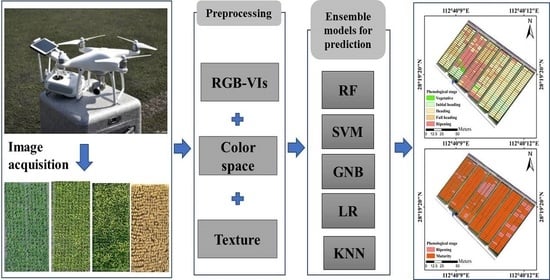
Improved Accuracy of Phenological Detection in Rice Breeding by Using Ensemble Models of Machine Learning Based on UAV-RGB Imagery
Abstract
:1. Introduction
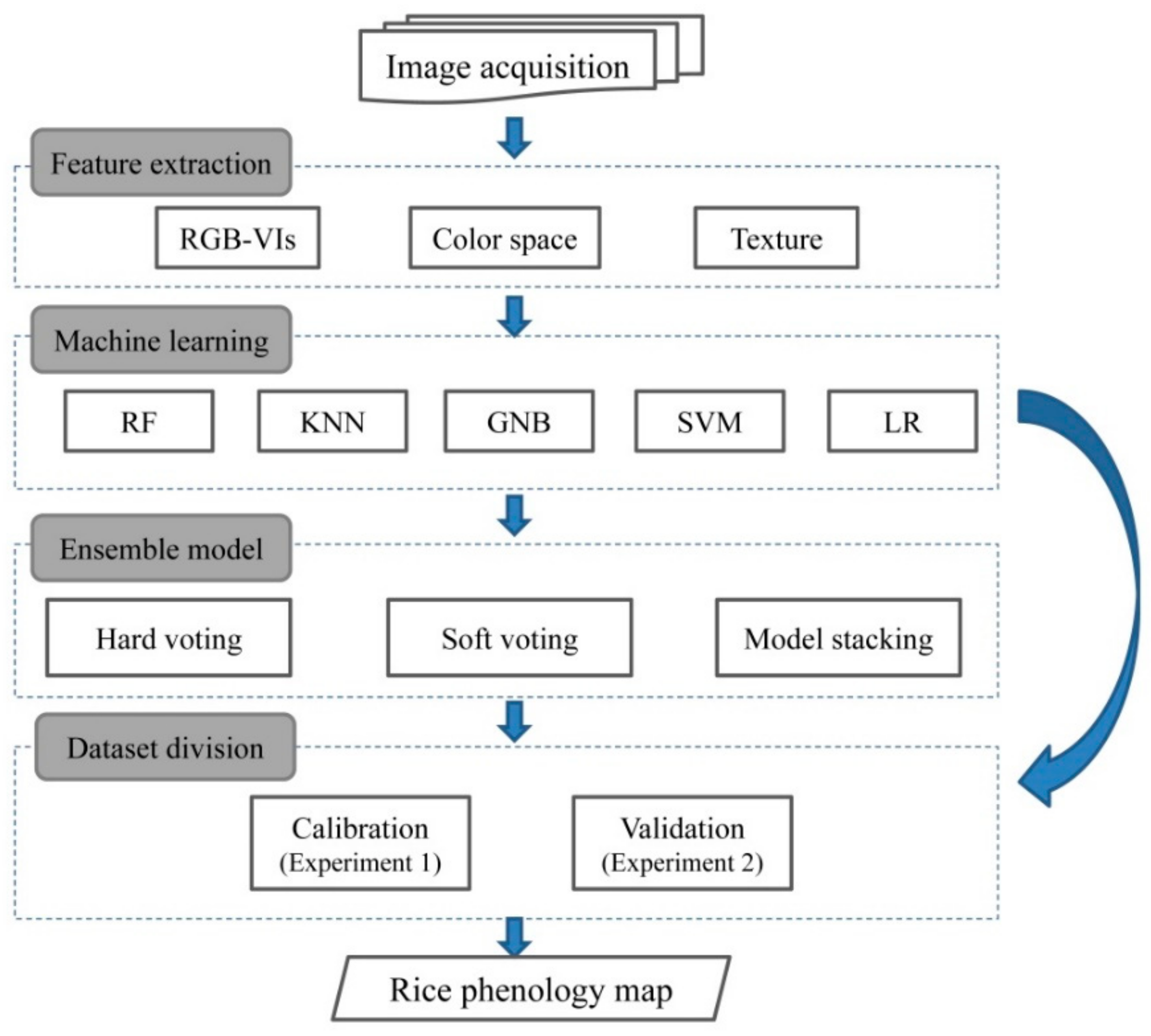
2. Materials and Methods
2.1. Study Area
2.2. Field-Based Observations of Rice Phenology
2.3. Image Prepocessing
2.3.1. Images Collection
2.3.2. Generation of Orthomosaic Maps
2.3.3. Calculation of VIs or Color Space
2.3.4. Calculation of Texture Features
2.4. Classifier Techniques
2.4.1. Multiple Machine Learning Algorithms
2.4.2. Ensemble Models
2.5. Evaluation of Model Accuracy
3. Results
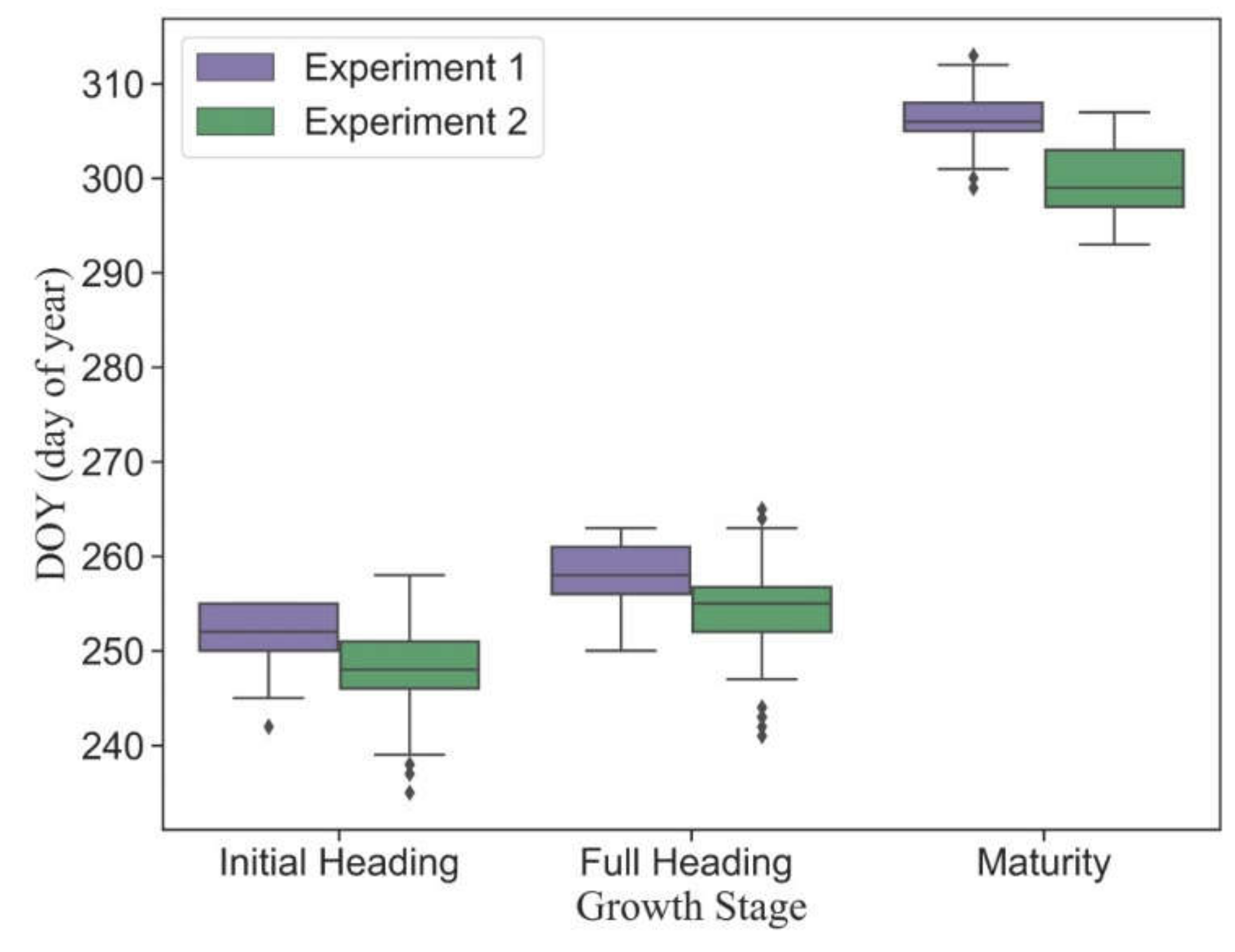
3.1. Variation of Observed Phenology in Different Plots
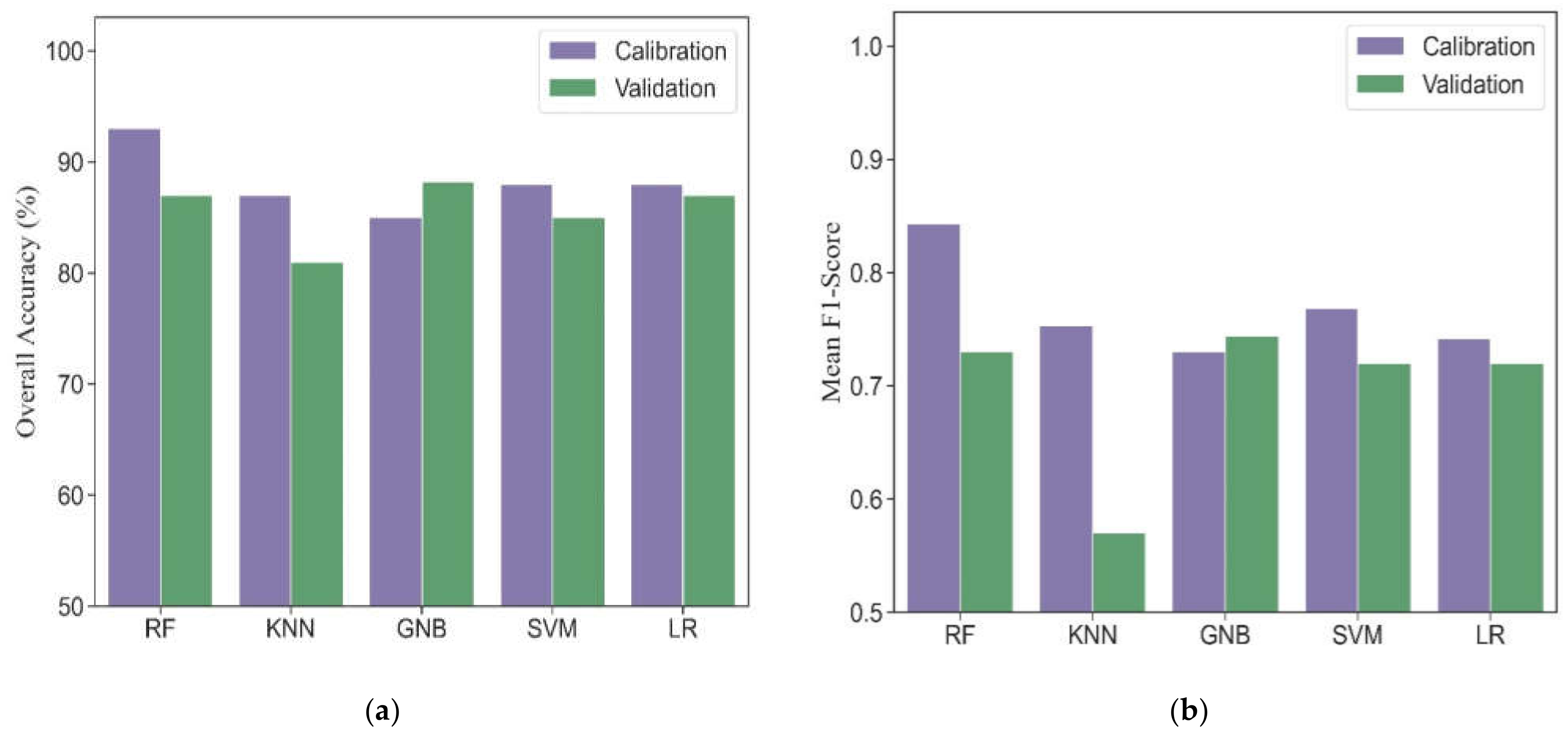
3.2. Model Performance by Multiple Machine Learning
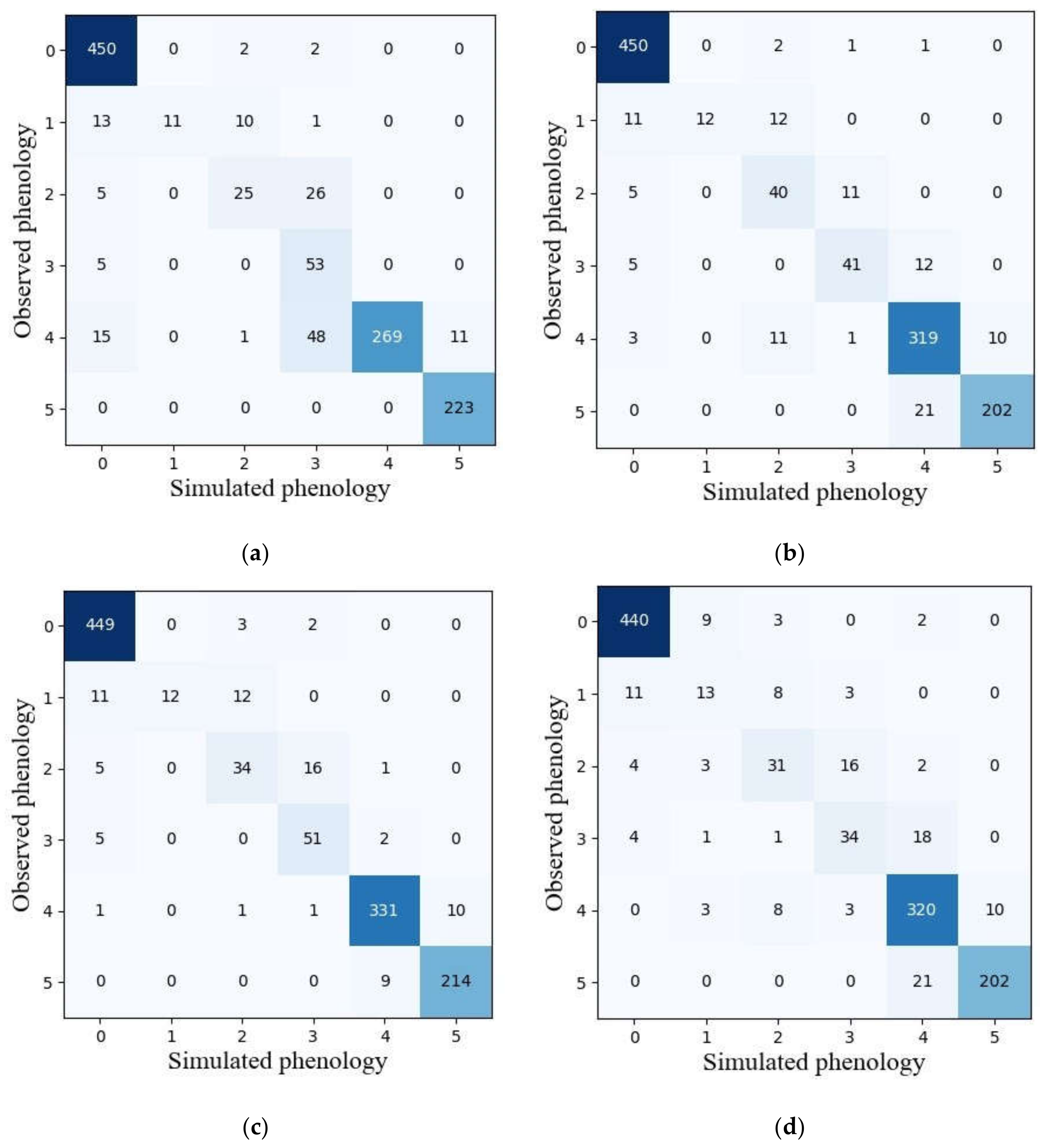
3.3. Model Performance by Ensemble Models
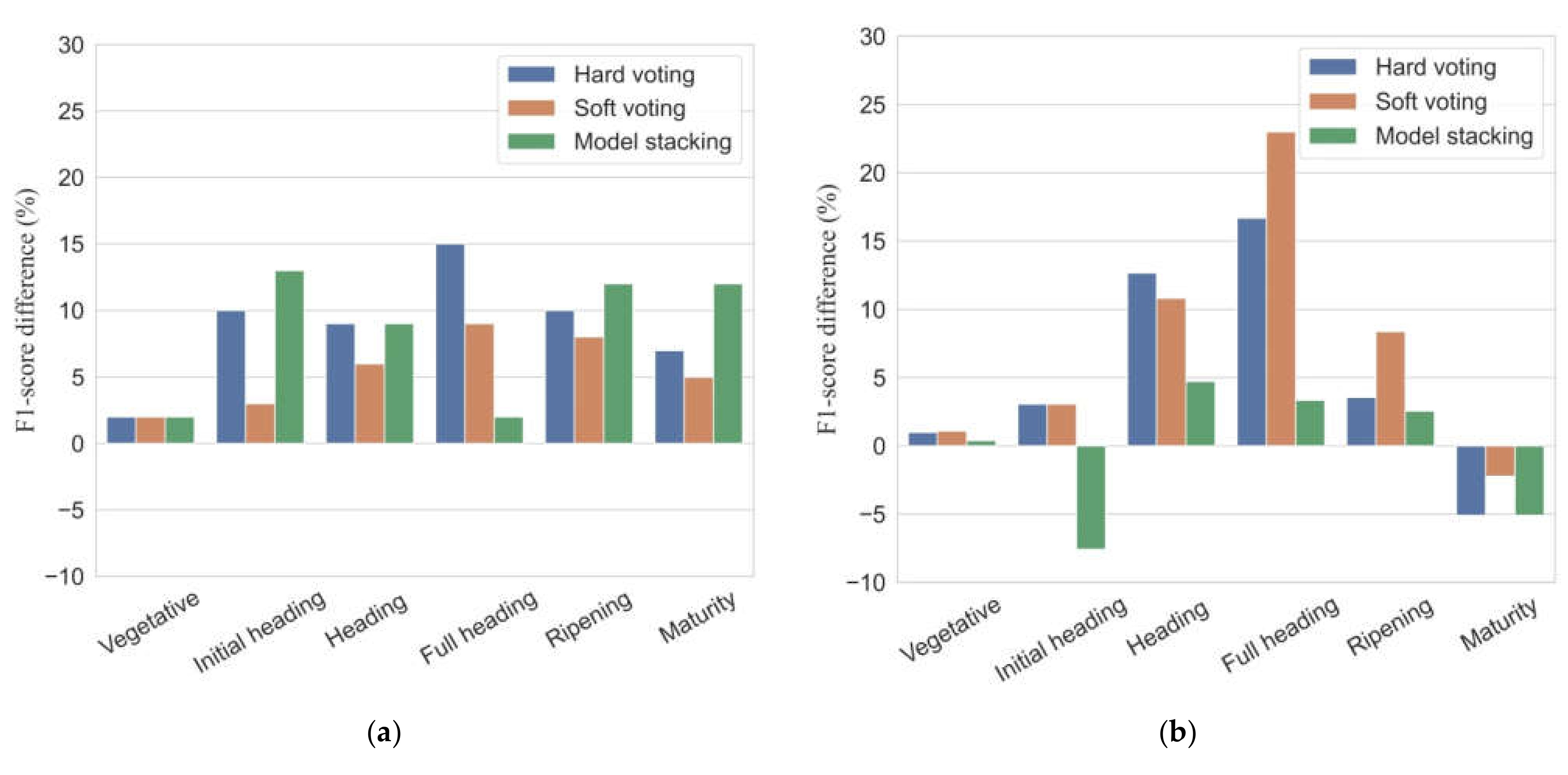
3.4. Single Best Model vs. Ensemble Models
4. Discussion
4.1. Feasibility of Ensemble Models for Phenology Detection
4.2. Combination of Feature Variables
4.3. Potential of UAV-Based Imagery in Rice Breeding Trails
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Model | Parameters | Range | The Optimal Value |
|---|---|---|---|
| RF | max_depth | 10–100 (interval: 10) | 50 |
| n_estimators | 3–15 (interval: 1) | 6 | |
| KNN | n_neighbors | 1–5 | 5 |
| GNB | -- | -- | default |
| SVM | C | 1–11 | 2 |
| kernel | ‘linear’ and ‘rbf’ | ‘linear’ | |
| LR | C | 1-11 | 4 |
| penalty | ‘L1′ and ‘L2′ | ‘L1′ | |
| solver | ‘liblinear’, ‘lbfgs’, ‘newton-cg’,’sag’ | ‘liblinear’ |
References
- Ma, Y.; Jiang, Q.; Wu, X.; Zhu, R.; Gong, Y.; Peng, Y.; Duan, B.; Fang, S. Monitoring hybrid rice phenology at initial heading stage based on low-altitude remote sensing data. Remote Sens. 2021, 13, 86. [Google Scholar] [CrossRef]
- Stoeckli, R.; Rutishauser, T.; Dragoni, D.; O’Keefe, J.; Thornton, P.E.; Jolly, M.; Lu, L.; Denning, A.S. Remote sensing data assimilation for a prognostic phenology model. J. Geophys. Res. Biogeosci. 2008, 113, G04021. [Google Scholar] [CrossRef]
- Mongiano, G.; Titone, P.; Pagnoncelli, S.; Sacco, D.; Tamborini, L.; Pilu, R.; Bregaglio, S. Phenotypic variability in Italian rice germplasm. Eur. J. Agron. 2020, 120, 126131. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Han, J.; Yu, J.; Huang, K. A near real-time deep learning approach for detecting rice phenology based on UAV images. Agric. For. Meteorol. 2020, 287, 107938. [Google Scholar] [CrossRef]
