generated with the formation of camera positioning [66]. Gradually, dense point clouds were generated using a dense stereo-matched algorithm with the structure from motion (SfM) technique and exported for crop height extraction in the Lidar 360 software platform (GreenValley International, Berkeley, CA, USA). The average point density of the SfM captures was 824 and 945 pt/m2 for the MS and the HS site, respectively. Furthermore, an *.LAS file (Lidar data exchange) was created and converted to a multipoint structure for measuring crop height. Lastly, by taking a horizontal cross-sectional profile of the point cloud for each plot (Figure 5), the vertical difference (‘z’ direction) between the reference soil surface and the apex point (average top of the terminal spikelet) was measured [67], and plot-wise crop heights were calculated by taking the mean of at least the 5000 highest cloud data points in each plot (Supplementary Figure S3).
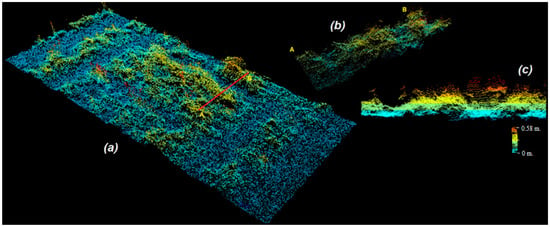
Figure 5.
The 3D point cloud extraction: (a) 3D point clouds of a representative plot with a representative cross-sectional line from ‘A’ to ‘B’ (red straight line); (b) cross-sectional area of the plot; (c) horizontal profile view and crop heights from the horizontal soil surface (blue base) of the cross-sectional area. The color bar (right corner) indicates actual crop heights from the soil surface of the cross-sectional plot area.
2.4. Vegetation Indices Derived from UAV Multispectral and Proximal Hyperspectral Data
Several vegetation indices (NDVI, OSAVI, and EVI) were derived from UAV multispectral imagery and proximal hyperspectral data to evaluate crop characteristics (Table 2). The NDVI was calculated as the simple normalization process between near-infrared (NIR) and red (R) spectral bands using Equation (1). We also computed OSAVI as an improved VI over the NDVI. The OSAVI enhances canopy reflectance by subduing the soil reflectance using an optimized soil adjusted coefficient (L = 0.16) in Equation (2). The EVI was computed using Equation (3) for its ability to optimize crop reflectance with improved sensitivity for the biomass yield prediction.

Table 2.
Vegetation indices evaluated in this study.
2.5. Statistical Analyses
2.5.1. Regression Analysis
A linear regression model was fitted in MATLAB R2020a (The Mathworks® Inc., Natick, MA, USA) to predict biomass and grain yield in response to individual VIs and crop heights using a 10-fold cross-validation method. A test of significance at p < 0.05 for a difference from ‘0’ was used to determine significance of biomass and grain yield prediction. The accuracy of the models was assessed by measuring the root-mean-square error (RMSE) (Equation (4)) and coefficient of determination (R2) Equation (5) in the cross-validation. In general, small RMSE and high R2 values can be used to determine the accuracy of a model.
where n is the total number of data points, represents the observed value at, is the predicted value at , represents an average observed value, and represents an average predicted value.
2.5.2. Analysis of Variance (ANOVA)
We performed two-way ANOVA using the SPSS® Statistics 25 software platform (IBM, New York, NY, USA) to determine whether any significant differences existed among the site means of wheat genotypes based on VIs, 3D point cloud-derived crop height, biomass yield (g/m2), and grain yield (g/m2) at the MS and HS sites. The testing of significant differences between levels within each factor was performed using Fisher’s protected least significant difference (LSD) test at the 5% significance level. The variables were treated as nested effects, and replicates were fitted as a random effect.
2.5.3. Machine Learning
We used four multivariate supervised regression learning techniques in ML, i.e., MLR, SVM, GPR, and ANN, to predict biomass yield and grain yield for both MS and HS sites as a function of site-specific VIs and 3D point cloud-derived crop heights in MATLAB R2020a environment (The Mathworks® Inc., Natick, MA, USA). These ML algorithms were selected specifically, since previous studies on non-sodic soils (see Table 1) have identified the diversified usefulness of these techniques in agricultural applications, including yield prediction, and they can also be applied on small to moderate datasets, for example, at a small field scale. A principal component analysis (PCA) was used for model feature selection to understand explained data variance in the models before applying the ML algorithms for prediction. The ML architecture employed in this study is illustrated in Figure 6 with a stepwise process including the design of training and testing datasets, feature selection, cross-validation, model performance evaluation, and decision making.
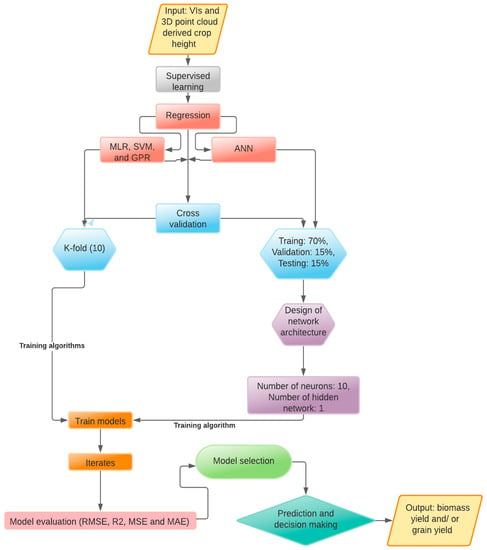
Figure 6.
Machine learning architecture employed for the prediction of biomass and grain yields in this study.
Multitarget Linear Regression
We used and compared linear, robust linear, and stepwise algorithms under this category. In general, a multi-robust linear regression algorithm is less sensitive to outliers and also provides flexibility in fitting [72]. The stepwise regression uses variables (VIs, crop height, biomass yield, and grain yield) at each step and removes variables that are not significant (p > 0.05) for the prediction [73].
Support Vector Machine Regression
We used and compared different kernel functions in SVM, such as linear, Gaussian, cubic, and quadratic for their ability to accurately predict the outcomes. The SVM is considered a reliable supervised learning technique that is used for classification, pattern recognition, regression, and prediction [74]. The linear kernel is used for a linearly separable dataset [75], whereas the Gaussian SVM is used for complex relationships [76]. Quadratic and cubic functions can provide flexibility in fitting [72].
Gaussian Process Regression
The GPR can predict the outcomes accurately. However, it is often difficult to interpret the outcomes. As recommended by The MathWorks Inc. [72], the preset flexibility in model type gallery was maintained in the MATLAB R2020a environment to avoid overfitting and to produce a small error in training. The kernel functions, such as exponential, squared exponential, matern 5/2, and rational quadratic were used and compared in their ability to predict the outcomes.
Artificial Neural Network
A multilayer perceptron model (MLP) is the most used network model in ANN. The backpropagation algorithm of MLP is used to search for the lowest error function in the weight space using gradient-based methods [77]. To avoid overfitting in the MLP result, it is important to train the network model with an appropriate number of hidden layers and neurons [78]. In this network model, a sum of inputs and bias passes through a transfer function is used to deliver the output to the activation point. The perceptron network is composed of artificial nodes or neurons. These nodes or neurons are the units, which can process information in layers and are connected by synaptic weights. They can transfer information in a supervised way for building a prediction model after the classification of data. The classified data are stored in a memory. Typically, the model is interconnected by the nodes or neurons in a three-layer network structure (input, hidden, and output layer). Each node or neuron of a layer is linked to the next layer. The input data carry information on the network structure, which is multiplied by weights while entering the hidden layer. Furthermore, the hidden layer processes this information, and the output layer classifies and predicts an outcome. The weights are the preassigned numbers, which are in the form of an argument that is passed to a nonlinear mathematical function and an activation function to return numbers between 0 and 1 [79].
