Detection of Banana Plants Using Multi-Temporal Multispectral UAV Imagery
Abstract
1. Introduction
2. Materials and Methods
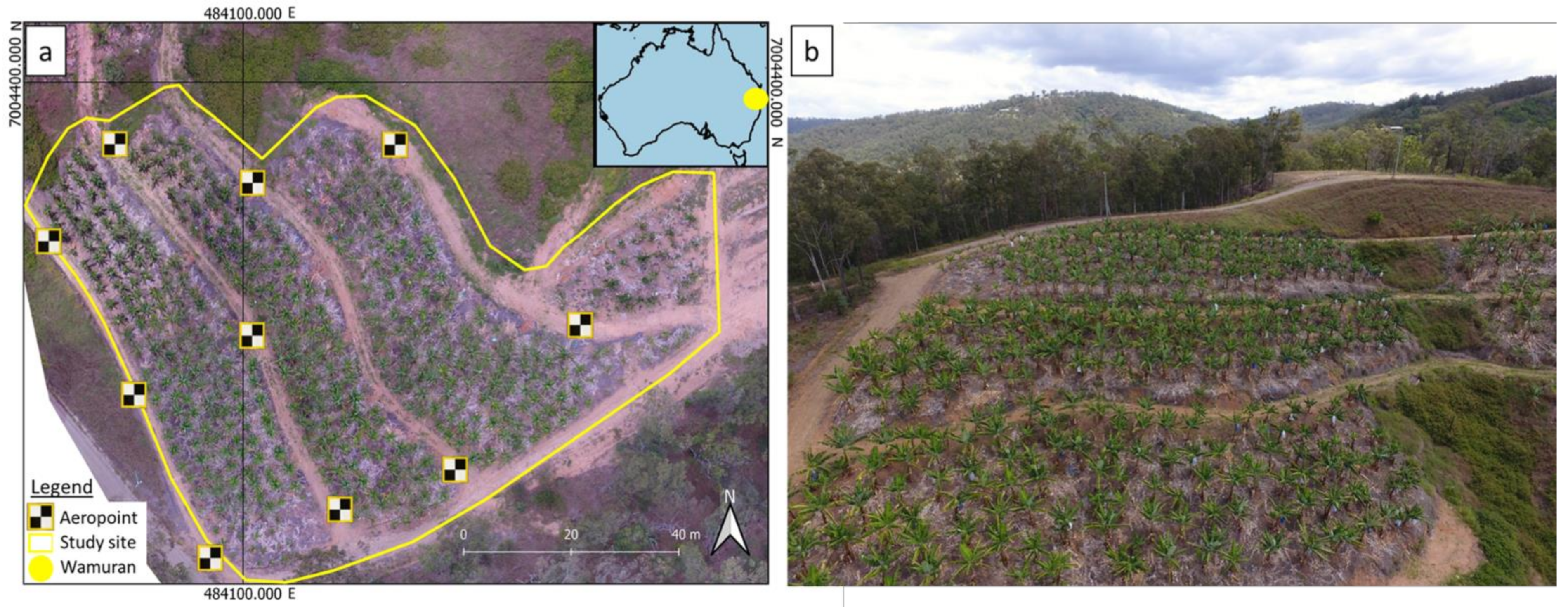
2.1. Study Location
2.2. UAV Data Collection and Ground Validation
2.3. UAV Data Pre-Processing
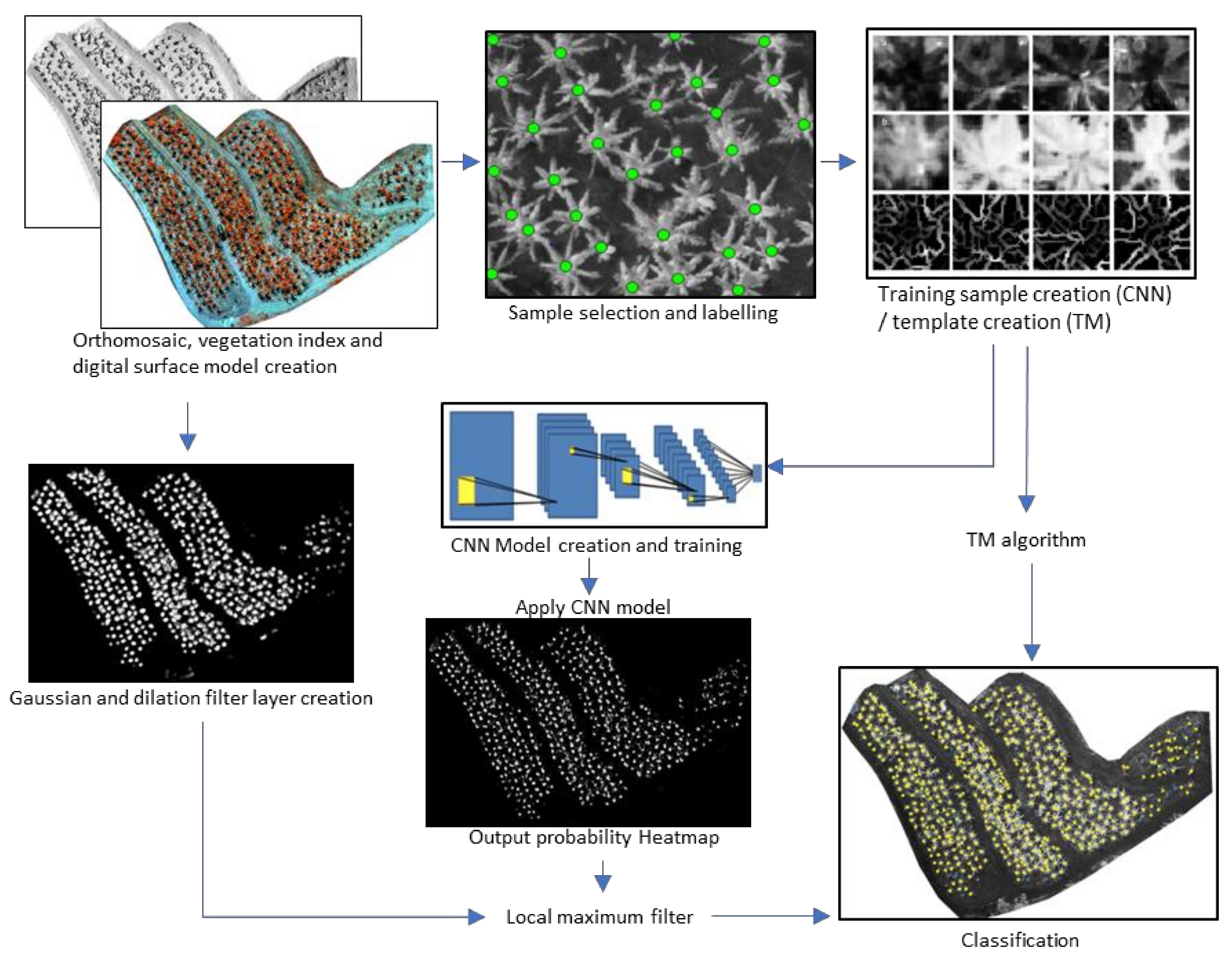
2.4. Image Classification
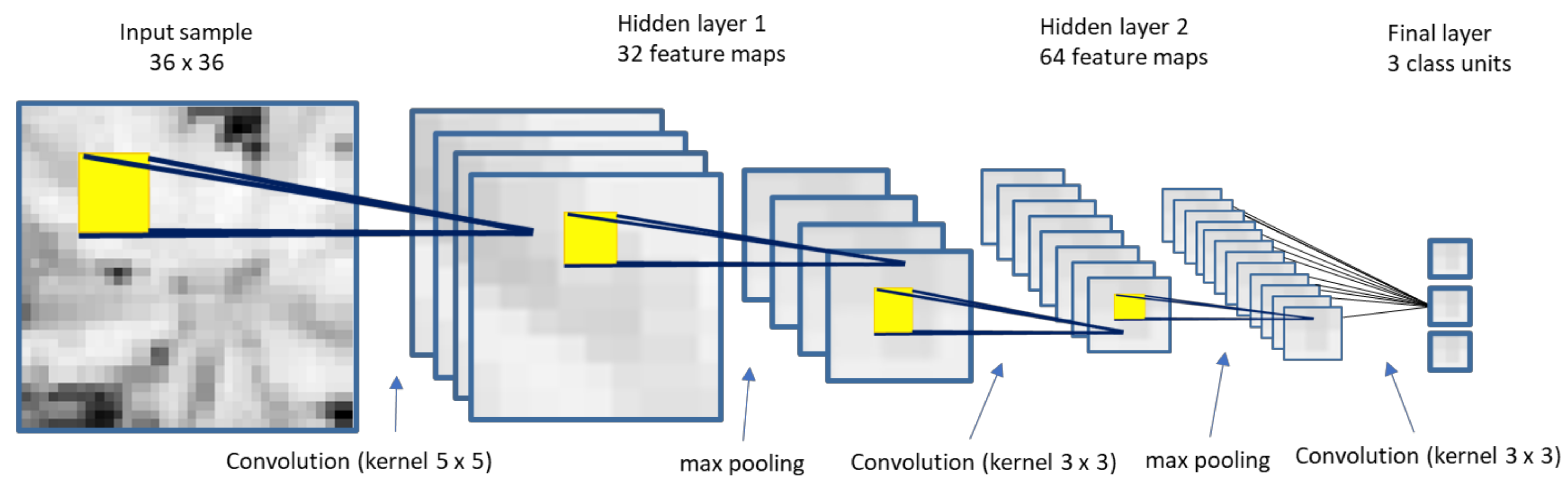
2.4.1. CNN
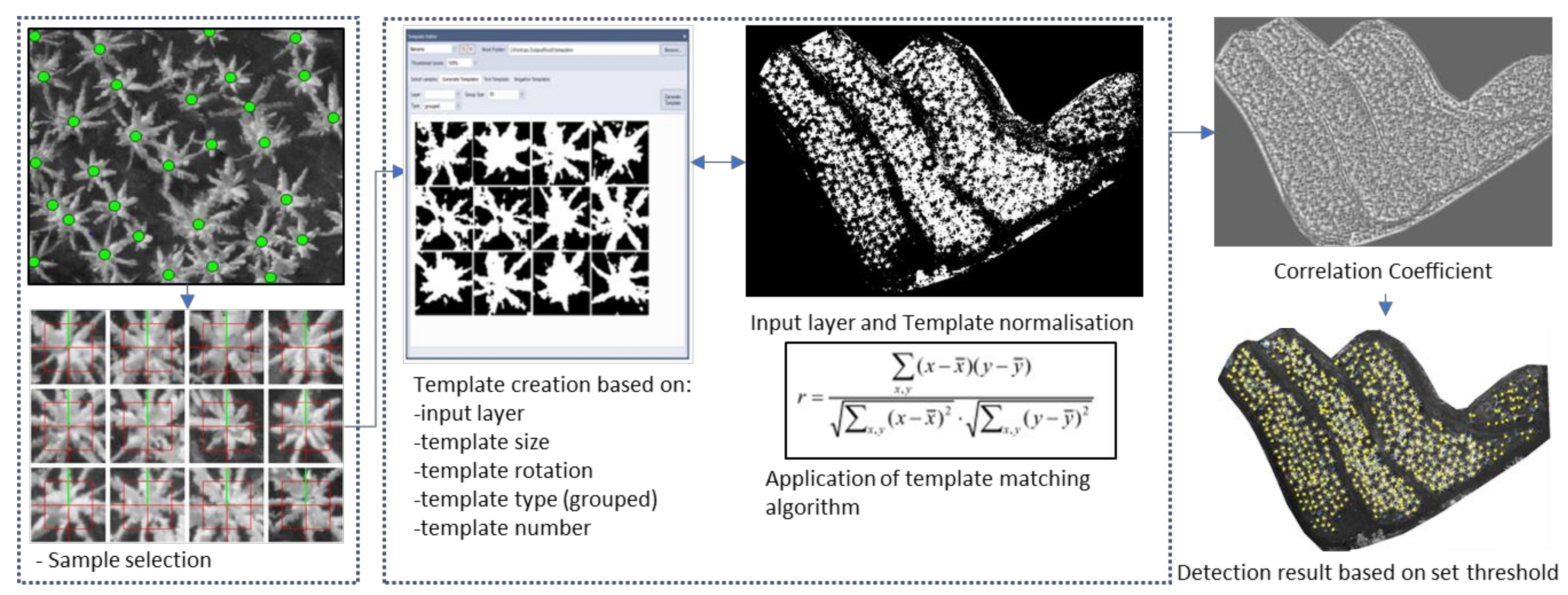
2.4.2. Template Matching
2.4.3. Local Maxima Filter
