1. Introduction
Recently, hyper spectral image (HSI) classification has attracted the attention of researchers owing to its numerous applications such as land change monitoring, urban development, resource management, disaster prevention, and scene interpretation [
1]. Generally, in HSI classification, a specific category is assigned to each pixel in an image. However, since most multispectral, ultraspectral, and hyperspectral images generate a large number of high-dimensional pixels with many category labels, it is a challenging task to effectively separate these pixels with similar land cover spectral properties. First, because of the large number of HSI pixels, numerous schemes applied in HSI classification were based on supervised learning [
1], including the support vector machine (SVM) [
2,
3], nearest feature line [
4,
5], random forest [
6], manifold learning [
7,
8], sparse representation [
9], and deep learning (DL) [
1]. Second, because HSI contains high-dimensional information, many studies have focused on dimension reduction (DR) and reported the importance of DR in HSI classification [
10].
Principal component analysis (PCA) [
11] is the most popular DR algorithm; it subtracts the mean of the population from each sample to obtain the covariance matrix and extracts a transformation matrix by maximizing the scatter of samples. PCA also plays a role in the pre-processing for other advanced DR algorithms to remove noise and mitigate overfitting [
1,
10]. Linear discriminant analysis (LDA) [
12], and discriminant common vectors [
13] are advanced versions of PCA. Since the PCA algorithm is based on linear measurement, it is ineffective in revealing the local structure of samples when samples are distributed in a manifold structure. Many methods based on manifold learning and kernel methods have been proposed to overcome the abovementioned problem. Manifold learning was proposed to preserve the topology of the locality of training samples. He et al. [
14] proposed a locality preserving projection (LPP) scheme to preserve the local topology of training data for face recognition. Because the sample scatter obtained through LPP is based on the relationship between neighbors, the local manifold information of samples is preserved, and therefore, the performance of the LPP scheme was shown to be higher than that of linear measurement methods. Tu et al. [
7] presented the Laplacian eigenmaps (LE) method, which uses polarimetric synthetic aperture radar data for land cover classification. The LE scheme maintains the high-dimensional polarimetric manifold information in an intrinsic low-dimensional space. Wang and He [
15] also used LPP to pre-process data for HSI classification. Kim et al. [
16] presented a manifold-based method called locally linear embedding (LLE) to reduce the dimensionality of HSI. Li et al. [
8,
17] presented the local Fisher discriminant analysis (LFDA) scheme, which takes into account the merits of both LDA and LPP to reduce the dimensionality of HSI. Luo et al. [
18] proposed a supervised neighborhood preserving embedding method to extract the salient features for HSI classification. Zhang et al. [
19] employed a sparse low-rank approximation scheme to regularize a manifold structure, and HSI data were treated as cube data for classification.
Generally, these methods based on manifold learning all preserve the topology of the locality of training samples and outperform the traditional linear measurement methods. However, according to Boots and Gordon [
20], nonlinear information cannot be extracted through manifold learning, and the effectiveness of manifold learning is limited by noise. Therefore, the kernel tricks were employed to obtain a nonlinear feature space and improve the extraction of nonlinear information. Because the use of the kernel tricks improves the performance of a given method [
21], the kernelization approach was adopted in both linear measurement methods and manifold learning methods to improve HSI classification. Boots and Gordon [
20] employed the kernelization method to alleviate the noise effect in manifold learning. Scholkopf et al. [
22] proposed a kernelization PCA scheme, which makes use of kernel tricks to find a high-dimensional Hilbert space and extract non-linear salient features missed by PCA. In addition, Lin et al. [
23] presented a framework for DR based on multiple kernel learning. The multiple kernels were first unified to generate a high dimensional space by a weighted summation, and then these multiple features were projected to a low dimensional space. However, using their proposed framework, they also attempted to determine proper weights for a combination of kernels and DR simultaneously, which increased the complexity of the method. Hence, Nazarpour and Adibi [
24] presented a novel weight combination algorithm that was used to only extract good kernels from some basic kernels. Although the proposed idea was a simple and effective idea for multiple kernel learning, it used kernel discriminant analysis based on linear measurement for classification, and thus would not preserve the manifold topology of multiple kernels in high dimension space. Li et al. [
25] proposed a kernel integration algorithm that linearly assembles multiple kernels to extract both spatial and spectral information. Chen et al. [
26] used a kernelization method based on sparse representation for HSI classification. In their approach, a query sample was represented by all training data in a generated kernel space, and all training samples were also represented in a linear combination of their neighboring pixels. Resembling the idea of multiple kernel learning, Zhang et al. [
27] presented an algorithm for multiple-feature integration based on multiple kernel learning and employed it to classify HSI data; their proposed algorithm assembles shape, texture, and spectral information to improve the performance of HSI classification. In addition to obtaining a salient feature space for HSI classification, DR can be a critical pre-processing step for DL. Zhu et al. [
10] proposed an HSI classification method based on 3D generative adversarial networks (GANs) and PCA. Their experimental results demonstrated that the performance of GANs is adversely affected if there is no PCA pre-processing step. However, as mentioned earlier, because PCA is a linear measurement method, it may miss some useful manifold information. Therefore, a DR algorithm that can extract manifold information should be used to improve the performance of GANs.