A multilayer feedforward network model was performed in this study for prediction of biomass and for prediction of grain yield (Figure 7). Each multilayer feedforward network model consisted of a number of input, hidden, and output layers. The network model was selected by optimizing hidden layers, neurons, and functions using multiple runs and cross-validated by assigning 70% of the dataset to training, 15% to testing, and the remaining 15% to model validation. The network was trained using a Levenberg-Marquardt backpropagation algorithm. This algorithm is known to be used for its quick convergence in backpropagation [80,81]. The perceptron determines the prediction outputs by evaluating the input weights.
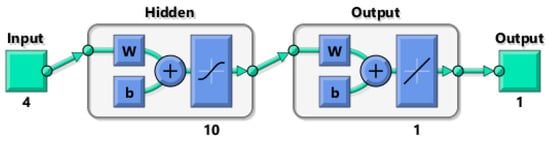
Figure 7.
The optimized ANN architecture in MATLAB 2020a environment used in this study for prediction of biomass yield and grain yield at both MS and HS site. The input layer (4) (VIs and 3D point cloud-derived crop height), hidden layer with 10 neurons (w = weight, b = bias), and the output layer (biomass yield and/or grain yield) are depicted in the network structure.
The performance of the MLs was evaluated by measuring RMSE (Equation (4)), R2 (Equation (5)), mean squared error (MSE) (Equation (6)), and mean absolute error (MAE) (Equation (7)) in the cross-validation. As mentioned earlier in Section 2.5.1, a small RMSE, MSE, and MAE for biomass and grain yield prediction was used to determine the optimum performance of the ML models.
where n is the total number of data points, represents the observed value at, is the predicted value at , represents an average observed value, and represents an average predicted value.
3. Results
3.1. Soil Constraints and Agro-Climatic Conditions
The depth-wise distribution of soil physicochemical properties (Figure 2) showed that both sites have very similar clay contents (41–58%) (Figure 2e) and, thus, are likely to have a similar water holding capacity. Both sites also have a higher ESP in the subsoil (18.2% for the MS and 24% for the HS site, at 100–150 cm) than the surface soil (2.7% for the MS and 14% for the HS site, at 0–10 cm) (Figure 2d). In addition, both sites have high Cl concentrations in the subsoil at 100–150 cm, with the HS site (>2300 mg/kg) having substantially higher Cl than the MS site (700–750 mg/kg) (Figure 2b).
Initial (prior to sowing) volumetric soil moisture data (Figure 2f) showed that both the sites had adequate and mostly similar soil moisture content stored at 0–150 cm depth (a mean of ~36.6% for the MS and ~38.8% for the HS site), which helped in the germination of crops despite low in-crop rainfall during sowing to emergence (~10 mm for both MS and HS sites) (Figure 1). Agro-climatic conditions at both sites were similar (p > 0.01) from May to October 2018 (Figure 1), suggesting that crops at both sites received similar rainfall during the growing season.
3.2. Sensor Performances
3.2.1. GreenSeeker® NDVI
Crop stagewise in situ monitoring from the measurement of GreenSeeker® NDVI during crop development indicated that maximum canopy development occurred at approximately 110–112 DAS (just before flowering) at both sites (Supplementary Figure S4). At this time, the NDVI for healthy cultivars reached a peak of ~0.74 for the MS and ~0.53 for the HS site; thus, this was identified as a suitable time to differentiate between cultivars. Higher NDVI values at the MS site also indicated more favorable crop growth and canopy greenness compared to the HS site (Supplementary Figure S4). Previous studies reported that, in non-constrained soils, the NDVI values for healthy vegetation gradually increase from the beginning of the crop season and reach a peak at the middle of the season or before flowering [82,83]. The data from this study also suggest that plants growing on a constrained soil follow a similar pattern of development, with close to flowering (110–112 DAS) being the most suitable time for obtaining remote sensing data of crops to evaluate and differentiate crop conditions for forecasting biomass and grain yields. Furthermore, biomass and grain yield were both significantly and positively correlated with GreenSeeker® NDVI near flowering at both sites (Table 3).

Table 3.
Cross-validation of parameters for biomass yield and grain yield as a function of GreenSeeker®-measured NDVI near flowering on a moderately sodic (MS) and a highly sodic (HS) site. Coefficient of determination (R2) values are presented with different asterisks to depict probabilities of a significant difference from ‘0’ with a significant level of p < 0.05 *, p < 0.001 **, and p < 0.0001 ***; n = 72.
3.2.2. UAV Multispectral Imaging
The VIs derived from UAV multispectral data were also significantly and positively correlated with biomass and grain yield at both sites (Figure 8). When comparing the UAV multispectral VIs, the EVI was more closely associated with biomass yield at both sites (R2 = 0.82; RMSE = 41.6 g/m2 for the MS and R2 = 0.67; RMSE = 40.0 g/m2 for the HS site) compared to the OSAVI and NDVI (Figure 8 and Table 4), whereas the NDVI was more closely correlated with grain yield at both sites (R2 = 0.75; RMSE = 17.7 g/m2 for the MS and R2 = 0.53, RMSE = 25.4 g/m2 for the HS site) compared to the EVI and OSAVI (Figure 8 and Table 4).
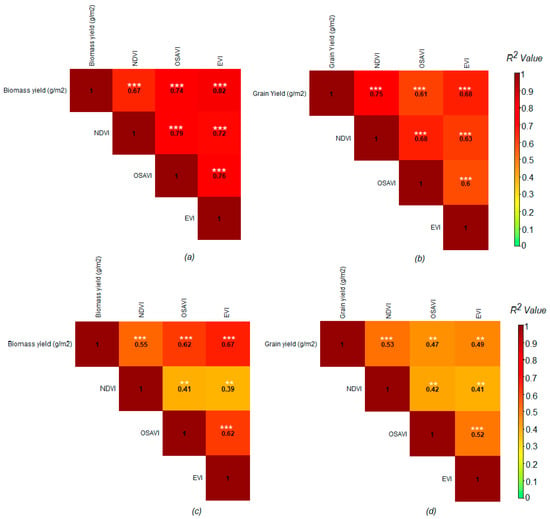
Figure 8.
Relationship between UAV-multispectral imagery-derived VIs with biomass yield and grain yield at a moderately sodic (MS) (a,b) and highly sodic (HS) (c,d) site; the matrix shows the coefficient of determination (R2) values between the variables, as also indicated by the color scale (right). Coefficient of determination (R2) values are presented with different asterisks to depict probabilities of a significant difference from ‘0’ with a significant level of p < 0.001 ** and p < 0.0001 ***; n = 72.

Table 4.
Cross-validation of root-mean-square error (RMSE) of biomass yield and grain yield as a function of individual VIs, derived from both UAV multispectral and ground-based proximal hyperspectral data at a moderately sodic (MS) and a highly sodic soil site; n = 72.
3.2.3. Proximal Hyperspectral Sensing
Non-imaging and ground-based proximal hyperspectral sensing-derived VIs showed a significant positive association with biomass and grain yield at both sites (Figure 9). Similar to the UAV multispectral VIs, the hyperspectral data-derived EVI was more closely associated with biomass yield at both sites (R2 = 0.56; RMSE = 65.4 g/m2 for the MS and R2 = 0.42; RMSE = 53.3 g/m2 for the HS site) compared to the OSAVI and NDVI (Figure 9 and Table 4), whereas the NDVI had a closer agreement with grain yield at both sites (R2 = 0.48; RMSE = 25.5 g/m2 for the MS and R2 = 0.37; RMSE = 29.2 g/m2 for the HS site) than the EVI and OSAVI (Figure 9 and Table 4).
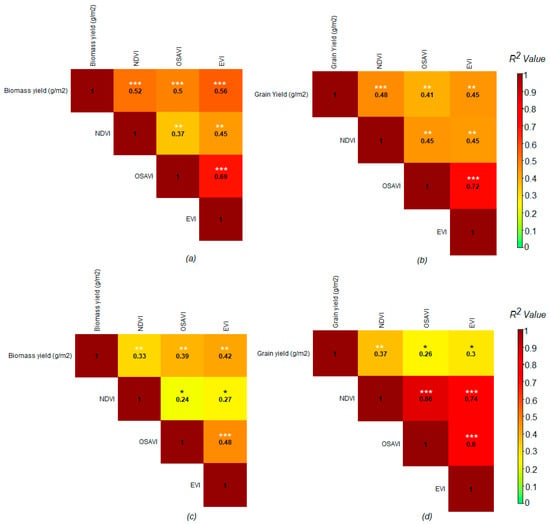
Figure 9.