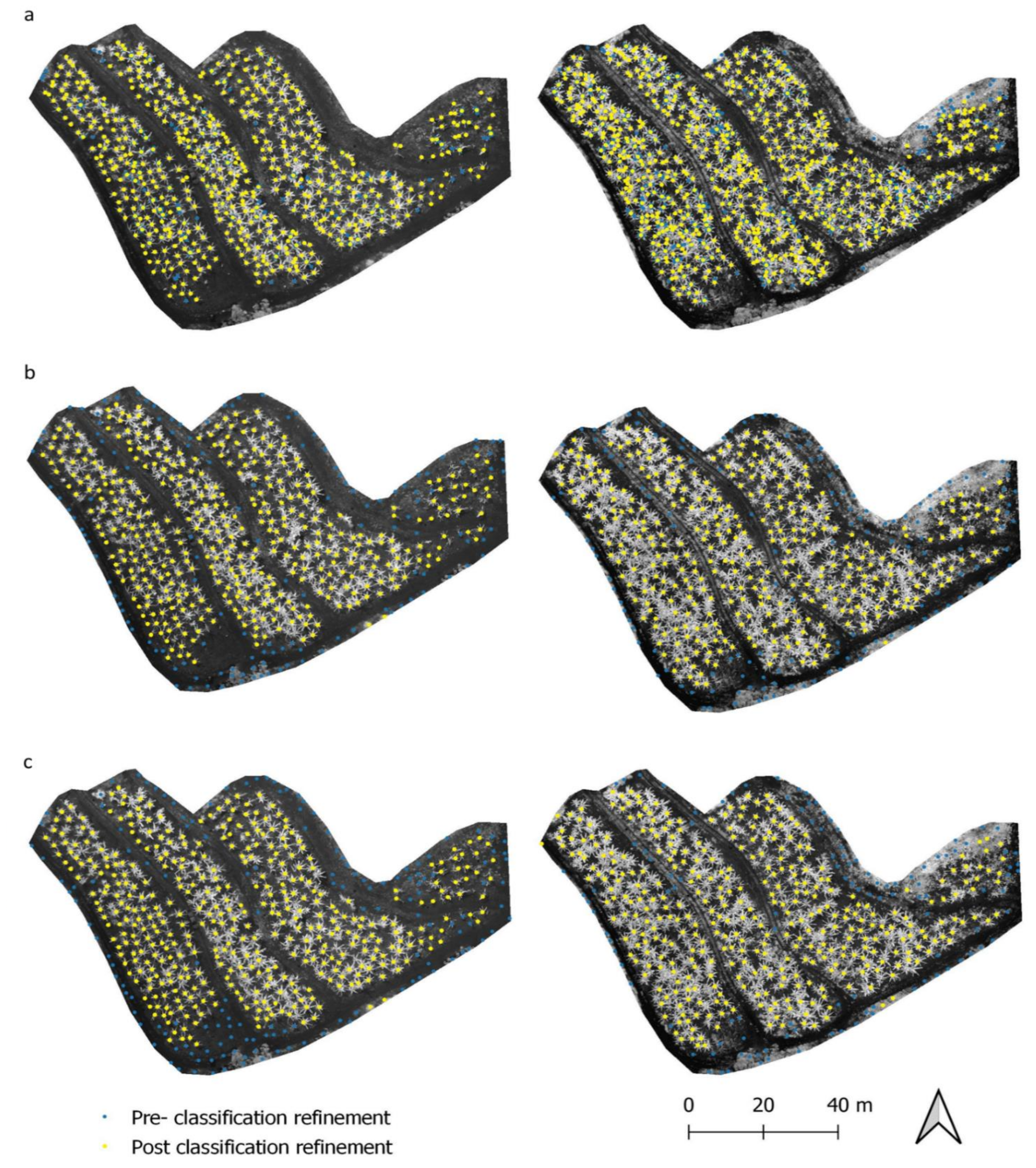
2.4.4. Classification Refinement
2.4.5. Accuracy Assessment
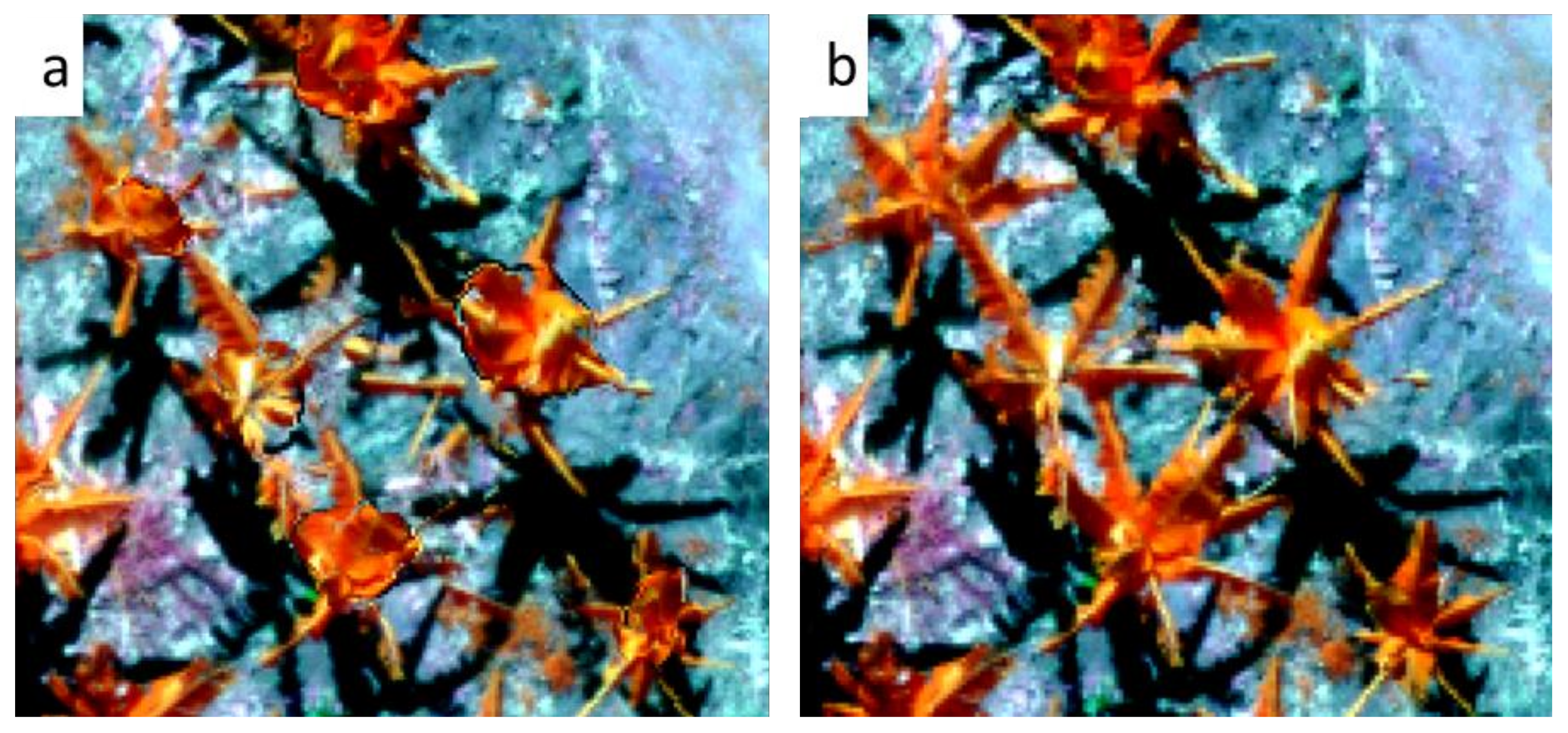
2.5. Relation of Plant Morphology and Detection Rate
3. Results
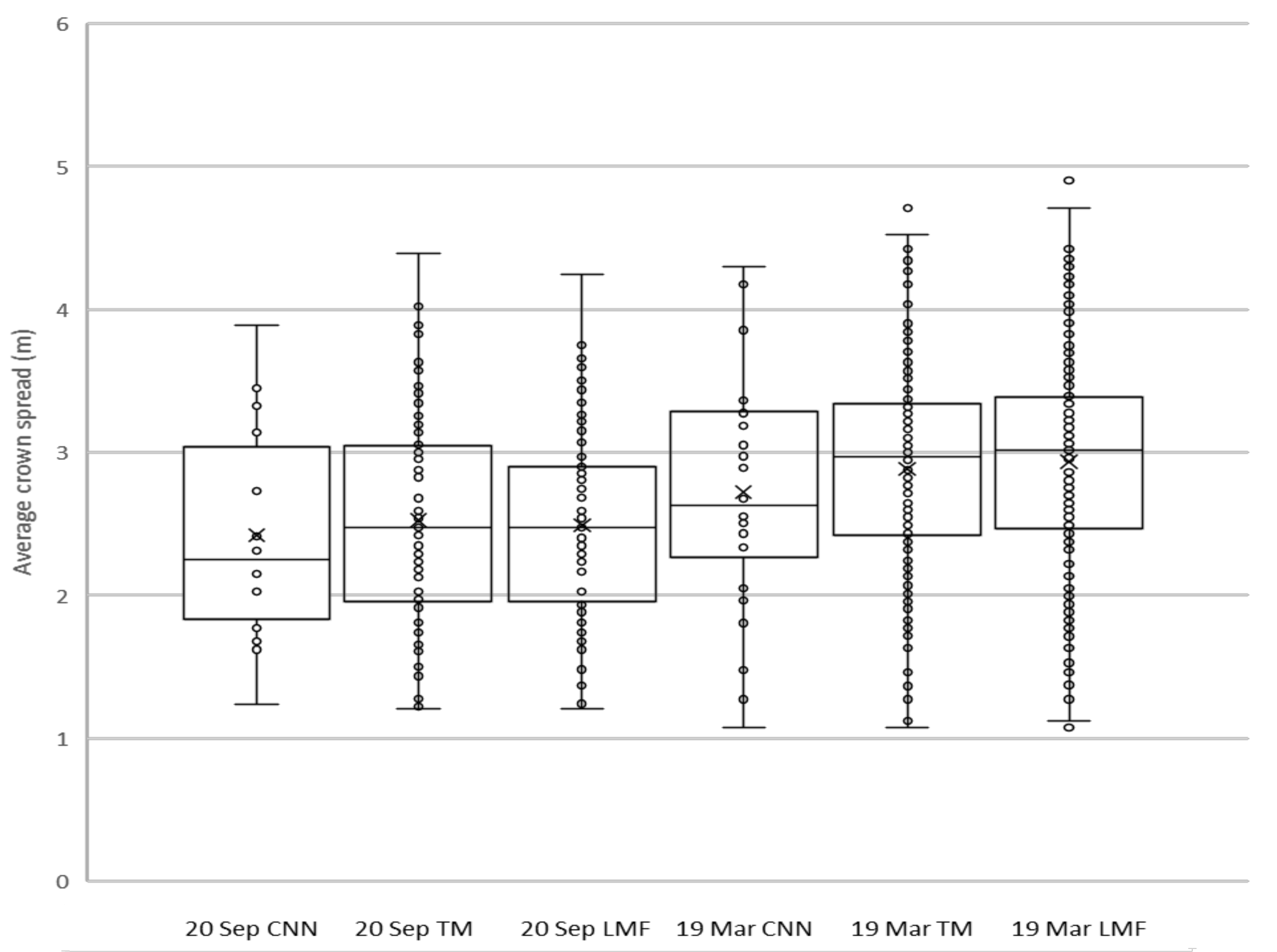
3.1. Detection Rate of Banana Plants
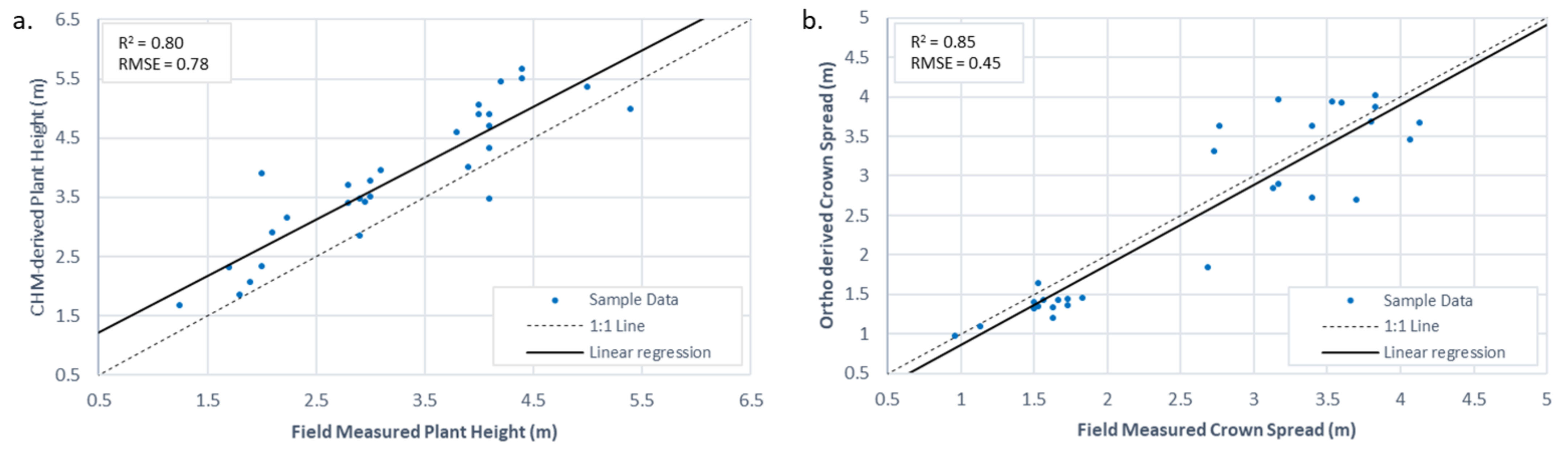
3.2. Individual Plant Morphology Estimation
3.3. Effects of Crown Morphologies on Banana Plant Detection Rates
4. Discussion