- Haghshenas, A.; Emam, Y. Image-based tracking of ripening in wheat cultivar mixtures: A quantifying approach parallel to the conventional phenology. Comput. Electron. Agric. 2019, 156, 318–333. [Google Scholar] [CrossRef]
- Pan, Z.; Huang, J.; Zhou, Q.; Wang, L.; Cheng, Y.; Zhang, H.; Blackburn, G.A.; Yan, J.; Liu, J. Mapping crop phenology using NDVI time-series derived from HJ-1 A/B data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 188–197. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, T.; Wardlow, B.D.; Gitelson, A.A.; Verma, S.B.; Suyker, A.E.; Arkebauer, T.J. A two-step filtering approach for detecting maize and soybean phenology with time-series MODIS data. Remote Sens. Environ. 2010, 114, 2146–2159. [Google Scholar] [CrossRef]
- Sakamoto, T.; Yokozawa, M.; Toritani, H.; Shibayama, M.; Ishitsuka, N.; Ohno, H. A crop phenology detection method using time-series MODIS data. Remote Sens. Environ. 2005, 96, 366–374. [Google Scholar] [CrossRef]
- Thompson, D.R.; Wehmanen, O.A. Using Landsat digital data to detect moisture stress. Photogramm. Eng. Remote Sens. 1979, 45, 201–207. [Google Scholar]
- Qiu, B.; Luo, Y.; Tang, Z.; Chen, C.; Lu, D.; Huang, H.; Chen, Y.; Chen, N.; Xu, W. Winter wheat mapping combining variations before and after estimated heading dates. ISPRS J. Photogramm. Remote Sens. 2017, 123, 35–46. [Google Scholar] [CrossRef]
- Wu, X.; Yang, W.; Wang, C.; Shen, Y.; Kondoh, A. Interactions among the phenological events of winter wheat in the north China plain-based on field data and improved MODIS estimation. Remote Sens. 2019, 11, 2976. [Google Scholar] [CrossRef] [Green Version]
- Singha, M.; Wu, B.; Zhang, M. An object-based paddy rice classification using multi-spectral data and crop phenology in Assam, northeast India. Remote Sens. 2016, 8, 479. [Google Scholar] [CrossRef] [Green Version]
- Onojeghuo, A.O.; Blackburn, G.A.; Wang, Q.; Atkinson, P.M.; Kindred, D.; Miao, Y. Rice crop phenology mapping at high spatial and temporal resolution using downscaled MODIS time-series. GIsci. Remote Sens. 2018, 55, 659–677. [Google Scholar] [CrossRef] [Green Version]
- Singha, M.; Sarmah, S. Incorporating crop phenological trajectory and texture for paddy rice detection with time series MODIS, HJ-1A and ALOS PALSAR imagery. Eur. J. Remote Sens. 2019, 52, 73–87. [Google Scholar] [CrossRef] [Green Version]