Relationship between non-imaging, ground-based proximal hyperspectral data-derived VIs with biomass yield and grain yield at a moderately sodic (MS) (a,b) and highly sodic (HS) (c,d) site; the matrix shows the coefficient of determination (R2) values between the variables, as also indicated by the color scale (right). Coefficient of determination (R2) values are presented with different asterisks to depict probabilities of a significant difference from ‘0’ with a significant level of p < 0.05 *, p < 0.001 **, and p < 0.0001 ***; n = 72.
The VIs derived from handheld, proximal hyperspectral sensing exhibited a range of generally higher values compared to UAV multispectral VIs, indicating a slightly greater variability in handheld hyperspectral data within a site compared to those using UAV. For example, the maximum peak of the proximal hyperspectral NDVI value reached ~0.92 and ~0.71 at the MS and HS site, while the UAV-derived multispectral NDVI reached the maximum peak of ~0.72 and ~0.49 at the MS and HS site (110–112 DAS). This might be due to the changes in prevailing environmental conditions in the field during a longer period of survey using a handheld, proximal sensor that might have caused larger data variability compared to the rapid and automated UAV-based sensing [84,85]. Furthermore, UAV multispectral VIs were more closely associated with both biomass and grain yield than ground-based proximal GreenSeeker® NDVI or ASD FieldSpec hyperspectral VIs, as indicated by the R2 values.
3.2.4. UAV RGB Sensor-Based 3D Point Cloud Techniques
Biomass and grain yield were significantly and positively correlated with 3D point cloud-derived crop height, which explained 71% and 50% of the variability of biomass yield for the MS and HS site and 56% and 39% of the variability in grain yield for the MS and HS sites, respectively (Table 5), which was a slightly better association compared to manual, ground-based measurements of crop height at both sites (Table 5). On the other hand, a significant association (p < 0.01) was observed between ground-measured crop height and 3D point cloud crop height for both sites (R2 = 0.53 and 0.44 for the MS and HS site, respectively) (Supplementary Figure S5); a lower association between ground-measured crop height and biomass and/or grain yield suggests a larger data variability in ground-measured crop height compared to 3D point cloud crop height. This is sensible, since ground-measured crop height using a ruler relies on manual eye estimation of the field observer, whereas high-throughput UAV measurements, automated data processing, and extraction of crop height using software algorithms might have improved data quality and precision. This suggests that there may be a reduced need for extensive, labor-intensive, and manual measurements of crop height in the field. There is little or no suggestion that, for indicating biomass and yield potential, manual height measurements are superior to data obtained by UAV.

Table 5.
Cross-validation of model parameters for biomass yield and grain yield as a function of crop heights at a moderately sodic (MS) and a highly sodic (HS) site. Coefficient of determination (R2) values with different asterisks indicate different probabilities that the correlation differed from ‘0’ with a significant level of p < 0.05 *, p < 0.001 **, and p < 0.0001 ***; n = 72.
3.3. Comparing ML Algorithms for Prediction of Biomass and Grain Yield on Rain-Fed Sodic Soil
We compared different ML algorithms and different kernel functions integrated with UAV multispectral VIs and 3D point cloud crop height to test if these improve the estimation of biomass and grain yield of wheat on rain-fed sodic soil. The smallest prediction error likely suggests the best-performing model.
Under multitarget regression, the stepwise kernel was found to be effective in the prediction of both biomass and grain yield and have the lowest error estimates compared to the multiple linear and/or multi-robust linear kernels (Table 6 and Table 7). In SVM, the linear kernel function achieved superior prediction accuracy (lowest error) in comparison to quadratic, cubic, and Gaussian kernel functions at both MS and HS sites (Table 6 and Table 7). In GPR, the squared sequential, matern 5/2, and rational quadratic functions predicted biomass yield with slightly less error than an exponential function. However, the exponential kernel achieved superior accuracy in grain yield prediction in comparison to the other GPR kernels (Table 5 and Table 6). Overall, the MLP kernel in ANN achieved greatest estimated accuracy in the prediction of both biomass and grain yields at the two sites compared to all ML algorithms and kernels used in this study. The VIs and crop height combinedly explained 89% and 82% of the variability of biomass yield at the MS and HS site and 88% and 74% of the variability of grain yield at the MS and HS site using an MLP kernel in ANN (Table 6 and Table 7). The best-fitted models using ANN are illustrated in Supplementary Figures S6 and S7. The results suggest that canopy spectral information and crop height are useful indicators of aboveground biomass and/or grain yield of wheat on rain-fed sodic soil, and they can improve our understanding and ability to forecast biomass yield and grain yield variability in-field.

Table 6.
Performance evaluation between ML algorithms and kernels based on biomass yield prediction as a function of combined UAV multispectral VIs and 3D point cloud crop height on sodic soil; n = 72.

Table 7.
Performance evaluation between ML algorithms and kernels based on grain yield prediction as a function of combined UAV multispectral VIs and 3D point cloud crop height on sodic soil; n = 72.
3.4. Comparing Crop Growth, Biomass, and Grain Yields on Sodic Soils
The ANOVA outcome showed that the VIs and 3D point cloud-derived crop height was significantly different (p < 0.001) at MS and HS sites. The mean NDVI, OSAVI, EVI, and crop height were significantly higher at the MS site than at the HS site (Figure 10). As they are important indicators of growth and health of crops, significantly lower values at the HS site suggest that crops grown at the HS site experienced more stress than at the MS site. This stress was primarily due to sodic soil constraints rather than environmental factors, as the in-season crop rainfall, air temperature, and initial moisture stored in the soil profile were similar at both sites (Figure 1 and Figure 2).
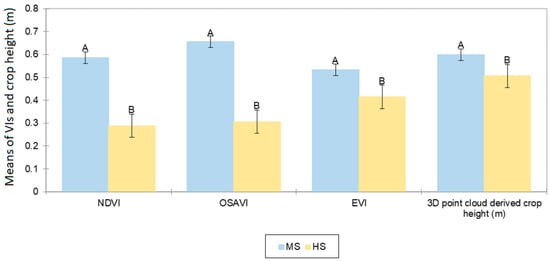
Figure 10.
Means of UAV multispectral-based VI and 3D point cloud-derived crop height at a moderately sodic (MS) and a highly sodic (HS) site measured close to flowering. The error bars represent standard errors of the mean of the respective crop parameters for the 72 yield plots. Significant differences between the sites are indicated using different letters at p < 0.001.
When comparing the performance of wheat genotype biomass yield between sites, the current study indicated that most of the genotypes had significantly higher predicted biomass (p < 0.05) at the MS site than the HS site close to flowering (Figure 11), whereas, at maturity, genotypic grain yield performance was substantially lower (p < 0.01) at the HS site compared to the MS site (Figure 12). The results suggest that crop stress most likely increased with development and had a significant negative influence on grain yield at the maturity stage, especially for the genotypes at the HS site.
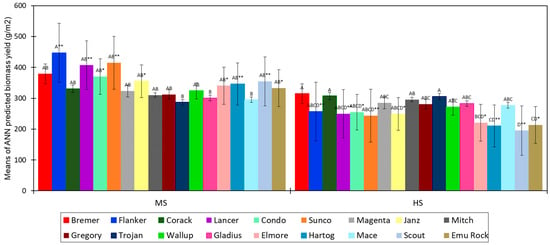
Figure 11.
Means of ANN-predicted biomass yield for 18 wheat genotypes at a moderately sodic (MS) and a highly sodic (HS) site close to flowering. Significant interactions (p < 0.05) between the genotypes at each site are indicated using different letters. Same letters between the genotypes indicate that interactions are nonsignificant (p > 0.05). Significant differences between the sites (MS and HS) for each genotype are indicated using asterisks at p < 0.01 **, p < 0.05 *. The error bars represent the standard error of the mean of four replications for each genotype.