4.1. Evaluation of the Different Classification Methods
4.2. Evaluation of Classification Refinement
4.3. Application Effectiveness for Multi-Temporal Detections
4.4. Evalution of Crown Morphology on Detection Success
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scott, G.J. A review of root, tuber and banana crops in developing countries: Past, present and future. Int. J. Food Sci. Technol. 2021, 56, 1093–1114. [Google Scholar] [CrossRef] [PubMed]
- FAO. Banana Facts and Figures. Available online: http://www.fao.org/economic/est/est-commodities/bananas/bananafacts/en/#.XXh7-Sgzb-h (accessed on 30 November 2020).
- FAO. Banana Market Review February 2020 Snapshot. Available online: http://www.fao.org/3/ca9212en/ca9212en.pdf (accessed on 30 November 2020).
- Picq, C.; Fouré, E.; Frison, E.A. Bananas and Food Security; Bioversity International: Maccarese-Stazione, Italy, 1999; Available online: https://www.bioversityinternational.org/fileadmin/user_upload/online_library/publications/pdfs/709.pdf (accessed on 30 November 2020).
- Horticulture Innovation. Australian Horticulture Statistics Handbook; Hort Innovation: Sydney, Australia, 2020; Available online: https://www.horticulture.com.au/growers/help-your-business-grow/research-reports-publications-fact-sheets-and-more/grower-resources/ha18002-assets/australian-horticulture-statistics-handbook/ (accessed on 12 May 2021).