- Kibret, K.S.; Marohn, C.; Cadisch, G. Use of MODIS EVI to map crop phenology, identify cropping systems, detect land use change and drought risk in Ethiopia—An application of Google Earth Engine. Eur. J. Remote Sens. 2020, 53, 176–191. [Google Scholar] [CrossRef]
- Ren, J.; Campbell, J.B.; Shao, Y. Estimation of SOS and EOS for midwestern US corn and soybean crops. Remote Sens. 2017, 9, 722. [Google Scholar] [CrossRef] [Green Version]
- Wardlow, B.D.; Kastens, J.H.; Egbert, S.L. Using USDA crop progress data for the evaluation of greenup onset date calculated from MODIS 250-meter data. Photogramm. Eng. Remote Sens. 2006, 72, 1225–1234. [Google Scholar] [CrossRef] [Green Version]
- Vrieling, A.; Meroni, M.; Darvishzadeh, R.; Skidmore, A.K.; Wang, T.; Zurita-Milla, R.; Oosterbeek, K.; O’Connor, B.; Paganini, M. Vegetation phenology from Sentinel-2 and field cameras for a Dutch barrier island. Remote Sens. Environ. 2018, 215, 517–529. [Google Scholar] [CrossRef]
- Zhou, M.; Ma, X.; Wang, K.; Cheng, T.; Tian, Y.; Wang, J.; Zhu, Y.; Hu, Y.; Niu, Q.; Gui, L.; et al. Detection of phenology using an improved shape model on time-series vegetation index in wheat. Comput. Electron. Agric. 2020, 173, 105398. [Google Scholar] [CrossRef]
- Wang, H.; Magagi, R.; Goita, K.; Trudel, M.; McNairn, H.; Powers, J. Crop phenology retrieval via polarimetric SAR decomposition and Random Forest algorithm. Remote Sens. Environ. 2019, 231, 111234. [Google Scholar] [CrossRef]
- Fikriyah, V.N.; Darvishzadeh, R.; Laborte, A.; Khan, N.I.; Nelson, A. Discriminating transplanted and direct seeded rice using Sentinel-1 intensity data. Int. J. Appl. Earth Obs. Geoinf. 2019, 76, 143–153. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Li, S.; Wang, Y.; Dai, L.; Lin, S. Monitoring rice phenology based on backscattering characteristics of multi-temporal RADARSAT-2 datasets. Remote Sens. 2018, 10, 340. [Google Scholar] [CrossRef] [Green Version]
- Duan, T.; Chapman, S.C.; Guo, Y.; Zheng, B. Dynamic monitoring of NDVI in wheat agronomy and breeding trials using an unmanned aerial vehicle. Field Crop. Res. 2017, 210, 71–80. [Google Scholar] [CrossRef]
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Park, J.Y.; Muller-Landau, H.C.; Lichstein, J.W.; Rifai, S.W.; Dandois, J.P.; Bohlman, S.A. Quantifying leaf phenology of individual trees and species in a tropical forest using Unmanned Aerial Vehicle (UAV) images. Remote Sens. 2019, 11, 1534. [Google Scholar] [CrossRef] [Green Version]