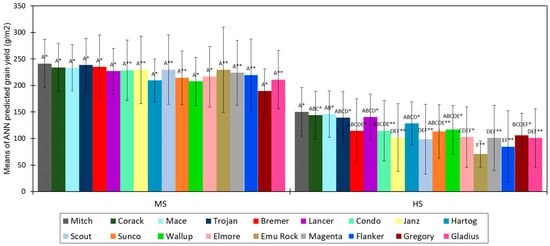
Figure 12.
Means of estimates grain yield from multispectral VIs and 3D point cloud crop height data using an ANN algorithm for 18 wheat genotypes at a moderately sodic (MS) and a highly sodic (HS) site. Significant interactions (p < 0.05) between the genotypes at each site are indicated using different letters. Same letters between the genotypes indicate that interactions are nonsignificant (p > 0.05) within a site. Significant differences between the sites (MS and HS) for each genotype are indicated using asterisks at p < 0.01 **, p < 0.05 *. The error bars represent the standard error of the mean of four replications for each genotype.
When considering a single site, the discrimination between genotypes was more prominent at the HS site than the MS site (Figure 12), suggesting variable effects of high sodic soil constraints on tolerance and performance of wheat genotypes. This also indicates that the techniques used in this study can be useful to differentiate between cultivars in more sodic soil conditions. The study identified that wheat genotypes Mitch, Corack, Mace, Trojan, Lancer, and Bremer were more tolerant to sodic soil constraints than Emu Rock, Janz, Flanker, and Gladius. However, seasonal changes and changes in soil constraint level may have diversified influences over genotypic performance. Hence, implementation of these strategies over different seasonal and sodic soil conditions, using various of crops and/or cultivars, is recommended for future studies. Overall, the results support the suggestion that integrated UAV optical remote sensing and ML techniques have the ability to improve estimation of crop growth and yield on sodic soils that can be used to identify cultivars tolerant to sodic soil environments.
4. Discussion
The present study aimed to develop an integrated optical remote sensing and ML-based framework to quantify wheat genotypic traits to assist the estimation of growth and yield on sodic soils. We tested if integrated optical remote sensing and ML-based techniques can accurately forecast yield and help to distinguish tolerant genotypes on sodic soil, which can assist agronomists/researchers to select appropriate techniques/traits for phenotyping on sodic soils by reducing the need for extensive, labor-intensive, and manual methods of phenotyping to examine the traits and crop growth.
4.1. Traits and Sensor Performances
Fern et al. [86] reported that the OSAVI can better estimate green biomass and vegetative cover than NDVI in non-constrained semi-arid rangeland soil. Similarly, our study indicated that both EVI and OSAVI are slightly better indicators of biomass yield than NDVI in rain-fed sodic soil environment (Figure 8 and Figure 9 and Table 4). However, a number of studies reported that NDVI can be a useful indicator of grain yield performance on non-sodic soils [35,87,88,89]. Likewise, the present research found that NDVI is a more useful indicator of wheat grain yield than OSAVI and EVI in the presence of sodic soil constraints (Figure 8 and Figure 9 and Table 4). Crop rainfall and ambient air temperature can have a large influence on canopy vigor and greenness, particularly in rain-fed conditions. Adequate rainfall between late tillering and flowering crop growth stages (Figure 1) in the current study could have allowed us to obtain sufficient canopy spectral cues that were closely correlated with biomass and grain yield.
When comparing sensors, the high-resolution UAV RedEdge-M multispectral sensor showed a slightly greater performance in estimating crop growth and yield than ground-based proximal hyperspectral and GreenSeeker® sensors (Figure 8 and Figure 9). A systematic and rapid UAV flight plan and operation might have potentially helped to achieve a good quality of imaging data with the multispectral sensor when compared to handheld proximal sensors, as UAV multispectral imagery-derived traits were more closely associated with both biomass and grain yields than either proximal GreenSeeker or ASD FieldSpec sensor-derived VIs (Table 3 and Figure 8 and Figure 9). Saura et al. [90] also reported that a high precision of UAV data can be achieved with a systematic flight plan in automatic mode, and they demonstrated the potential benefits of using it for improved crop yield prediction.
The accuracy of the handheld spectroradiometer used to acquire the hyperspectral data might have been reduced due to weather and prevailing environmental conditions. For example, the longer time taken to obtain handheld measurements compared to the flight time of the drone would have allowed the environment to change more during the period of measurement. The spectral data combine a mixture of diffuse and specular reflectance because of different particle sizes, scattering, and multicollinearity [91]. Variation in spectral reflectance is often caused by differences in the size of particles in the object. This causes a deviation of light at various angles as a function of the wavelengths and creates scattering effects [84]. Ana Belén, Enoc, Víctor Marcelo, Marta, and José Ramón [84] also reported that results did not satisfactorily improve even after normalization and preprocessing of hyperspectral spectroradiometer data for a vineyard study. However, our study was unable to pinpoint the exact reason for variations in the data. The problem might be overcome by using UAV hyperspectral sensing with an automated flight plan, as UAV sensing can capture information rapidly and, thus, reduce the influence of environmental variability. Pádua et al. [85] observed that unmanned aerial systems (UAS) with specific imaging sensors achieve better results than handheld devices.
Yang and Jinfei [92] estimated canopy height using UAV 3D point cloud data and demonstrated the ease with which the technique can be used to obtain crop growth information at the field level. A few studies reported that UAV-based crop height measurement may be integrated with cropping traits to improve the estimation of the aboveground biomass of barley and/or winter wheat on non-constrained soil [53,93,94]. Hence further research is warranted to employ these integrated sensor-based techniques and traits to improve estimation of biomass and/or yield on rain-fed sodic soils. UAV RGB 3D point cloud techniques show promise for the extraction of crop height, which is closely related to aboveground biomass and grain yield on sodic soils.
4.2. Yield Prediction on Rain-Fed Sodic Soils Using ML
We compared four ML algorithms (MLR, SVM, GPR, and ANN) for crop growth and yield prediction on sodic soils and observed that the ANN predicted aboveground biomass and grain yield with slightly less error than the others. The results suggest that the ANN has superior ability to account for site-specific complexities of agricultural traits that can be used to improve estimation of growth and yield, particularly on sodic soils. Recently, ANN has become popular for different applications in agriculture, including crop yield prediction. For example, Safa et al. [95] reported a better ability of an ANN model relative to MLR for estimating wheat yield using a heterogenous dataset in multiple arable farms on non-sodic soils in New Zealand. Mokarram and Bijanzadeh [96] also reported a better prediction accuracy using the MLP (ANN) network model in comparison to MLR for barley biomass yield (R2 = 0.89) and grain yield (R2 = 0.92) on non-constrained soils. Recently, Dhakal, Gautam, and Bhattarai [76] reported that an ANN model with 10 neurons in the hidden layer performed comparatively better in training with R2 = 0.84 and RMSE = 1.5 MJ·m−2·day−1 when estimating global solar radiation. Furthermore, their study also found that the stepwise kernel has good potential for estimation with less error (R2 = 0.88 and RMSE = 1.5 MJ·m−2·day−1). Ekici et al. [97] reported that the Matern 5/2 kernel in GPR produced better results in comparison to other supervised ML algorithms and kernels for a hybrid power system study. However, there are a limited number of studies on agricultural biomass and grain yield prediction using UAV- and ground-based optical sensor-derived VIs, using crop height as input crop parameters with comprehensive ML approaches, particularly on sodic soil. The performance of the models can be influenced by sample size, site characteristics, areal coverage, and the traits used for the growth and yield prediction study. On sodic soils, the main challenge lies in accurate plant sampling and extraction of useful cropping traits due to larger spatial variability and incomplete crop cover caused by soil constraints compared to non-constrained soils [48]. Hence, the appropriate selection of traits and the accurate extraction of crop information using high-throughput sensing and techniques were essential to feed the ML models for improved prediction accuracy. It was also necessary to test and compare model performance individually at the MS and HS site with varying levels of soil constraints to investigate the performance of complex traits for evaluating crop growth and yield variation. Crop production on sodic soil has previously been reported to be negatively affected due to high Cl concentration in subsoil, which decreased water and nutrient uptake by root [5,98]. However, those were labor-intensive and time-consuming techniques to determine the relative performance of crops/cultivars and tolerance to soil constraints [5,98]. Hence, the present study demonstrated the efficacy of optical remote sensing and statistical modeling approaches to help identify appropriate traits. Overall, our results support the suggestion that integrated optical UAV remote sensing and ML techniques have the potential to improve estimation of crop growth and yield on sodic soils. This information can also be important in the discrimination between the performance of cultivars/genotypes.