Finally, because of the numerous category labels of HSI, a more powerful classifier is required to improve the performance of HSI classification. Recently, DL has been viewed as the most powerful tool in pattern recognition. Zhu et al. [
10] used GANs for HSI classification and obtained a favorable result. Liu et al. [
28] proposed a Siamese convolutional neural network to extract salient features for improving the performance of HSI classification. He et al. [
29] proposed multiscale covariance maps to improve CNN and integrate both spatial and spectral information in a natural manner. Hu et al. [
30] proposed a convolutional long short-term memory method to effectively extract both spectral and spatial features and improve the performance of HSI classification. Deng et al. [
31] proposed a deep metric learning-based feature embedding model that can overcome the problem of having only a limited number of training samples. Deng et al. [
32] also proposed a unified deep network integrated with active transfer learning, which also overcomes this problem. Chen et al. [
1] proposed fine-grained classification based on DL; a densely connected CNN was employed for supervised HSI classification and GAN for semi-supervised HSI classification.
From the presented introduction, the challenges in HSI classification can be summarized as follows:
Owing to the high dimensions of HSI, an effective DR method is required to improve classification performance and overcome the overfitting problem.
Owing to the numerous category labels of HSI, a powerful classifier is required to improve the classification performance.
Because DR improves HSI classification considerably and overcomes the overfitting problem, in this study, a modification of the feature line embedding (FLE) algorithm [
4,
5] based on SVM [