- Burkart, A.; Hecht, V.L.; Kraska, T.; Rascher, U. Phenological analysis of unmanned aerial vehicle based time series of barley imagery with high temporal resolution. Precis. Agric. 2017, 19, 134–146. [Google Scholar] [CrossRef]
- Chen, J.; Jonsson, P.; Tamura, M.; Gu, Z.H.; Matsushita, B.; Eklundh, L. A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky-Golay filter. Remote Sens. Environ. 2004, 91, 332–344. [Google Scholar] [CrossRef]
- Jonsson, P.; Eklundh, L. Seasonality extraction by function fitting to time-series of satellite sensor data. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Z.; Tao, F.; Zhang, L.; Luo, Y.; Zhang, J.; Han, J.; Xie, J. Integrating multi-source data for rice yield prediction across China using machine learning and deep learning approaches. Agric. For. Meteorol. 2021, 297, 108275. [Google Scholar] [CrossRef]
- Carbonneau, P.E.; Dugdale, S.J.; Breckon, T.P.; Dietrich, J.T.; Fonstad, M.A.; Miyamoto, H.; Woodget, A.S. Adopting deep learning methods for airborne RGB fluvial scene classification. Remote Sens. Environ. 2020, 251, 112107. [Google Scholar] [CrossRef]
- Li, J.; Wu, Z.; Hu, Z.; Li, Z.; Wang, Y.; Molinier, M. Deep learning based thin cloud removal fusing vegetation red edge and short wave infrared spectral information for Sentinel-2A imagery. Remote Sens. 2021, 13, 157. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Lozada, D.N.; Zhang, Z.; Pumphrey, M.O.; Carter, A.H. Deep Learning for Predicting Complex Traits in Spring Wheat Breeding Program. Front. Plant Sci. 2021, 11, 613325. [Google Scholar] [CrossRef]
- Hieu, P.; Olafsson, S. On Cesaro Averages for Weighted Trees in the Random Forest. J. Classif. 2020, 37, 223–236. [Google Scholar] [CrossRef]
- Hieu, P.; Olafsson, S. Bagged ensembles with tunable parameters. Comput. Intell. 2019, 35, 184–203. [Google Scholar] [CrossRef]
- Shahhosseini, M.; Hu, G.; Archontoulis, S.V. Forecasting Corn Yield With Machine Learning Ensembles. Front. Plant Sci. 2020, 11, 1120. [Google Scholar] [CrossRef]
- Masiza, W.; Chirima, J.G.; Hamandawana, H.; Pillay, R. Enhanced mapping of a smallholder crop farming landscape through image fusion and model stacking. Int. J. Remote Sens. 2020, 41, 8739–8756. [Google Scholar] [CrossRef]
- Fageria, N.K. Yield physiology of rice. J. Plant Nutr. 2007, 30, 843–879. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Li, D.; Zhou, X.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Evaluation of RGB, color-infrared and multispectral images acquired from unmanned aerial systems for the estimation of nitrogen accumulation in rice. Remote Sens. 2018, 10, 824. [Google Scholar] [CrossRef] [Green Version]
