1. Introduction
Greenhouse production in Canada in 2019 comprised about 838 specialized commercial greenhouses covering more than 17.6 million m
2 (mainly in Ontario), generating more than
$1.1 billion in revenue, and employing more than 12,429 persons [
1]. Cucumber is one of the main vegetables produced, with a harvested land area of 4.8 million m
2 and a production of about 240,451 metric tons or CAD 485 million [
1]. As with other crops, fungal diseases can affect greenhouse crops and be a significant limiting production factor [
2]. Powdery mildew is one cucumber plant disease that can cause yield losses of 30–50% [
3]. It is all due to
Podosphaera xanthii, a biotrophic pathogen that interacts with the host without killing the host cells to obtain nutrients [
4].
To control cucumber mildew, approaches based on temperature and relative humidity have been proposed as early warning methods [
5,
6,
7]. Other methods include periodic visual inspection of the plants by technicians. This approach is time-consuming, costly, and does not collect spatial information. It can also be unreliable as leaves that look healthy can be infected. Detecting a disease when a plant wilts or collapses is too late, as in a greenhouse, diseases can spread quickly. Therefore, early disease detection is needed for greenhouse crop management to reduce pesticide usage and ensure quality and the safety in the production chain.
Digital imaging can be a great help in finding diseases in greenhouses, and several studies are testing it to detect crop diseases. RGB images were used to detect canker severities over citrus leaves [
8,
9], powdery mildew over greenhouse cucumber plants (when symptoms were already visible) [
10,
11,
12], and potato leaves infected with early blight, late blight, or powdery mildew [
13,
14]. Multispectral imagery was tested to detect rice dwarf virus (RDV), rice blast (
Magnaporthe oryzae), and glume blight (
Phyllosticta glumarum) [
15], Hyperspectral data and imagery were acquired to detect
Cercospora leaf spot, powdery mildew, and leaf rust over sugar beet leaves [
16] as well as decay lesions caused by
Penicillium digitatum in citrus fruit [
17] and
Scletorinia sclerotiorum on oilseed rape stems [
18]. Powdery mildew over cucumber plants was studied with hyperspectral data acquired between 450 and 1100 [
19] and 400 to 900 nm [
20]. Late-blight disease on potato or tomato leaves was detected using reflectance in the green and red bands [
21,
22,
23], the red-edge band [
21,
24,
25,
26,
27], and the near-infrared (NIR) and shortwave infrared (SWIR) bands [
22,
25]. Chlorophyll fluorescence was also used with various sensors including portable fluorescence sensors, fiber-optic fluorescence spectrometers, and multispectral fluorescence imaging devices [
28,
29,
30] because pathogens induce the production of waxes or resistant-specific compounds, such as lignin, both of which affect leaf chlorophyll fluorescence [
29] emitted in two bands (400–600 and 650–800 nm) [
31].
Fluorescence can also be emitted by compounds from secondary metabolism in the blue and green regions of the spectrum [
32]. Hyperspectral imagery combined with chlorophyll fluorescence and thermograms was used by Berdugo et al. [
33] to study cucumber leaves infected with powdery mildew. Multicolor fluorescence imagery was tested to detect leaf powdery mildew in the case of zucchini (
Cucurbita pepo L.) [
34] and melon (
Cucumis melo L.) [
35]. However, fluorescence-based methods have some limitations, the major drawback being the need for darkness during image acquisition. [
30]. This implies that images should be acquired during the night [
28,
36,
37], but applying light at this time in greenhouses may be problematic because it can prevent plants from entering the flowering, fruiting, or stem extension stage [
37]. Furthermore, fluorescence systems require UV light (315–390 nm) to excite the plants, so for practical reasons fluorescence-based technology was not considered in the study. All the aforementioned studies including, fluorescence, were mainly pilot studies that used laboratory measurements, mainly at the leaf level, and could not be applied directly to greenhouses.
Detecting diseases in greenhouses using multispectral images presents several challenges. Multispectral cameras acquire images using more than one sensor, and most UAV multispectral cameras are frame-types, which means each band is captured independently by each sensor. To obtain a multispectral image, all band images need to be aligned because each sensor has a different position and point of view. The images are also acquired at close range to the plants and are not georeferenced; hence, the GPS-based method used in commercial software such as Pix4D (Lausanne, Switzerland) cannot work accurately for band alignment and image stitching. Finally, the images are acquired under natural conditions, and illumination corrections should be performed to reduce differences during image acquisition.