- Broadley, R.; Chay-Prove, P.; Rigden, P.; Daniells, J.; Treverrow, N.; Akehurst, A.; Newley, P.; Harris, D.; Pattison, T.; McCarthy, P.; et al. Subtropical Banana Information Kit Your Growing Guide to Better Farming Guide; Manual. Agrilink Series QI04011; Department of Primary Industries, Queensland Horticulture Institute: Brisbane, QLD, Australia, 2004. [Google Scholar]
- Twyford, I.T. Banana nutrition: A review of principles and practice. J. Sci. Food Agric. 1967, 18, 177. [Google Scholar] [CrossRef] [PubMed]
- Memon, N.; Memon, K.; Shah, Z.-u.-h. Plant Analysis as a diagnostic tool for evaluating nutritional requirements of bananas. Int. J. Agric. Biol. 2005, 7, 824–831. [Google Scholar]
- Bouwman, A.F.; Beusen, A.H.W.; Lassaletta, L.; van Apeldoorn, D.F.; van Grinsven, H.J.M.; Zhang, J.; van Ittersum, M.K. Lessons from temporal and spatial patterns in global use of N and P fertilizer on cropland. Sci. Rep. 2017, 7, 40366. [Google Scholar] [CrossRef]
- Good, A.G.; Beatty, P.H. Fertilizing nature: A tragedy of excess in the commons. PLoS Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef]
- Joyce Dória Rodrigues, S.; Pasqual, M.; Rodrigues, F.A.; Lacerda, W.S.; Sergio Luiz Rodrigues, D.; Sebastião de Oliveira, S.; Paixão, C.A. Correlation between morphological characters and estimated bunch weight of the Tropical banana cultivar. Afr. J. Biotechnol. 2012, 11, 10682. [Google Scholar] [CrossRef]
- Swarupa, V.; Ravishankar, K.V.; Rekha, A. Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta 2014, 239, 735–751. [Google Scholar] [CrossRef]
- Barker, W.; Steward, F. Growth and Development of the Banana Plant II. The Transition from the Vegetative to the Floral Shoot in Musa acuminata cv. Gros Michel. Ann. Bot. 1962, 26, 413–423. [Google Scholar] [CrossRef]
- Lamour, J.; Leroux, C.; Le Moguédec, G.; Naud, O.; Léchaudel, M.; Tisseyre, B. Disentangling the sources of chlorophyll-content variability in banana fields. In Precision Agriculture ’19; Wageningen Academic Publishers: Wageningen, The Netherlands, 2019; pp. 407–417. [Google Scholar]
- Lamour, J.; Le Moguédec, G.; Naud, O.; Lechaudel, M.; Taylor, J.; Tisseyre, B. Evaluating the drivers of banana flowering cycle duration using a stochastic model and on farm production data. Precis. Agric. 2020. [Google Scholar] [CrossRef]
- Thomas, D.S.; Turner, D.W. Banana (Musa sp.) leaf gas exchange and chlorophyll fluorescence in response to soil drought, shading and lamina folding. Sci. Hortic. 2001, 90, 93–108. [Google Scholar] [CrossRef]
- Taylor, S.E.; Sexton, O.J. Some Implications of Leaf Tearing in Musaceae. Ecology 1972, 53, 143–149. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Tu, Y.-H.; Johansen, K.; Phinn, S.; Robson, A. Measuring Canopy Structure and Condition Using Multi-Spectral UAS Imagery in a Horticultural Environment. Remote Sens. 2019, 11, 269. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; Serrano, N.; Arquero, O.; Peña, J.M. High-throughput 3-D monitoring of agricultural-tree plantations with unmanned aerial vehicle (UAV) technology. PLoS ONE 2015, 10, e0130479. [Google Scholar] [CrossRef]
- Wu, D.; Johansen, K.; Phinn, S.; Robson, A. Suitability of airborne and terrestrial laser scanning for mapping tree crop structural metrics for improved orchard management. Remote Sens. 2020, 12, 1647. [Google Scholar] [CrossRef]
- Watts, A.C.; Ambrosia, V.G.; Hinkley, E.A. Unmanned Aircraft Systems in Remote Sensing and Scientific Research: Classification and Considerations of Use. Remote Sens. 2012, 4, 1671. [Google Scholar] [CrossRef]
- Basso, B.; Fiorentino, C.; Cammarano, D.; Schulthess, U. Variable rate nitrogen fertilizer response in wheat using remote sensing. Precis. Agric 2016, 17, 168–182. [Google Scholar] [CrossRef]
- Wang, J.; Shen, C.; Liu, N.; Jin, X.; Fan, X.; Dong, C.; Xu, Y. Non-Destructive Evaluation of the Leaf Nitrogen Concentration by In-Field Visible/Near-Infrared Spectroscopy in Pear Orchards. Sensors 2017, 17, 538. [Google Scholar] [CrossRef]
- Bolton, D.K.; Friedl, M.A. Forecasting crop yield using remotely sensed vegetation indices and crop phenology metrics. Agric. For. Meteorol. 2013, 173, 74–84. [Google Scholar] [CrossRef]
- Viña, A.; Gitelson, A.A.; Rundquist, D.C.; Keydan, G.; Leavitt, B.; Schepers, J. Monitoring Maize (L.) Phenology with Remote Sensing. Agron. J. 2004, 96, 1139. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2015, 100, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Susič, N.; Žibrat, U.; Širca, S.; Strajnar, P.; Razinger, J.; Knapič, M.; Vončina, A.; Urek, G.; Stare, B.G. Discrimination between abiotic and biotic drought stress in tomatoes using hyperspectral imaging. Sens. Actuators B Chem. 2018. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Johansen, K.; Sohlbach, M.; Sullivan, B.; Stringer, S.; Peasley, D.; Phinn, S. Mapping banana plants from high spatial resolution orthophotos to facilitate plant health assessment. Remote Sens. 2014, 6, 8261–8286. [Google Scholar] [CrossRef]