- Lucieer, A.; Turner, D.; King, D.H.; Robinson, S.A. Using an Unmanned Aerial Vehicle (UAV) to capture micro-topography of Antarctic moss beds. Int. J. Appl. Earth Obs. Geoinf. 2014, 27, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.; Li, L.; Schmitz, N.; Tiaz, L.F.; Greenberg, J.A.; Diers, B.W. Development of methods to improve soybean yield estimation and predict plant maturity with an unmanned aerial vehicle based platform. Remote Sens. Environ. 2016, 187, 91–101. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Han, T.; Yu, X.; Ye, H.; Hu, H.; Hu, Z. Evaluation of grain yield based on digital images of rice canopy. Plant Methods 2019, 15, 28. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Vonbargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. Am. Soc. Agric. Biol. Eng. 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Walker, C.N.; Angus, J.F. Estimating the nitrogen status of crops using a digital camera. Field Crop. Res. 2010, 118, 221–227. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Li, Z.; Song, X.; Li, Z.; Xu, X.; Wang, P.; Zhao, C. Winter wheat nitrogen status estimation using UAV-based RGB imagery and gaussian processes regression. Remote Sens. 2020, 12, 3778. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Hall-Beyer, M. Practical guidelines for choosing GLCM textures to use in landscape classification tasks over a range of moderate spatial scales. Int. J. Remote Sens. 2017, 38, 1312–1338. [Google Scholar] [CrossRef]
- Li, S.; Yuan, F.; Ata-Ui-Karim, S.T.; Zheng, H.; Cheng, T.; Liu, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cao, Q. Combining color indices and textures of UAV-based digital imagery for rice LAI estimation. Remote Sens. 2019, 11, 1763. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.; Yang, G.; Tian, Q.; Feng, H.; Xu, K.; Zhou, C. Estimate of winter-wheat above-ground biomass based on UAV ultrahigh-ground-resolution image textures and vegetation indices. ISPRS J. Photogramm. Remote Sens. 2019, 150, 226–244. [Google Scholar] [CrossRef]
- Zheng, H.; Ma, J.; Zhou, M.; Li, D.; Yao, X.; Cao, W.; Zhu, Y.; Cheng, T. Enhancing the nitrogen signals of rice canopies across critical growth stages through the integration of textural and spectral information from Unmanned Aerial Vehicle (UAV) multispectral imagery. Remote Sens. 2020, 12, 957. [Google Scholar] [CrossRef] [Green Version]