4.3. Crop Growth and Yield Vary with Changes in Sodic Soil Constraints
Recent studies have suggested that constraints in sodic soil can increase crop water stress and negatively affect the physiological performance of wheat [15,48]. A study reported that the UAV thermal imaging-derived crop water stress index had a close negative agreement with wheat growth and yield at critical crop development stages, such as close to flowering and maturity on rain-fed sodic soils [48], suggesting that wheat grown on sodic soils appears water-stressed. In this study, we also observed a significant variation in spectral indices and crop growth close to flowering across different sodic soil conditions (Figure 10) and the variation was most likely due to water limitations. High subsoil ESP and Cl concentrations have a strong potential to restrict root water and nutrient uptake from the soil and reduce rooting depth [98,99]. Thus, in both soils, crops would be expected to be stressed and have reduced growth, with profound adverse impacts at the HS site, which supports the suggestions made by an earlier study that high sodic soil constraints can reduce wheat physiological growth [15]. Studies have also reported that canopy temperature is an important trait to evaluate wheat physiological growth on sodic soils under water-limited environments [15,48]. Results from the current study indicated that optical remote sensing-based spectral VIs and crop height information are also strong indicators of wheat biomass and grain yields on rain-fed sodic soils and can be used to differentiate crop growth, stress, and yield performance between different levels of sodicity. Overall, the results confirm the adverse impacts of soil constraints, which can have a large influence on biomass and/or grain yield (Figure 11 and Figure 12). Genotypic biomass and yield performance variations/differences were observed within a site, and it was clearer between the sites with the changes in soil constraint level. The results closely correspond to the findings of a recently published study on sodic soil [48], where the authors demonstrated that water stress can further increase with the seasonal development of wheat crop in highly sodic soil environments compared to moderately sodic soils and can adversely affect wheat yield at maturity. Overall, the results support the suggestion that wheat genotypic growth and yield performance vary with sodic soil constraints, and that genotypic tolerance to soil constraints can be determined using the appropriate selection of traits and integrated high-throughput approaches, including UAV optical remote sensing and ML.
5. Conclusions
This study successfully evaluated the potential of optical remote sensing and ML algorithms to improve the estimation of growth and yield of wheat genotypes, which is useful to differentiate genotypic tolerance to sodic soils. A range of approaches were tested and proposed to demonstrate their application to crops grown in sodic soil. The potential of a number of sensors and ML algorithms was assessed for their ability to accurately forecast wheat biomass and grain yields for identifying constraint tolerant cultivars on sodic soils.
Different VIs derived from a UAV multispectral (RedEdge-M) sensor predicted biomass yield and grain yield at least as well as, and possibly better than, the handheld proximal hyperspectral spectroradiometer and/or GreenSeeker® sensors. Although hyperspectral sensing was reported to be superior in previous studies conducted on non-constrained soils, we could not achieve high accuracy using a handheld non-imaging hyperspectral sensor. Crop height extracted from UAV RGB-based 3D point cloud was found to have a closer association with both biomass and grain yield than ground-based manually measured crop height, suggesting the adaptation of this technique on sodic soil. Furthermore, ML algorithms integrated with optical remote sensing-derived traits were tested and compared for their ability to improve forecasting of crop growth and yield on sodic conditions. Overall, the present study demonstrated the efficacy of optical remote sensing and ML-based techniques in providing real-time and detailed information on appropriate trait selection for evaluating crop growth and yield variation to identify constraint-tolerant cultivars in a rain-fed sodic soil environment. Follow-up studies should focus on implementing these techniques with a greater diversity of genotypes and at multiple sodic sites to compare the results. This study can greatly assist agronomists/plant physiologists in the selection of appropriate techniques/traits for phenotyping on sodic soils by reducing the need for extensive, labor-intensive, and manual methods of phenotyping to examine the relative performance of crop traits, growth, and yield. In addition, this may help guide decisions by farmers and breeders in selecting tolerant varieties in sodic soil environments to sustain agricultural production.
The key research findings are the following:
- High sodic soil constraints negatively affected crop growth and development and reduced yield.
- A number of the methods were able to discriminate differences between sites and some between genotypes within a site.
- The UAV multispectral (RedEdge-M) sensor performed with slightly less error than the ground-based handheld proximal hyperspectral and/or GreenSeeker® sensors for the measurements of crop traits.
- The UAV RGB-based 3D point cloud technique is promising for crop height measurements and suggests there is a reduced need for the manual, labor-intensive, and tedious process of crop height measurements in the field.
- The EVI was in more close association with biomass yield and the NDVI with grain yield on sodic soils.
- Integrated VIs and crop height were useful indicators of biomass and grain yield performance of wheat genotypes on rain-fed sodic soil.
- The ANN performed slightly better than multitarget regression, SVM, and GPR in estimating biomass yield and grain yield on sodic soils.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/rs13173482/s1, Figure S1: Drone image of a representative plot illustrating a typical destructive biomass sampling area (‘yellow’ square box indicating 0.5 × 3 middle rows area) and plant reflectance measurements area using GreenSeeker and ASD FieldSpec (‘orange’ rectangular box including 3 middle rows) within the plot. One of the propeller markers for GPS location calibration is shown in the lower left hand of the image, Figure S2: A representative site image showing classified and separated soil and canopy pixels using Otsu’s algorithm, Figure S3: A representation of crop height extraction method using RGB-3D point cloud techniques, (A) aerial view of the plot-wise 3D point clouds, (B) triangulated irregular network (TIN), (C) cross-sectional profile of a single plot for measurement of crop height, (D) crop volume measurement, Figure S4: Seasonal GreenSeeker NDVI values of the moderately sodic (MS) and the highly sodic (HS) site, Figure S5: Correlation between ground-measured crop height and 3D point cloud derived crop height; (a) moderately sodic site, (b) highly sodic site; n = 72, Figure S6: Cross-validation of correlation between ground-measured biomass yield and ANN predicted biomass yield as a function of UAV-multispectral VIs and 3D point cloud crop height; (a) moderately sodic (MS) and (b) highly sodic site (HS); n = 72, Figure S7: Cross-validation of correlation between ground-measured grain yield and ANN predicted grain yield as a function of UAV-multispectral VIs and 3D point cloud crop height; (a) moderately sodic (MS) and (b) highly sodic site (HS); n = 72, Table S1: Wheat genotypes for 2018 experiment sites.
Author Contributions
Conceptualization, M.R.C., S.D., J.C., A.A., S.C., N.W.M. and Y.P.D.; methodology, M.R.C.; software, M.R.C. and S.D.; validation, M.R.C. and S.D.; formal analysis, M.R.C.; investigation, M.R.C.; resources, Y.P.D.; data curation, M.R.C., J.C. and A.A.; writing—original draft preparation, M.R.C.; writing—review and editing, S.D., J.C., A.A., N.W.M. and Y.P.D.; visualization, M.R.C. and S.D.; supervision, J.C., S.C., N.W.M. and Y.P.D.; project administration, Y.P.D.; funding acquisition, Y.P.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Australian Grains Research and Development Corporation (GRDC) (Grant No.-UA00159) and The University of Queensland, Australia.