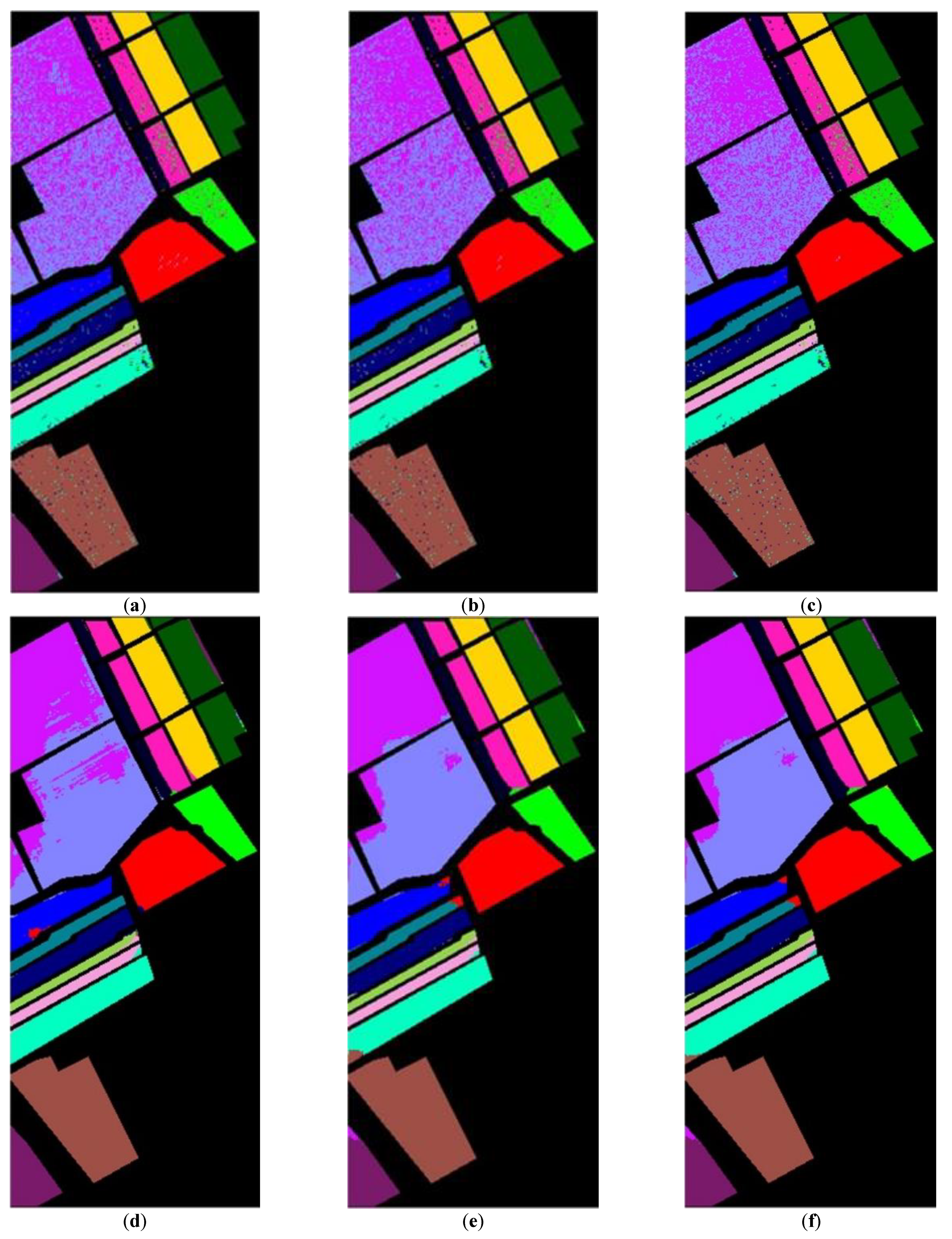
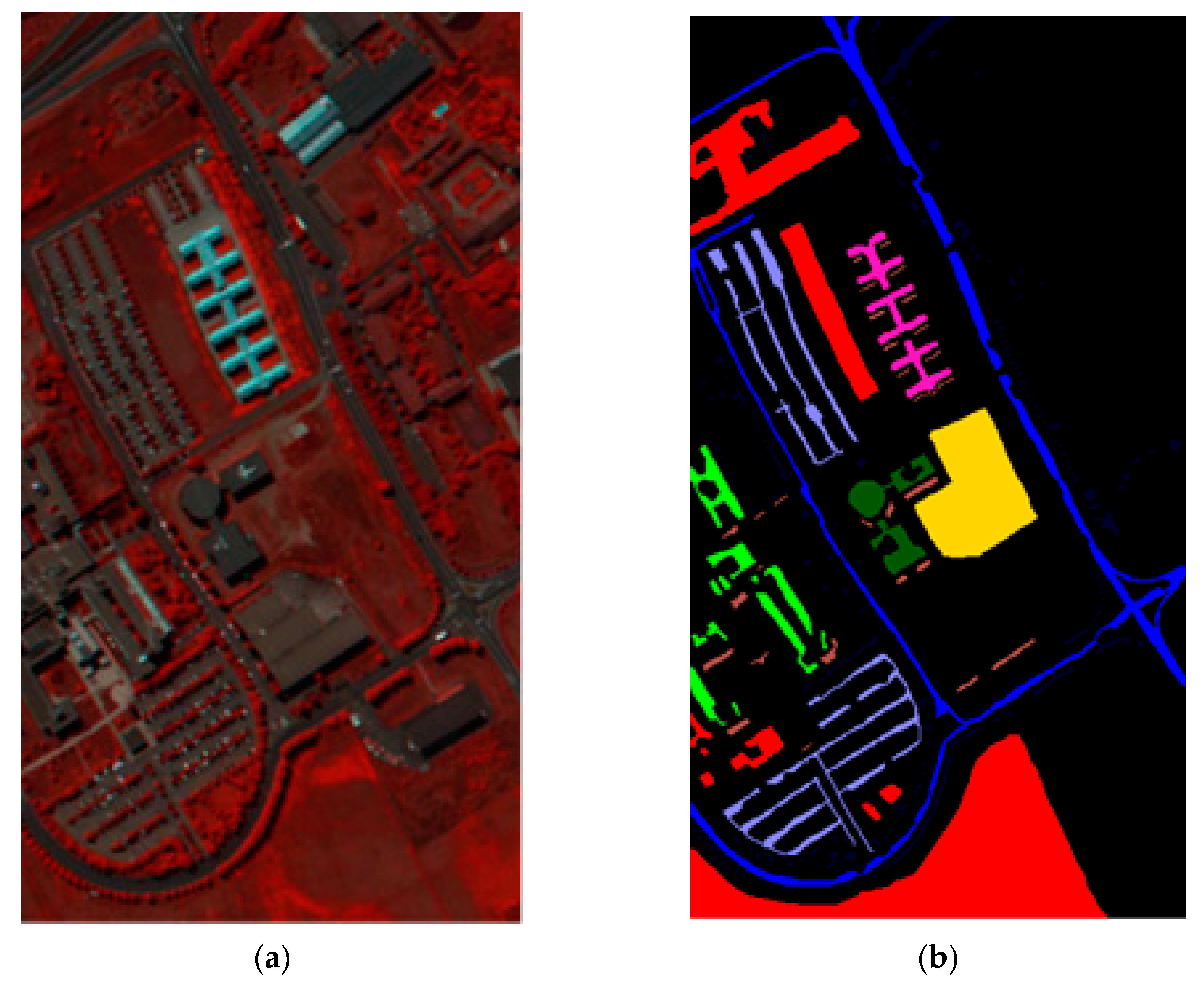
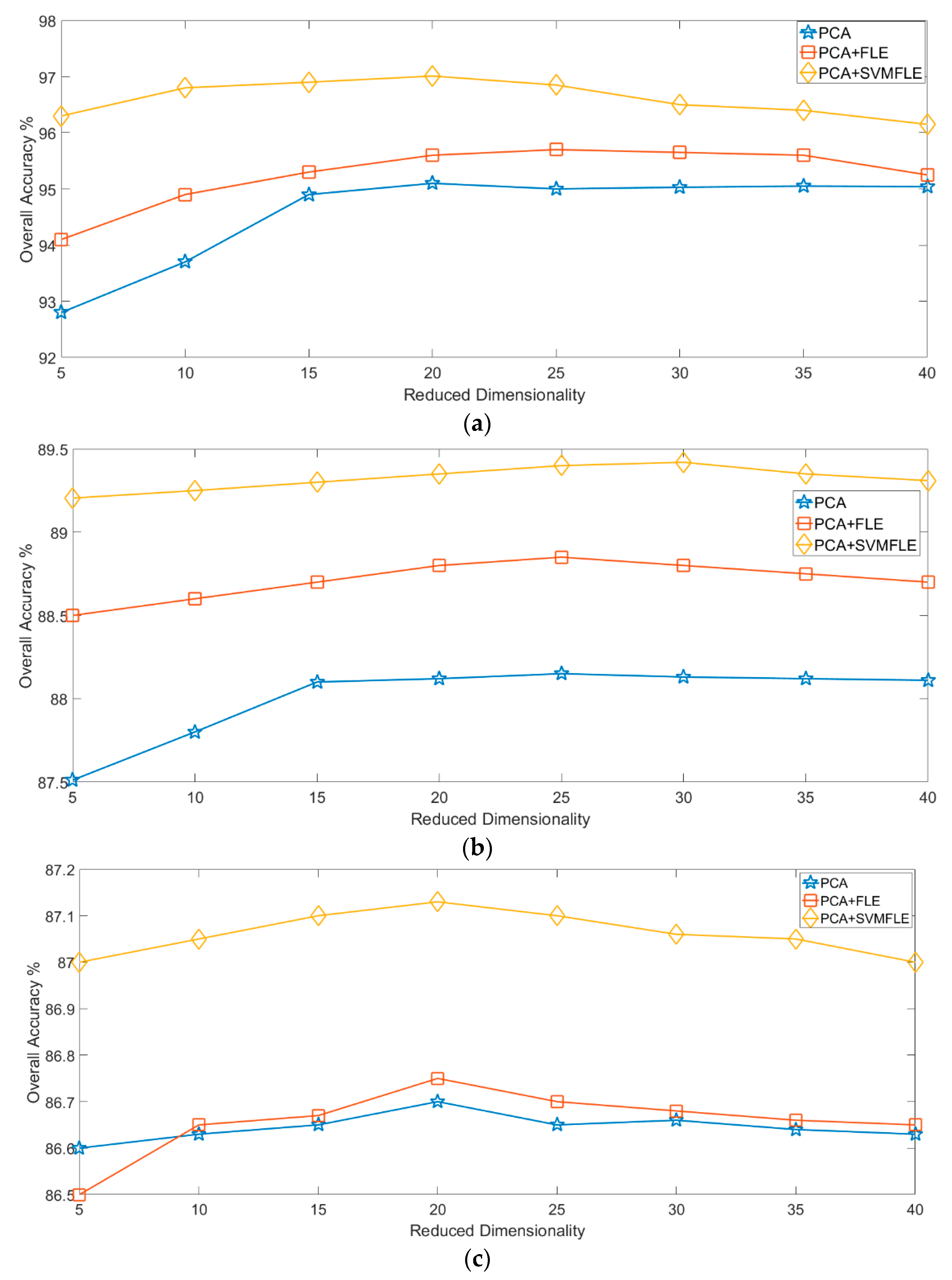
33], referred to as SVMFLE, was proposed for DR. In this algorithm, SVM is first employed for selecting the boundary samples and calculating the between-scatter matrix to enhance the discriminant ability. As we know, the scatter calculated from the boundary samples among classes could reduce the noise impact and improve the classification results. Second, the dispersion degree among samples is devised to automatically determine a better weight of the between-class scatter. By doing so, the reduced space with more discriminant power is obtained. Three benchmark data sets were used to evaluate the algorithm proposed. The experimental results demonstrated that the proposed SVMFLE method effectively improves the performance of both nearest neighbor (NN) and GAN classifiers.