- Wood, E.M.; Pidgeon, A.M.; Radeloff, V.C.; Keuler, N.S. Image texture as a remotely sensed measure of vegetation structure. Remote Sens. Environ. 2012, 121, 516–526. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Aguilar, R.; Zurita-Milla, R.; Izquierdo-Verdiguier, E.; de By, R.A. A cloud-based multi-temporal ensemble classifier to map smallholder farming systems. Remote Sens. 2018, 10, 729. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.C.; Gao, R.; Wang, L.M. Bayesian network classifiers based on Gaussian kernel density. Expert Syst. Appl. 2016, 51, 207–217. [Google Scholar] [CrossRef]
- Buhlmann, P.; Yu, B. Additive logistic regression: A statistical view of boosting—Discussion. Ann. Stat. 2000, 28, 377–386. [Google Scholar] [CrossRef]
- Pena, J.M.; Gutierrez, P.A.; Hervas-Martinez, C.; Six, J.; Plant, R.E.; Lopez-Granados, F. Object-based image classification of summer crops with machine learning methods. Remote Sens. 2014, 6, 5019–5041. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, P.A.; Lopez-Granados, F.; Pena-Barragan, J.M.; Jurado-Exposito, M.; Hervas-Martinez, C. Logistic regression product-unit neural networks for mapping Ridolfia segetum infestations in sunflower crop using multitemporal remote sensed data. Comput. Electron. Agric. 2008, 64, 293–306. [Google Scholar] [CrossRef]
- Polikar, R. Essemble based systems in decision making. IEEE Circuits Syst. Mag. 2006, 6, 21–45. [Google Scholar] [CrossRef]
- Wozniak, M.; Grana, M.; Corchado, E. A survey of multiple classifier systems as hybrid systems. Inf. Fusion 2014, 16, 3–17. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Cheng, T.; Yao, X.; Deng, X.; Tian, Y.; Cao, W.; Zhu, Y. Detection of rice phenology through time series analysis of ground-based spectral index data. Field Crop. Res. 2016, 198, 131–139. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, X.; Wei, H.; Wang, D.; Chen, R.; Wang, L.; Gu, W. Predicting the invasive trend of exotic plants in China based on the ensemble model under climate change: A case for three invasive plants of Asteraceae. Sci. Total Environ. 2021, 756, 143841. [Google Scholar] [CrossRef]