- Johansen, K.; Phinn, S.; Witte, C.; Philip, S.; Newton, L. Mapping banana plantations from object-oriented classification of SPOT-5 imagery. Photogramm. Eng. Remote Sens. 2009, 75, 1069–1081. [Google Scholar] [CrossRef]
- Clark, A.; McKechnie, J. Detecting Banana Plantations in the Wet Tropics, Australia, Using Aerial Photography and U-Net. Appl. Sci. 2020, 10, 2017. [Google Scholar] [CrossRef]
- Pinz, A. A computer vision system for the recognition of trees in aerial photographs. Multisource Data Integr. Remote Sens. 1991, 3099, 111–124. [Google Scholar]
- Pouliot, D.; King, D.; Bell, F.; Pitt, D. Automated tree crown detection and delineation in high-resolution digital camera imagery of coniferous forest regeneration. Remote Sens. Environ. 2002, 82, 322–334. [Google Scholar] [CrossRef]
- Pollock, R. Model-Based Approach to Automatically Locating Tree Crowns in High Spatial Resolution Images. In Proceedings of the SPIE Image and Signal Processing for Remote Sensing, Rome, Italy, 30 December 1994; Volume 2315. [Google Scholar] [CrossRef]
- Larsen, M.; Rudemo, M. Optimizing templates for finding trees in aerial photographs. Pattern Recognit. Lett. 1998, 19, 1153–1162. [Google Scholar] [CrossRef]
- Lamar, W.R.; McGraw, J.B.; Warner, T.A. Multitemporal censusing of a population of eastern hemlock (Tsuga canadensis L.) from remotely sensed imagery using an automated segmentation and reconciliation procedure. Remote Sens. Environ. 2005, 94, 133–143. [Google Scholar] [CrossRef]
- Bunting, P.; Lucas, R. The delineation of tree crowns in Australian mixed species forests using hyperspectral Compact Airborne Spectrographic Imager (CASI) data. Remote Sens. Environ. 2006, 101, 230–248. [Google Scholar] [CrossRef]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of Citrus Trees from Unmanned Aerial Vehicle Imagery Using Convolutional Neural Networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef]
- Li, W.; Dong, R.; Fu, H.; Yu, L. Large-Scale Oil Palm Tree Detection from High-Resolution Satellite Images Using Two-Stage Convolutional Neural Networks. Remote Sens. 2018, 11, 11. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. A review of the use of convolutional neural networks in agriculture. J. Agric. Sci. 2018, 156, 312–322. [Google Scholar] [CrossRef]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, M.; Wang, C.; Luo, L.; Li, J.; Lian, G.; Zou, X. Recognition and Localization Methods for Vision-Based Fruit Picking Robots: A Review. Front Plant Sci. 2020, 11, 510. [Google Scholar] [CrossRef]
- Harto, A.B.; Prastiwi, P.A.D.; Ariadji, F.N.; Suwardhi, D.; Dwivany, F.M.; Nuarsa, I.W.; Wikantika, K. Identification of Banana Plants from Unmanned Aerial Vehicles (UAV) Photos Using Object Based Image Analysis (OBIA) Method (A Case Study in Sayang Village, Jatinangor District, West Java). HAYATI J. Biosci. 2019, 26, 7. [Google Scholar]
- Handique, B.K.; Goswami, C.; Gupta, C.; Pandit, S.; Gogoi, S.; Jadi, R.; Jena, P.; Borah, G.; Raju, P.L.N. Hierarchical classification for assessment of horticultural crops in mixed cropping pattern using UAV-borne multi-spectral sensor. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, XLIII-B3-2020, 67–74. [Google Scholar] [CrossRef]
- Kestur, R.; Angural, A.; Bashir, B.; Omkar, S.N.; Anand, G.; Meenavathi, M.B. Tree Crown Detection, Delineation and Counting in UAV Remote Sensed Images: A Neural Network Based Spectral–Spatial Method. J. Indian Soc. Remote Sens. 2018, 46, 991–1004. [Google Scholar] [CrossRef]
- Neupane, B.; Horanont, T.; Hung, N.D. Deep learning based banana plant detection and counting using high-resolution red-green-blue (RGB) images collected from unmanned aerial vehicle (UAV). PLoS ONE 2019, 14, e0223906. [Google Scholar] [CrossRef] [PubMed]
- Gomez Selvaraj, M.; Vergara, A.; Montenegro, F.; Alonso Ruiz, H.; Safari, N.; Raymaekers, D.; Ocimati, W.; Ntamwira, J.; Tits, L.; Omondi, A.B.; et al. Detection of banana plants and their major diseases through aerial images and machine learning methods: A case study in DR Congo and Republic of Benin. ISPRS J. Photogramm. Remote Sens. 2020, 169, 110–124. [Google Scholar] [CrossRef]
- Zou, X.; Mõttus, M. Sensitivity of Common Vegetation Indices to the Canopy Structure of Field Crops. Remote Sens. 2017, 9, 994. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of red edge position and chlorophyll content by reflectance measurements near 700 nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Bureau of Meteorology. Climate Statistics for Australian Locations: Beerburrum Weather Station. Available online: http://www.bom.gov.au/climate/averages/tables/cw_040284.shtml (accessed on 18 March 2018).
- Queensland Government. Queensland Spatial Catalogue—QSpatial. Available online: https://qldspatial.information.qld.gov.au/catalogue/custom/search.pag (accessed on 25 March 2021).