Institutional Review Board Statement
“Not applicable” for studies not involving human or animals.
Acknowledgments
This research was undertaken as part of the Australian Grains Research and Development Corporation (GRDC) project entitled ‘Improving wheat yields on sodic, magnesic, and dispersive soils’. We gratefully acknowledge the funding support from the GRDC and The University of Queensland, Australia. The growers provided an important contribution to the field research through trial cooperation. The authors convey their sincere thanks. In addition, the authors thank Scott Diefenbach and Daniel Smith for their support during field data collection, as well as Lachlan Fowler, Mel Schneemilch, and Katherine Raymont for helping with useful laboratory resources.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
References
- Rengasamy, P.; Walters, L. Introduction to Soil Sodicity; CRC: Boca Raton, FL, USA, 1994. [Google Scholar]
- Rengasamy, P. Salt-Affected Soils in Australia; GRDC: Barton, Australia, 2016; pp. 1–63. [Google Scholar]
- Dalal, R.; Blasi, M.; So, H. High Sodium Levels in Subsoil Limits Yields and Water Use in Marginal Cropping Areas; GRDC Final Report; Grains Research and Development Corporation: Canberra, Australia, 2002. [Google Scholar]
- Dang, Y.P.; Dalal, R.C.; Buck, S.R.; Harms, B.; Kelly, R.; Hochman, Z.; Schwenke, G.D.; Biggs, A.J.W.; Ferguson, N.J.; Norrish, S.; et al. Diagnosis, extent, impacts, and management of subsoil constraints in the northern grains cropping region of Australia. Aust. J. Soil Res. 2010, 48, 105. [Google Scholar] [CrossRef]
- Dang, Y.P.; Dalal, R.C.; Routley, R.; Schwenke, G.D.; Daniells, I. Subsoil constraints to grain production in the cropping soils of the north-eastern region of Australia: An overview. Aust. J. Exp. Agric. 2006, 46, 19–35. [Google Scholar] [CrossRef]
- Dang, Y.P.; Christopher, J.; Dalal, R.C. Genetic diversity in barley and wheat for tolerance to soil constraints. Agronomy 2016, 6, 55. [Google Scholar] [CrossRef]
- Johannsen, W.L. The Genotype Conception of Heredity. Am. Nat. 1911, 45, 129–159. [Google Scholar] [CrossRef]
- Pask, A.; Pietragalla, J.; Mullan, D.; Reynolds, M.E. (Eds.) Physiological Breeding II: A Field Guide to Wheat Phenotyping; CIMMYT: Mexico City, Mexico, 2012; pp. 1–132. [Google Scholar]
- Reynolds, M.; Pask, A.; Mullan, D. (Eds.) Physiological Breeding I: Interdisciplinary Approaches to Improve Crop Adaptation; CIMMYT: Mexico City, Mexico, 2012; pp. 1–174. [Google Scholar]
- Fiorani, F.; Schurr, U. Future Scenarios for Plant Phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Montagnoli, A.; Terzaghi, M.; Fulgaro, N.; Stoew, B.; Wipenmyr, J.; Ilver, D.; Rusu, C.; Scippa, G.S.; Chiatante, D. Non-destructive Phenotypic Analysis of Early Stage Tree Seedling Growth Using an Automated Stereovision Imaging Method. Front. Plant Sci. 2016, 7, 1644. [Google Scholar] [CrossRef]
- Candiago, S.; Remondino, F.; De Giglio, M.; Dubbini, M.; Mario, G. Evaluating Multispectral Images and Vegetation Indices for Precision Farming Applications from UAV Images. Remote Sens. 2015, 7, 4026–4047. [Google Scholar] [CrossRef]
- Gnädinger, F.; Schmidhalter, U. Digital Counts of Maize Plants by Unmanned Aerial Vehicles (UAVs). Remote Sens. 2017, 9, 544. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Christopher, J.; Apan, A.; Choudhury, M.R.; Chapman, S.; Menzies, N.W.; Dang, Y.P. UAV-Thermal imaging and agglomerative hierarchical clustering techniques to evaluate and rank physiological performance of wheat genotypes on sodic soil. ISPRS J. Photogramm. Remote Sens. 2021, 173, 221–237. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W. The use of imaging spectrometry for agricultural applications. ISPRS J. Photogramm. Remote Sens. 1999, 54, 299–304. [Google Scholar] [CrossRef]
- Madec, S.; Baret, F.; de Solan, B.; Thomas, S.; Dutartre, D.; Jezequel, S.; Hemmerlé, M.; Colombeau, G.; Comar, A. High-Throughput Phenotyping of Plant Height: Comparing Unmanned Aerial Vehicles and Ground LiDAR Estimates. Front. Plant Sci. 2017, 8, 2002. [Google Scholar] [CrossRef]
- Roy Choudhury, M.; Christopher, J.; Apan, A.A.; Chapman, S.C.; Menzies, N.W.; Dang, Y.P. Integrated high-throughput phenotyping with high resolution multispectral, hyperspectral and 3D point cloud techniques for screening wheat genotypes under sodic soils. In Proceedings of the TROPAG: International Tropical Agriculture Conference, Brisbane, Australia, 11–13 November 2019. [Google Scholar]
- Boomsma, C.; Santini, J.; West, T.; Brewer, J.; McIntyre, L.; Vyn, T. Maize grain yield responses to plant height variability resulting from crop rotation and tillage system in a long-term experiment. Soil Tillage Res. 2010, 106, 227–240. [Google Scholar] [CrossRef]
- Jimenez-Berni, J.A.; Deery, D.M.; Rozas-Larraondo, P.; Condon, A.G.; Rebetzke, G.J.; James, R.A.; Bovill, W.D.; Furbank, R.T.; Sirault, X.R.R. High Throughput Determination of Plant Height, Ground Cover, and Above-Ground Biomass in Wheat with LiDAR. Front. Plant Sci. 2018, 9, 237. [Google Scholar] [CrossRef]
- Carly, S.; Michael, J.S.; Norman, E.; Michael, B.; Murilo, M.M.; Tianxing, C. Unmanned aircraft system-derived crop height and normalized difference vegetation index metrics for sorghum yield and aphid stress assessment. J. Appl. Remote Sens. 2017, 11, 026035. [Google Scholar] [CrossRef]
- Guo, Y.; Shi, Z.; Huang, J.; Wang, L.; Cheng, Y.; Zheng, G. Mapping Horizontal and Vertical Spatial Variability of Soil Salinity in Reclaimed Areas. In Digital Soil Mapping Across Paradigms, Scales and Boundaries; Springer Environmental Science and Engineering; Zhang, G.L., Ed.; Springer: Singapore, 2016; pp. 33–45. [Google Scholar]
- Dang, Y.P.; Dalal, R.C.; Pringle, M.J.; Biggs, A.J.W.; Darr, S.; Sauer, B.; Moss, J.; Payne, J.; Orange, D. Electromagnetic induction sensing of soil identifies constraints to the crop yields of north-eastern Australia. Soil Res. 2011, 49, 559–571. [Google Scholar] [CrossRef]
- Thessler, S.; Kooistra, L.; Teye, F.; Huitu, H.; Bregt, A. Geosensors to Support Crop Production: Current Applications and User Requirements. Sensors 2011, 11, 6656–6684. [Google Scholar] [CrossRef] [PubMed]
- Potuckova, M.; Červená, L.; Kupková, L.; Lhotakova, Z.; Lukeš, P.; Hanus, J.; Novotny, J.; Albrechtová, J. Comparison of Reflectance Measurements Acquired with a Contact Probe and an Integration Sphere: Implications for the Spectral Properties of Vegetation at a Leaf Level. Sensors 2016, 16, 1801. [Google Scholar] [CrossRef] [PubMed]
- Suarez, L.; Apan, A.; Werth, J. Hyperspectral sensing to detect the impact of herbicide drift on cotton growth and yield. ISPRS J. Photogramm. Remote Sens. 2016, 120, 65–76. [Google Scholar] [CrossRef]
- Feng, A.; Zhou, J.; Vories, E.D.; Sudduth, K.A.; Zhang, M. Yield estimation in cotton using UAV-based multi-sensor imagery. Biosyst. Eng. 2020, 193, 101–114. [Google Scholar] [CrossRef]
- Stein, M.; Bargoti, S.; Underwood, J. Image based mango fruit detection, localisation and yield estimation using multiple view geometry. Sensors 2016, 16, 1915. [Google Scholar] [CrossRef]
- Marino, S.; Cocozza, C.; Tognetti, R.; Alvino, A. Use of proximal sensing and vegetation indexes to detect the inefficient spatial allocation of drip irrigation in a spot area of tomato field crop. Int. J. Adv. Precis. Agric. 2015, 16, 613–629. [Google Scholar] [CrossRef]
- Basso, B.; Cammarano, D.; Cafiero, G.; Marino, S.; Alvino, A. Cultivar discrimination at different site elevations with remotely sensed vegetation indices. Ital. J. Agron. 2010, 6, e1. [Google Scholar] [CrossRef]
- Stefano, M.; Arturo, A. Detection of spatial and temporal variability of wheat cultivars by high-resolution vegetation indices. Agronomy 2019, 9, 226. [Google Scholar] [CrossRef]