The rest of this paper is organized as follows: In
Section 2, previous and related works are discussed. In
Section 3, the proposed algorithm of SVM-based sample selection incorporated into the FLE is introduced. In
Section 4, the proposed method was compared with other state-of-the-art schemes for HSI classification to demonstrate its effectiveness. Finally, in
Section 5, the conclusions are drawn.
3. Feature Line Embedding Based on Support Vector Machine (SVMFLE)
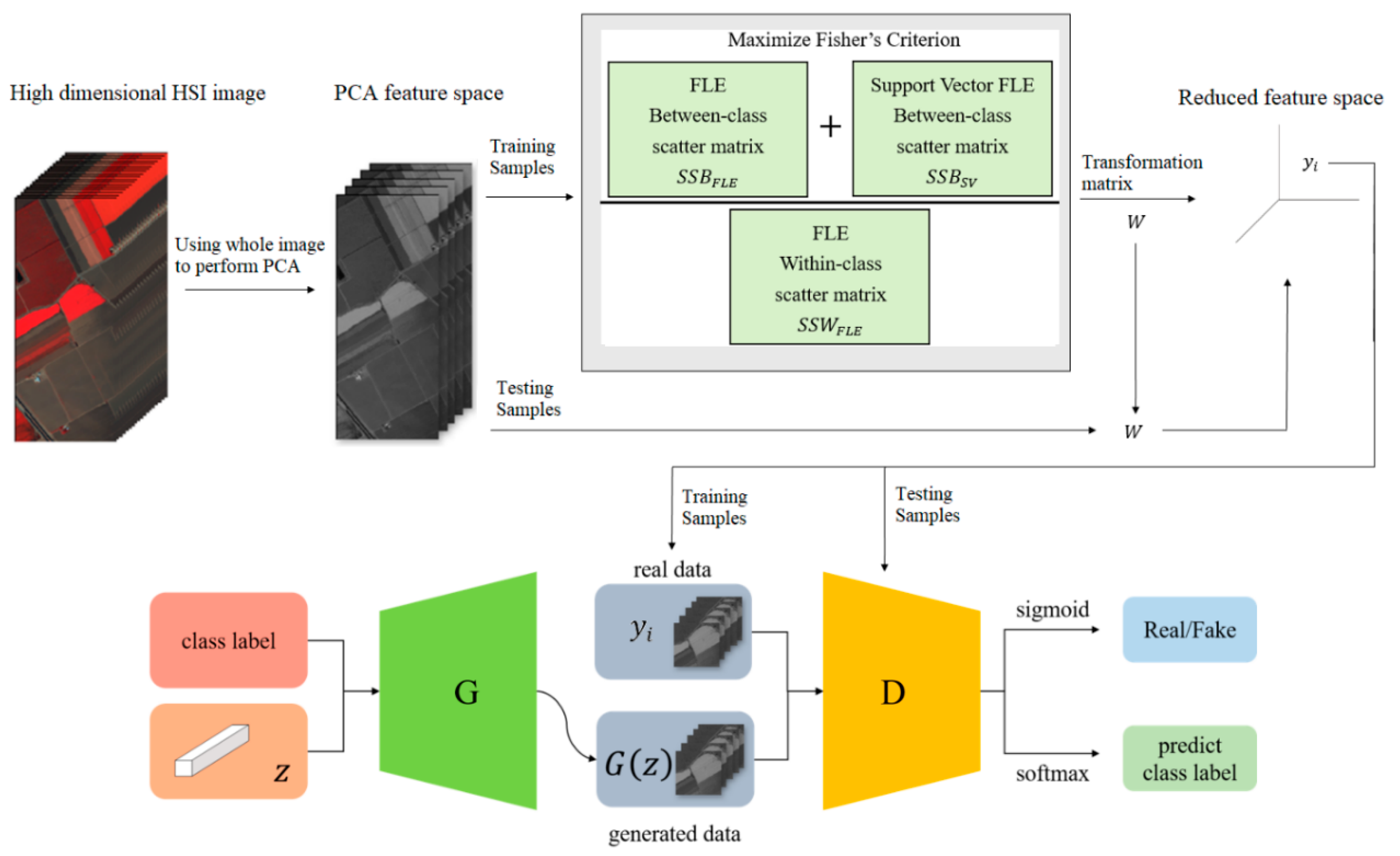
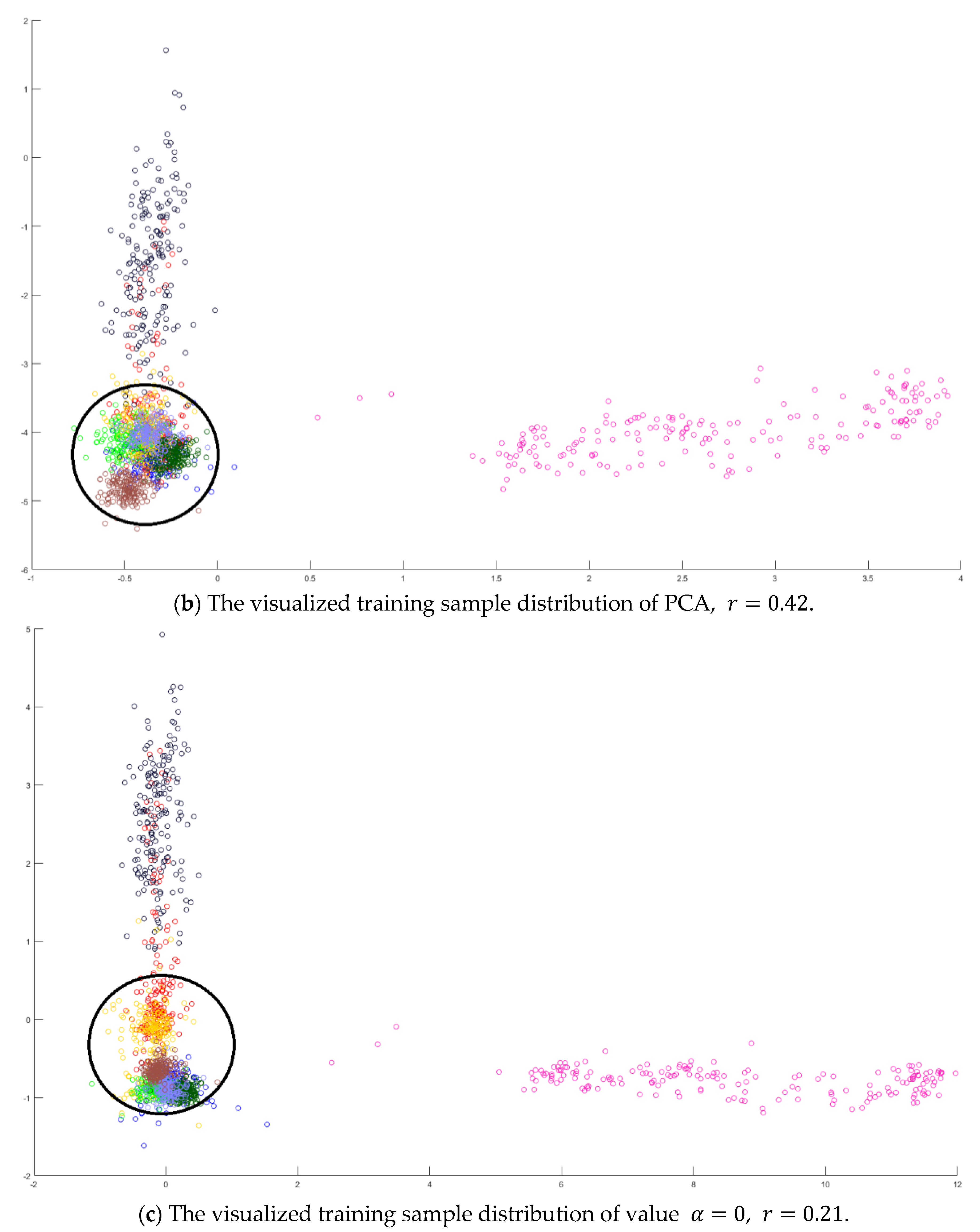
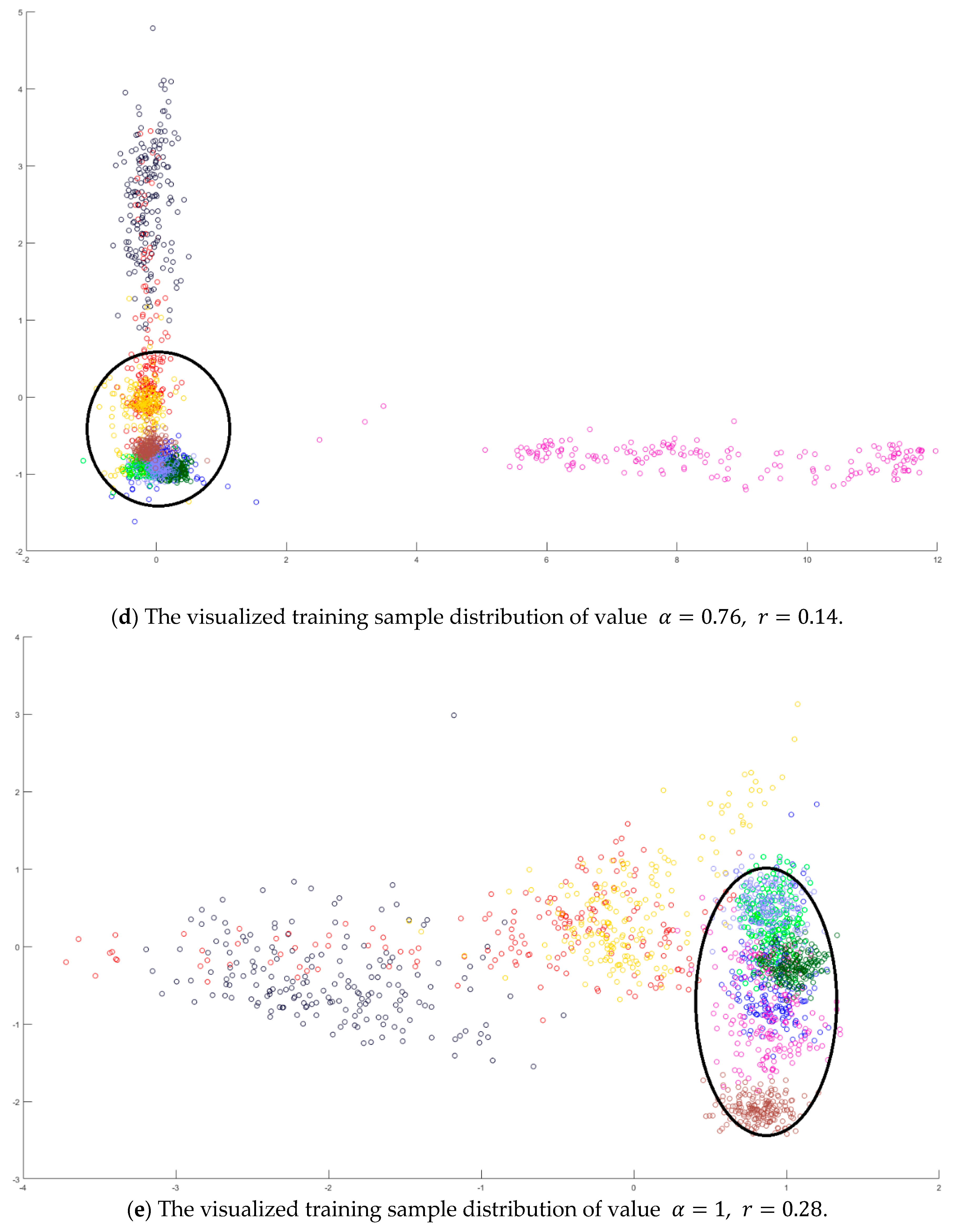
Hyperspectral images are the sensing data with high dimensions. Due to the high dimensional properties, the original pixel dimensions are first reduced by the PCA process. Generally, PCA can find the transformation matrix for the best representation in the reduced feature space. Moreover, PCA also solves the small sample size problem for the further supervised eigenspace-based DR methods, for example, LDA [