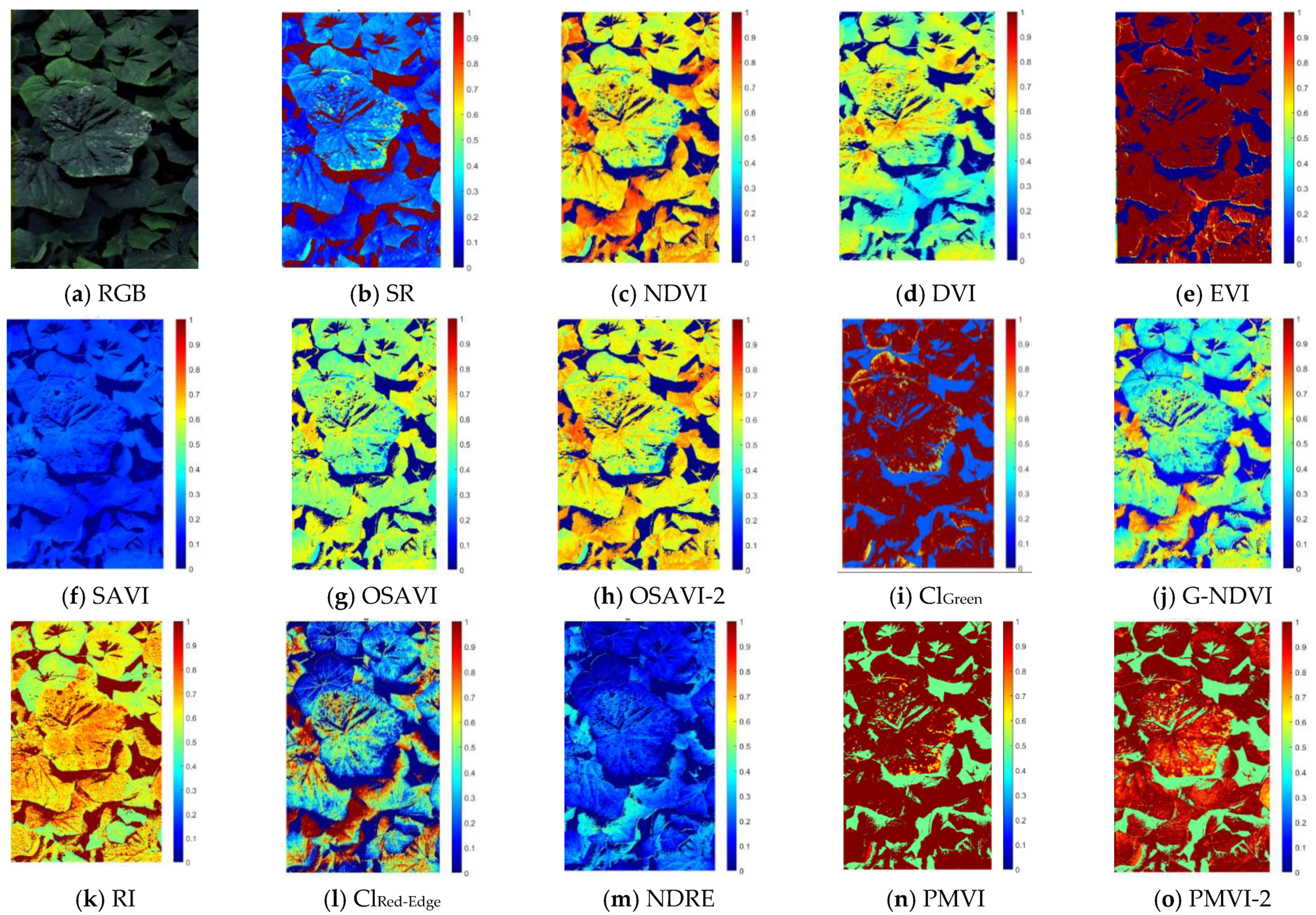
In this study, close-range non-georeferenced multispectral imagery acquired by the Micasense
® RedEdge camera (Micasense Inc., Seattle, Washington, USA), was mounted on a cart and tested to detect powdery mildew over greenhouse cucumber plants. The images were spatially and spectrally corrected and used to compute vegetation index images. The resulting images were entered into an SVM classifier to sort pixels as a function of their status (healthy or infected) as in Es-Saady et al. [
13], Islam et al. [
14], Shi et al. [
15], Folch et al. [
17], Kong et al. [
18], Berdugo et al. [
33], and Wang et al. [
38].
4. Discussion
In this study, we evaluated the feasibility of using multispectral images acquired at close-distance over greenhouse cucumber plants to detect powdery mildew using a Micasense
® RedEdge camera attached to a mechanic extension of a mobile cart. Images were obtained at 1.5 m from the top of the canopy but such a short distance between the camera and the plants meant that a GPS-based method for band alignment and image stitching would not work accurately [
67]. In this case, the images that needed to be registered (the moving images) were registered to a reference image (the fixed image) [
68]. The use of close-range distance imagery for plant disease detection, however, had been previously reported: Thomas et al. [
69] reported a distance of 80 cm from the top of the canopy of six barley cultivars in a study of their susceptibility to powdery mildew caused by
Blumeria graminis f. sp.
Hordei in a greenhouse environment. Under laboratory conditions, a distance of 40 cm from the plants was reported by Kong et al. [
18], who acquired hyperspectral images to detect Sclerotinia sclerotiorum on oilseed rape plants, and Fahrentrapp et al. [
67] used MicaSense RedEdge imagery to detect gray mold infection caused by Botrytis cinerea on tomato leaflets (
Solanum lycopersicum) from a distance of 66 cm from the plants.