- Lin, F.; Zhang, D.; Huang, Y.; Wang, X.; Chen, X. Detection of corn and weed species by the combination of spectral, shape and textural features. Sustainability 2017, 9, 1335. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, S.; Li, J.; Guo, X.; Wang, S.; Lu, J. Estimating biomass of winter oilseed rape using vegetation indices and texture metrics derived from UAV multispectral images. Comput. Electron. Agric. 2019, 166, 105026. [Google Scholar] [CrossRef]
- Volpato, L.; Pinto, F.; Gonzalez-Perez, L.; Thompson, I.G.; Borem, A.; Reynolds, M.; Gerard, B.; Molero, G.; Rodrigues, F.A., Jr. High throughput field phenotyping for plant height using uav-based rgb imagery in wheat breeding lines: Feasibility and validation. Front. Plant Sci. 2021, 12, 591587. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop phenomics: Current status and perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef] [Green Version]




| Date of UAV Flights | Vegetative | Initial Heading | Heading | Full Heading | Ripening | Maturity |
|---|---|---|---|---|---|---|
| 1 August 2020 | 437 | -- | -- | -- | -- | -- |
| 29 August 2020 | 417 | 12 | 4 | 4 | -- | -- |
| 11 September 2020 | 5 | 78 | 148 | 91 | 115 | -- |
| 30 September 2020 | -- | -- | -- | -- | 437 | -- |
| 1 November 2020 | -- | -- | -- | -- | 75 | 362 |
| Type | Variable | Formula/Description | References |
|---|---|---|---|
| VIs | Normalized red index (NRI) | R/(R + G + B) | [42] |
| Normalized green index (NGI) | G/(R + G + B) | [42] | |
| Normalized blue index (NBI) | B/(R + G + B) | [42] | |
| Normalized excess green index (ExG) | (2G − R − B)/(G + R + B) | [43] | |
| Normalized excess red index (ExR) | (1.4R − G)/(G + R + B) | [44] | |
| Green and red index (VIgreen) | (G−R)/(G+R) | [45] | |
| Color space | R band of UAV image (R) | DN values of R band | -- |
| G band of UAV image (G) | DN values of G band | -- | |
| B band of UAV image (B) | DN values of B band | -- | |
| Hue (H) | The DN values of hue | [46] | |
| Saturation (S) | The DN values of saturation | [46] | |
| Value (V) | The DN values of value | [46] |
| Data Set | Models | Phenological Stage | |||||
|---|---|---|---|---|---|---|---|
| Vegetative | Initial Heading | Heading | Full Heading | Ripening | Maturity | ||
| Calibration | RF | 1.00 | 0.64 | 0.79 | 0.75 | 0.95 | 0.93 |
| KNN | 0.98 | 0.63 | 0.70 | 0.49 | 0.86 | 0.86 | |
| GNB | 0.98 | 0.51 | 0.69 | 0.57 | 0.82 | 0.81 | |
| SVM | 1.00 | 0.53 | 0.74 | 0.62 | 0.89 | 0.83 | |
| LR | 1.00 | 0.59 | 0.74 | 0.39 | 0.89 | 0.84 | |
| Validation | RF | 0.97 | 0.52 | 0.59 | 0.53 | 0.85 | 0.89 |
| KNN | 0.92 | 0.27 | 0.42 | 0.14 | 0.79 | 0.88 | |
| GNB | 0.96 | 0.50 | 0.54 | 0.57 | 0.88 | 0.98 | |