12], LPP [
14], …, etc. The within-class and between-class scatters are two essential matrices in eigenspace-based DR. The FLE-based class scatters have been shown to be the effective representation in HSI classification [
4]. Selecting the discriminant samples is the key issue in scatter computation. In [
4], the selection strategy, the first
k nearest neighbors for a specified sample, is adopted to preserve the local topological structure among samples during training. In this proposed DR method as shown in
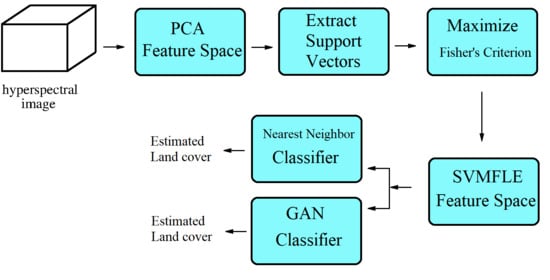
Figure 1, the samples with more discriminant power, that is, support vectors (SV), are selected for enhancement. SVMFLE calculates the between-class scatter using the SVs which are found by SVM. According to our experience, the scatter calculated from the boundary samples among classes could reduce the noise impact and improve the classification results.
In the proposed framework as shown in
Figure 1, all pixels on HSI are employed to obtain a transformation matrix
, and projected into a low dimensional PCA feature space. Next, some training samples are randomly selected, and SVs between classes are extracted by SVM in the PCA feature space. After that, FLE within-class scatter matrix
, FLE between-scatter matrix
, and SV-based FLE between-class scatter matrix
are calculated to maximize Fisher’s criterion for obtaining the transformation matrix
. The final projection matrix is
. Then, all pixels on HSI are projected into a five-dimensional feature space by the linear transformation
. Next, 200 training samples of dimension five in the SVMFLE feature space were randomly selected to train a GAN classifier. Finally, all pixels on HSI in this SVMFLE feature space are used for evaluating the performance of the GAN classifier.