- Mkhabela, M.S.; Bullock, P.; Raj, S.; Wang, S.; Yang, Y. Crop yield forecasting on the Canadian Prairies using MODIS NDVI data. Agric. For. Meteorol. 2011, 151, 385–393. [Google Scholar] [CrossRef]
- Magney, T.S.; Eitel, J.U.H.; Huggins, D.R.; Vierling, L.A. Proximal NDVI derived phenology improves in-season predictions of wheat quantity and quality. Agric. For. Meteorol. 2016, 217, 46–60. [Google Scholar] [CrossRef]
- Duan, T.; Chapman, S.C.; Guo, Y.; Zheng, B. Dynamic monitoring of NDVI in wheat agronomy and breeding trials using an unmanned aerial vehicle. Field Crop. Res. 2017, 210, 71–80. [Google Scholar] [CrossRef]
- Marti, J.; Bort, J.; Slafer, G.A.; Araus, J.L. Can wheat yield be assessed by early measurements of Normalized Difference Vegetation Index? Ann. Appl. Biol. 2007, 150, 253–257. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A rapid monitoring of NDVI across the wheat growth cycle for grain yield prediction using a multi-spectral UAV platform. Plant Sci. 2019, 282, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Gao, X.; Huete, A.R.; Ni, W.; Miura, T. Optical–biophysical relationships of vegetation spectra without background contamination. Remote Sens. Environ. 2000, 74, 609–620. [Google Scholar] [CrossRef]
- Gill, T.K.; Phinn, S.R.; Armston, J.D.; Pailthorpe, B.A. Estimating tree-cover change in Australia: Challenges of using the MODIS vegetation index product. Int. J. Remote Sens. 2009, 30, 1547–1565. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Shastry, K.A.; Sanjay, H.A.; Deshmukh, A. A parameter based customized artificial neural network model for crop yield prediction. J. Artif. Intell. 2016, 9, 23–32. [Google Scholar] [CrossRef][Green Version]
- Long, N.; Gianola, D.; Rosa, G.J.M.; Weigel, K.A. Application of support vector regression to genome-assisted prediction of quantitative traits. Theor. Appl. Genet. 2011, 123, 1065–1074. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, X.; Clinton, N.; Wang, N. An artificial neural network model for estimating crop yields using remotely sensed information. Int. J. Remote Sens. 2010, 25, 1723–1732. [Google Scholar] [CrossRef]
- Aghighi, H.; Azadbakht, M.; Ashourloo, D.; Shahrabi, H.S.; Radiom, S. Machine learning regression techniques for the silage maize yield prediction using time-series images of Landsat 8 OLI. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 4563–4577. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Liu, J.; Goering, C.E.; Tian, L. A neural network for setting target corn yields. Trans. ASAE 2001, 44, 705. [Google Scholar] [CrossRef]
- Miao, Y.; Mulla, D.; Robert, P. Identifying important factors influencing corn yield and grain quality variability using artificial neural networks. Int. J. Adv. Precis. Agric. 2006, 7, 117–135. [Google Scholar] [CrossRef]
- Das, S.; Christopher, J.; Apan, A.; Choudhury, M.R.; Chapman, S.; Menzies, N.W.; Dang, Y.P. Evaluation of water status of wheat genotypes to aid prediction of yield on sodic soils using UAV-thermal imaging and machine learning. Agric. For. Meteorol. 2021, 307, 108477. [Google Scholar] [CrossRef]
- Adak, A.; Murray, S.C.; Božinović, S.; Lindsey, R.; Nakasagga, S.; Chatterjee, S.; Anderson, S.L.; Wilde, S. Temporal vegetation indices and plant height from remotely sensed imagery can predict grain yield and flowering time breeding value in maize via machine learning regression. Remote Sens. 2021, 13, 2141. [Google Scholar] [CrossRef]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Yang, G.; Yang, X.; Fan, L. Estimation of the yield and plant height of winter wheat using UAV-based hyperspectral images. Sensors 2020, 20, 1231. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling maize above-ground biomass based on machine learning approaches using UAV remote-sensing data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Kyratzis, A.C.; Skarlatos, D.P.; Menexes, G.C.; Vamvakousis, V.F.; Katsiotis, A. Assessment of vegetation indices derived by UAV imagery for durum wheat phenotyping under a water limited and heat stressed Mediterranean Environment. Front. Plant Sci. 2017, 8, 1114. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Das, S.; Christopher, J.; Apan, A.; Roy Choudhury, M.; Chapman, S.; Menzies, N.W.; Dang, Y.P. UAV-thermal imaging: A robust technology to evaluate in-field crop water stress and yield variation of wheat genotypes. In Proceedings of the IEEE International India Geoscience and Remote Sensing Symposium 2020 (InGARSS 2020), Ahmedabad, India, 1–4 December 2020; pp. 138–141. [Google Scholar]
- ISO. General Requirements for the Competence of Testing and Calibration Laboratories; ISO 17025; ISO: Geneva, Switzerland, 2005. [Google Scholar]
- Shaw, R.J. (Ed.) Salinity and Sodicity; Queensland Department of Primary Industries: Brisbane, Australia, 1997; Volume QI 97035, pp. 79–96. [Google Scholar]
- Day, P.R. Particle Fractionation and Particle-Size Analysis; American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1965; pp. 545–567. [Google Scholar]
- Tucker, B. A proposed new reagent for the measurement of cation exchange properties of carbonate soils. Aust. J. Soil Res. 1985, 23, 633. [Google Scholar] [CrossRef]
- Roy Choudhury, M.; Mellor, V.; Das, S.; Christopher, J.; Apan, A.; Menzies, N.W.; Chapman, S.; Dang, Y.P. Improving estimation of in-season crop water use and health of wheat genotypes on sodic soils using spatial interpolation techniques and multi-component metrics. Agric. Water Manag. 2021, 255, 107007. [Google Scholar] [CrossRef]
- Suarez, L.A.; Apan, A.; Werth, J. Detection of phenoxy herbicide dosage in cotton crops through the analysis of hyperspectral data. Int. J. Remote Sens. 2017, 38, 6528–6553. [Google Scholar] [CrossRef]
- Beleites, C. HyperSpect Introduction. Spectroscopy—Imaging; University of Trieste, Leibniz Institute of Photonic Technology: Jena, Germany, 2015. [Google Scholar]
- Su, J.; Liu, C.; Coombes, M.; Hu, X.; Wang, C.; Xu, X.; Li, Q.; Guo, L.; Chen, W.-H. Wheat yellow rust monitoring by learning from multispectral UAV aerial imagery. Comput. Electron. Agric. 2018, 155, 157–166. [Google Scholar] [CrossRef]
- Taddia, Y.; Russo, P.; Lovo, S.; Pellegrinelli, A. Multispectral UAV monitoring of submerged seaweed in shallow water. Appl. Geomat. 2019, 12, 19–34. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Zhang, L.; Niu, Y.; Zhang, H.; Han, W.; Li, G.; Tang, J.; Peng, X. Maize canopy temperature extracted from uav thermal and rgb imagery and its application in water stress monitoring. Front. Plant Sci. 2019, 10, 1270. [Google Scholar] [CrossRef] [PubMed]
- Zietara, A.M. Creating Digital Elevation Model (DEM) Based on Ground Points Extracted from Classified Aerial Images Obtained from Unmanned Aerial Vehicle (UAV); Norwegian University of Science and Technology, Department of Civil and Environmental Engineering: Trondheim, Norway, 2017; Unpulished. [Google Scholar]
- Paulus, S.; Behmann, J.; Mahlein, A.-K.; Plümer, L.; Kuhlmann, H. Low-Cost 3D Systems: Suitable Tools for Plant Phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef] [PubMed]
- Rouse, J.J.W.; Haas, R.; Schell, J.; Deering, D. Monitoring vegetation systems in the great plains with erts. NASA Spec. Publ. 1974, 351, 309–1974. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Huete, A.; Justice, C.; Liu, H. Development of vegetation and soil indices for MODIS-EOS. Remote Sens. Environ. 1994, 49, 224–234. [Google Scholar] [CrossRef]
- Huete, A.R.; Liu, H.Q.; Batchily, K.; van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- The MathWorks Inc. Regression Learner App; The MathWorks Inc.: Natick, MA, USA, 2020. [Google Scholar]
- Frost, J. Guide to Stepwise Regression and Best Subsets Regression. 2021. Available online: https://statisticsbyjim.com/regression/guide-stepwise-best-subsets-regression/ (accessed on 3 March 2021).