Another problem with these images was the presence of an image background even though removing it is a standard preprocessing technique. As already noted by Wspanialy and Moussa [
10], disease detection techniques can be difficult with images having a high degree of background clutter in a greenhouse setting. In our study inside a commercial greenhouse, the first 700 columns of each image were cropped from left to right to remove the white greenhouse aisle. Cropping a section of the image was also used if there were a complex background in a real environment [
70]. We also noticed that due to the camera position, only the most exposed leaves were detected, which created dark areas inside the canopy vegetation that also presented some band misalignment. To remove the background cuttler noise as well as some structural wires present in the greenhouse, we created a binary mask using Bradley’s adaptative threshold method [
46], which computes a threshold for each pixel using the local mean intensity around the neighborhood of the pixel. This approach is useful due to the internal and external illumination differences among images. It gave better results than the global threshold, which was used in Otsu’s method [
71]. The resulting binary mask had a value of 0 for the background area and 1 for the vegetation.
The binary mask overlayed each respective aligned band to obtain band images with their background clutter noise removed and the foreground vegetation retained. The approach we used to remove the background is simpler than the algorithm proposed by Bai et al. [
72], which removes non-green background from images acquired over single cucumber leaves that had scabs. Indeed, in their algorithm, a correction method of abnormal points in a 3D histogram of the image was performed followed by rapid segmentation by decreasing histogram dimensions to construct a Gaussian optimization framework to obtain the optimal threshold to isolate the scabs. In addition, Bai et al. [
72] worked on images of single cucumber leaves, while in our case we removed the noise from images acquired over the whole plant canopy. Our approach did not need to acquire multiple images as did the method of Wspanialy and Moussa, [
10], who studied powdery mildew on greenhouse tomato plants. In this method, which focused on foreground leaves, two images of plants needed to be collected: one was illuminated with a red light to increase the contrast between the foreground leaves and the background. Then the images were converted to grayscale, and Otsu’s method was applied to create a binary mask. The noise was then removed by applying a 3 × 3 median filter to the mask. The filtered mask was then applied to the RGB images. Wspanialy and Moussa’s [
10] method was also applied to images acquired at 50 cm from the plant, while in our case the images were collected at 1.5 m above the plant canopy.
One challenge when working with close-range imagery inside a greenhouse is related to the differences in illumination during the image acquisition. Indeed, we observed a bluish-purple coloration over the aligned RGB images due to high illuminance in the blue band. To achieve color consistency, we applied a simple and efficient illumination estimation method developed by Cheng et al. [
47]. The method selects bright and dark pixels using a projection distance in the color distribution and then applies a principal component analysis to estimate the illumination direction. Such selection of bright and dark pixels in the image allows obtaining clusters of points with the largest color differences between these pixels.
We trained a fine Gaussian SVM to sort healthy and infected pixels using either individual band reflectance, the RGB composite, several vegetation indices, and all bands together. The overall classification accuracies for the RGB and all the band reflectances together were 93 and 99%, respectively, for the trained model. A major reduction in the overall classification accuracies (57%) was observed when all band reflectances were used. The validation of the SVM model produced the highest overall accuracy with the RGB composite (89%). It was higher than those of Ashhourloo et al. [
73], who applied a maximum likelihood classifier to sort healthy wheat leaves and those infected with
Puccinia triticina using various vegetation index images derived from hyperspectral data: narrow-band normalized difference vegetation index (83%), NDVI (81%), greenness index (77%), anthocyanin reflectance index (ARI) (75%), structural independent pigment index (73%), physiological reflectance index (71%), plant senescence reflectance index (69%), triangular vegetation index (69%), modified simple ratio (68%), normalized pigment chlorophyll ratio index (68%), and nitrogen reflectance index (67%). Our accuracy was also higher than those reported by Pineda et al. [
34], who tested an artificial neural network (70%), a logistic regression analysis (73%), and an SVM (46%) to classify multicolor fluorescence images acquired over healthy and infected zucchini leaves with powdery mildew.
However, our accuracies with both the training and validating datasets were lower than in Berdugo et al. [
33] (overall accuracy of 100%), who applied a stepwise discriminant analysis to discriminate healthy cucumber leaves from those infected with powdery mildew using the following features: effective quantum yield, SPAD 502 Plus Chlorophyll Meter (Spectrum Technologies, Inc., Aurora, IL, USA) values, maximum temperature difference (MTD), NDVI and ARI. Our overall classification accuracies were also lower than those reported by Thomas et al. [
69] (94%), who applied a non-linear SVM over a combination of pseudo-RGB representation and spectral information from hyperspectral images to classify powdery mildew caused by
Blumeria graminis f. sp.
Hordei in barley. Our overall classification accuracies were also lower than those of Kong et al. [
18], who applied a partial least square discriminant analysis (PLS–DA) (94%), a radial basis function neural network (RBFNN) (98%), an extreme learning machine (ELM) (99%), and a support vector machine (SVM) (99%) to hyperspectral images for classifying healthy pixels and those infected with Sclerotinia sclerotiorum on oilseed rape plants