| SVM | 0.95 | 0.47 | 0.55 | 0.57 | 0.85 | 0.91 | |
| LR | 0.97 | 0.53 | 0.65 | 0.44 | 0.87 | 0.83 | |
| Data Set | Models | Phenological Stage | |||||
|---|---|---|---|---|---|---|---|
| Vegetative | Initial Heading | Heading | Full Heading | Ripening | Maturity | ||
| Calibration | Hard voting | 1.00 | 0.61 | 0.78 | 0.72 | 0.92 | 0.88 |
| Soft voting | 1.00 | 0.54 | 0.75 | 0.66 | 0.90 | 0.86 | |
| Model stacking | 1.00 | 0.64 | 0.78 | 0.59 | 0.94 | 0.93 | |
| Validation | Hard voting | 0.97 | 0.53 | 0.67 | 0.74 | 0.92 | 0.93 |
| Soft voting | 0.97 | 0.53 | 0.65 | 0.80 | 0.96 | 0.96 | |
| Model stacking | 0.96 | 0.42 | 0.60 | 0.62 | 0.91 | 0.93 | |
| Type | Number | Features | Soft Voting | Hard Voting | Model Stacking |
|---|---|---|---|---|---|
| RGB-VIs | 1 | VIgreen | 9 | 11 | 8 |
| 2 | ExG | 8 | 8 | 6 | |
| 3 | ExR | 3 | 3 | 5 | |
| 4 | NRI | 7 | 7 | 7 | |
| 5 | NGI | 15 | 16 | 13 | |
| 6 | NBI | 19 | 19 | 18 | |
| Color space | 7 | R | 13 | 9 | 10 |
| 8 | G | 18 | 15 | 15 | |
| 9 | B | 5 | 6 | 2 | |
| 10 | H | 2 | 2 | 1 | |
| 11 | S | 4 | 4 | 4 | |
| 12 | V | 17 | 18 | 14 | |
| Texture | 13 | Contrast | 1 | 1 | 3 |
| 14 | Correlation | 20 | 17 | 20 | |
| 15 | Dissimilarity | 10 | 20 | 11 | |
| 16 | Entropy | 14 | 12 | 12 | |
| 17 | Homogeneity | 12 | 13 | 17 | |
| 18 | Mean | 16 | 14 | 19 | |
| 19 | Second moment | 11 | 10 | 16 | |
| 20 | Variance | 6 | 5 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, H.; Ma, F.; Li, Z.; Tan, Z.; Du, C. Improved Accuracy of Phenological Detection in Rice Breeding by Using Ensemble Models of Machine Learning Based on UAV-RGB Imagery. Remote Sens. 2021, 13, 2678. https://doi.org/10.3390/rs13142678
Ge H, Ma F, Li Z, Tan Z, Du C. Improved Accuracy of Phenological Detection in Rice Breeding by Using Ensemble Models of Machine Learning Based on UAV-RGB Imagery. Remote Sensing. 2021; 13(14):2678. https://doi.org/10.3390/rs13142678
Chicago/Turabian StyleGe, Haixiao, Fei Ma, Zhenwang Li, Zhengzheng Tan, and Changwen Du. 2021. "Improved Accuracy of Phenological Detection in Rice Breeding by Using Ensemble Models of Machine Learning Based on UAV-RGB Imagery" Remote Sensing 13, no. 14: 2678. https://doi.org/10.3390/rs13142678
APA StyleGe, H., Ma, F., Li, Z., Tan, Z., & Du, C. (2021). Improved Accuracy of Phenological Detection in Rice Breeding by Using Ensemble Models of Machine Learning Based on UAV-RGB Imagery. Remote Sensing, 13(14), 2678. https://doi.org/10.3390/rs13142678