- Shevade, S.K.; Keerthi, S.S.; Bhattacharyya, C.; Murthy, K.R.K. Improvements to the SMO algorithm for SVM regression. IEEE Trans. Neural Netw. 2000, 11, 1188–1193. [Google Scholar] [CrossRef]
- Bajaj, P. Creating Linear Kernel SVM in Python. 2018. Available online: https://www.geeksforgeeks.org/creating-linear-kernel-svm-in-python/ (accessed on 8 April 2021).
- Dhakal, S.; Gautam, Y.; Bhattarai, A. Evaluation of temperature-based empirical models and machine learning techniques to estimate daily global solar radiation at Biratnagar airport, Nepal. Adv. Meteorol. 2020, 2020, 8895311. [Google Scholar] [CrossRef]
- Trenz, O.; Šťastný, J.; Konečný, V. Agricultural data prediction by means of neural network. Agric. Econ. 2011, 57, 356–361. [Google Scholar] [CrossRef]
- Ennouri, K.; Ben Ayed, R.; Triki, M.; Ottaviani, E.; Mazzarello, M.; Hertelli, F.; Zouari, N. Multiple linear regression and artificial neural networks for delta-endotoxin and protease yields modelling of Bacillus thuringiensis. 3 Biotech 2017, 7, 187. [Google Scholar] [CrossRef]
- Zacharis, N.Z. Predicting student academic performance in blended learning using artificial neural networks. Int. J. Artif. Intell. Appl. 2016, 7, 17–29. [Google Scholar] [CrossRef]
- Kayabasi, A. An application of ANN trained by ABC algorithm for classification of wheat grains. Int. J. Intell. Syst. Appl. Eng. 2018, 1, 85–91. [Google Scholar] [CrossRef]
- Amaratunga, V.; Wickramasinghe, L.; Perera, A.; Jayasinghe, J.; Rathnayake, U. Artificial neural network to estimate the paddy yield prediction using climatic data. Math. Probl. Eng. 2020, 2020, 8627824. [Google Scholar] [CrossRef]
- Turvey, C.G.; Mclaurin, M.K. Applicability of the normalized difference vegetation index (NDVI) in Index-based crop insurance design. Weather Clim. Soc. 2012, 4, 271–284. [Google Scholar] [CrossRef]
- Mkhabela, M.S.; Mashinini, N.N. Early maize yield forecasting in the four agro-ecological regions of Swaziland using NDVI data derived from NOAA’s-AVHRR. Agric. For. Meteorol. 2005, 129, 1–9. [Google Scholar] [CrossRef]
- Ana Belén, G.-F.; Enoc, S.-A.; Víctor Marcelo, G.; Marta, G.-F.; José Ramón, R.-P. Field spectroscopy: A non-destructive technique for estimating water status in vineyards. Agronomy 2019, 9, 427. [Google Scholar] [CrossRef]
- Pádua, L.; Vanko, J.; Hruška, J.; Adão, T.; Sousa, J.J.; Peres, E.; Morais, R. UAS, sensors, and data processing in agroforestry: A review towards practical applications. Int. J. Remote Sens. 2017, 38, 2349–2391. [Google Scholar] [CrossRef]
- Fern, R.R.; Foxley, E.A.; Bruno, A.; Morrison, M.L. Suitability of NDVI and OSAVI as estimators of green biomass and coverage in a semi-arid rangeland. Ecol. Indic. 2018, 94, 16–21. [Google Scholar] [CrossRef]
- Singh, R.; Semwal, D.P.; Rai, A.; Chhikara, R.S. Small area estimation of crop yield using remote sensing satellite data. Int. J. Remote Sens. 2002, 23, 49–56. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Kyratzis, A.; Skarlatos, D.; Fotopoulos, V.; Vamvakousis, V.; Katsiotis, A. Investigating Correlation among NDVI Index Derived by Unmanned Aerial Vehicle Photography and Grain Yield under Late Drought Stress Conditions. Procedia Environ. Sci. 2015, 29, 225–226. [Google Scholar] [CrossRef][Green Version]
- Saura, J.R.; Reyes-Menendez, A.; Palos-Sanchez, P. Mapping multispectral digital Images using a Cloud Computing software: Applications from UAV images. Heliyon 2019, 5, e01277. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Yang, S.; Jinfei, W. Winter wheat canopy height extraction from UAV-based point cloud data with a moving cuboid filter. Remote Sens. 2019, 11, 1239. [Google Scholar] [CrossRef]
- Juliane, B.; Andreas, B.; Simon, B.; Janis, B.; Silas, E.; Georg, B. Estimating biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Jibo, Y.; Guijun, Y.; Changchun, L.; Zhenhai, L.; Yanjie, W.; Haikuan, F.; Bo, X. Estimation of winter wheat above-ground biomass using unmanned aerial vehicle-based snapshot hyperspectral sensor and crop height improved models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef]
- Safa, M.; Samarasinghe, S.; Nejat, M. Prediction of wheat production using artificial neural networks and investigating indirect factors affecting it: Case study in Canterbury province, New Zealand. J. Agric. Sci. Technol. 2015, 17, 791–803. [Google Scholar]
- Mokarram, M.; Bijanzadeh, E. Prediction of biological and grain yield of barley using multiple regression and artificial neural network models. Aust. J. Crop Sci. 2016, 10, 895–903. [Google Scholar] [CrossRef]
- Ekici, S.; Unal, F.; Ozleyen, U. Comparison of different regression models to estimate fault location on hybrid power systems. IET Gener. Transm. Distrib. 2019, 13, 4756–4765. [Google Scholar] [CrossRef]
- Dang, Y.P.; Dalal, R.; Mayer, D.; McDonald, M.; Routley, R.; Schwenke, G.; Buck, Y. High subsoil chloride concentrations reduce soil water extraction and crop yield on Vertosols in north-eastern Australia. Aust. J. Agric. Res. 2008, 59, 321–330. [Google Scholar] [CrossRef]
- Dang, Y.P.; Christopher, J.; Anzooman, M.; Choudhury, M.R.; Menzies, N.W. Wheat varietal tolerance to sodicity with variable subsoil constraints. In Proceedings of the GRDC Grains Research Update, Goondiwindi, Australia, 5–6 March 2019; p. 58. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).