- Johansen, K.; Raharjo, T.; McCabe, M.F. Using Multi-Spectral UAV Imagery to Extract Tree Crop Structural Properties and Assess Pruning Effects. Remote Sens. 2018, 10, 854. [Google Scholar] [CrossRef]
- Wang, C.; Myint, S.W. A simplified empirical line method of radiometric calibration for small unmanned aircraft systems-based remote sensing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 1876–1885. [Google Scholar] [CrossRef]
- Blozan, W. Tree measuring guidelines of the eastern native tree society. Bull. East. Nativ. Tree Soc. 2006, 1, 3–10. [Google Scholar]
- Tu, Y.-H.; Phinn, S.; Johansen, K.; Robson, A. Assessing Radiometric Correction Approaches for Multi-Spectral UAS Imagery for Horticultural Applications. Remote Sens. 2018, 10, 1684. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.; Schell, J.; Deering, D. Monitoring vegetation systems in the Great Plains with ERTS. In Proceedings of the Third Earth Resources Technology Satellite-1 Symposium, Washington, DC, USA, 10–14 December 1973; Volume 1, pp. 309–317. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Trimble. eCognition Developer User Guide; Trimble Inc.: Munich, Germany, 2020. [Google Scholar]
- Jasvilis, G.; Weise, C.; Zenger-Landolt, B. Finding complex patterns using template matching. In Proceedings of the Geobia 2016: Solution and Synergies, University of Twente Faculty of Geo-Information and Earth Observation (ITC), Enschede, The Netherlands, 14–16 September 2016. [Google Scholar]
- Ke, Y.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Turner, D.; Lahav, E. The Growth of Banana Plants in Relation to Temperature. Funct. Plant Biol. 1983, 10, 43–53. [Google Scholar] [CrossRef]
- Turner, D.W.; Fortescue, J.A.; Thomas, D.S. Environmental physiology of the bananas (Musa spp.). Braz. J. Plant Physiol. 2007, 19, 463–484. [Google Scholar] [CrossRef]
- Erikson, M.; Olofsson, K. Comparison of three individual tree crown detection methods. Mach. Vis. Appl. 2005, 16, 258–265. [Google Scholar] [CrossRef]
- Norzaki, N.; Tahar, K.N. A comparative study of template matching, ISO cluster segmentation, and tree canopy segmentation for homogeneous tree counting. Int. J. Remote Sens. 2019, 40, 7477–7499. [Google Scholar] [CrossRef]
- Larsen, M.; Eriksson, M.; Descombes, X.; Perrin, G.; Brandtberg, T.; Gougeon, F.A. Comparison of six individual tree crown detection algorithms evaluated under varying forest conditions. Int. J. Remote Sens. 2011, 32, 5827–5852. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Wu, D.; Johansen, K.; Phinn, S.; Robson, A.; Tu, Y.-H. Inter-comparison of remote sensing platforms for height estimation of mango and avocado tree crowns. Int. J. Appl. Earth Obs. Geoinf. 2020, 89, 102091. [Google Scholar] [CrossRef]


| Author | UAV/Sensor | Detection/Implementation | Region/Crop Type | Purpose |
|---|---|---|---|---|
| Kestur et al. | Fixed wing and quadcopter/GoPro RGB video footage | K-means, ELM on single-date imagery frames | India/structured smallholder open-field crops with mixed-age plants | Demonstrate potential for UAV for tree-crown studies by detection and delineation |
| Neupane et al. | DJI Phantom 3 /RGB orthomosaic | Faster RCNN on single-date imagery | Thailand/structured open-field commercial crop with young plants | Detect and count (by bounding box and localization) |
| Gomez Selvaraj et al. | DJI Phantom 4 Pro/RGB orthomosaics | Retinanet on multi-date imagery | West and Central Africa/unstructured smallholder crops with mixed-age plants | Crown detection (by bounding box) |
| Current study | 3DR Solo/Multispectral (G, R, RE, NIR) orthomosaics | CNN, TM, and LMF with GEOBIA on multi-temporal imagery | Australia/structured open-field commercial crop with mixed-age plants | Detection of inner crown of individual plants over multiple dates |
| 28 August | 20 September | 19 March | |
|---|---|---|---|
| GCP root mean square error (RMSE) | 0.01 m | 0.13 m | 0.10 m |
| Re-projection error (pixels) | 0.396 | 0.487 | 0.524 |
| Dense cloud point density (ppm2) * | 209 | 246 | 248 |
| Ground-sampling distance (GSD) | 4.22 cm/pix | 4.02 cm/pix | 4.07 cm/pix |
| 20 September | 19 March | |||||
|---|---|---|---|---|---|---|
| CNN | TM | LMF | CNN | TM | LMF | |
| Precision | 0.89 | 0.99 | 0.98 | 0.79 | 0.98 | 0.97 |
| Recall | 0.97 | 0.77 | 0.77 | 0.92 | 0.60 | 0.59 |
| F-score | 0.93 | 0.86 | 0.86 | 0.85 | 0.74 | 0.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aeberli, A.; Johansen, K.; Robson, A.; Lamb, D.W.; Phinn, S. Detection of Banana Plants Using Multi-Temporal Multispectral UAV Imagery. Remote Sens. 2021, 13, 2123. https://doi.org/10.3390/rs13112123
Aeberli A, Johansen K, Robson A, Lamb DW, Phinn S. Detection of Banana Plants Using Multi-Temporal Multispectral UAV Imagery. Remote Sensing. 2021; 13(11):2123. https://doi.org/10.3390/rs13112123
Chicago/Turabian StyleAeberli, Aaron, Kasper Johansen, Andrew Robson, David W. Lamb, and Stuart Phinn. 2021. "Detection of Banana Plants Using Multi-Temporal Multispectral UAV Imagery" Remote Sensing 13, no. 11: 2123. https://doi.org/10.3390/rs13112123
APA StyleAeberli, A., Johansen, K., Robson, A., Lamb, D. W., & Phinn, S. (2021). Detection of Banana Plants Using Multi-Temporal Multispectral UAV Imagery. Remote Sensing, 13(11), 2123. https://doi.org/10.3390/rs13112123