The within-class scatter
is calculated from the discriminant vectors of the same class by using the FLE strategy. Consider a specified sample
, the discriminant vectors are chosen from the first
nearest features lines generated by its eight nearest neighbors of the same class as shown in Equation (2). On the other hand, the between-class scatter is obtained from two parts. One scatter
is similar to the approach in [
4]. The first
nearest feature lines are selected for the between-class scatter computation from six nearest neighbors of sample
with different class labels as shown in Equation (3). Here, parameters
and
are set as 24 and 12, respectively, in the experiments. The other part of the between-class scatter is calculated from the support vectors generated by SVM. The one-against-all strategy is adopted in SVM, for example, two-class classification. Consider the positive training samples of a specified class
c and the negative samples of the other classes. These training samples are inputted to SVM for a two-class classification. After the learning process, as we know, the decision boundary is determined from the weighted summation of support vectors. These support vectors are the specified samples near the boundaries among classes. Two SV sets, positive SV
and negative SV
sets, are obtained to calculate the between-class scatter
as shown in the following equation:
These two scatters
and
are integrated as:
Here, parameter
indicates the ratio between the scatter for all points
and the scatter for support vectors
. Since support vectors usually locate at the class boundary regions, they are the samples with more discriminant power for learning. The transformation matrix
is found by maximizing the Fisher criterion
, in which matrix
is composed of the eigenvectors with the corresponding largest eigenvalues. The projected sample in the low-dimensional space is calculated by the linear projection
. Furthermore, the reduced data were used to train the GAN classifier in [
10]. The reduced HSI pixels are tested by the discriminator
in GAN. The pseudo-codes of the SVMFLE DR algorithm are listed in
Table 2.
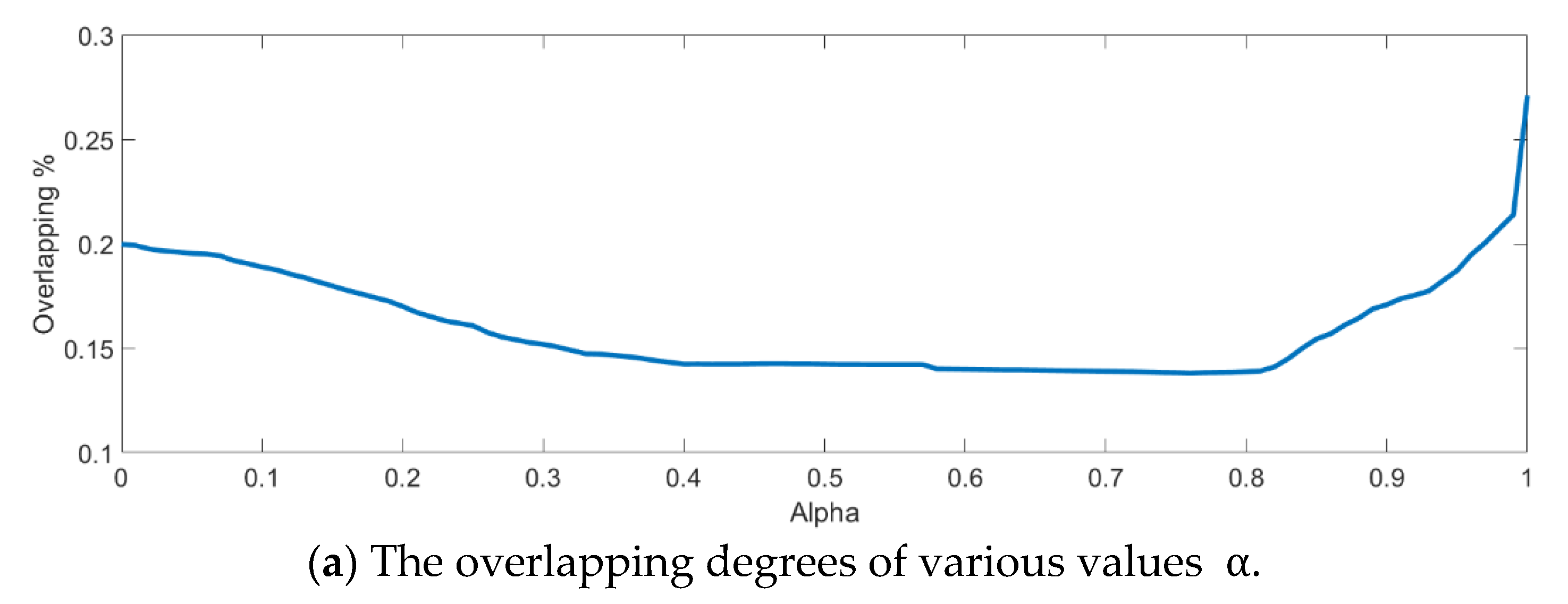
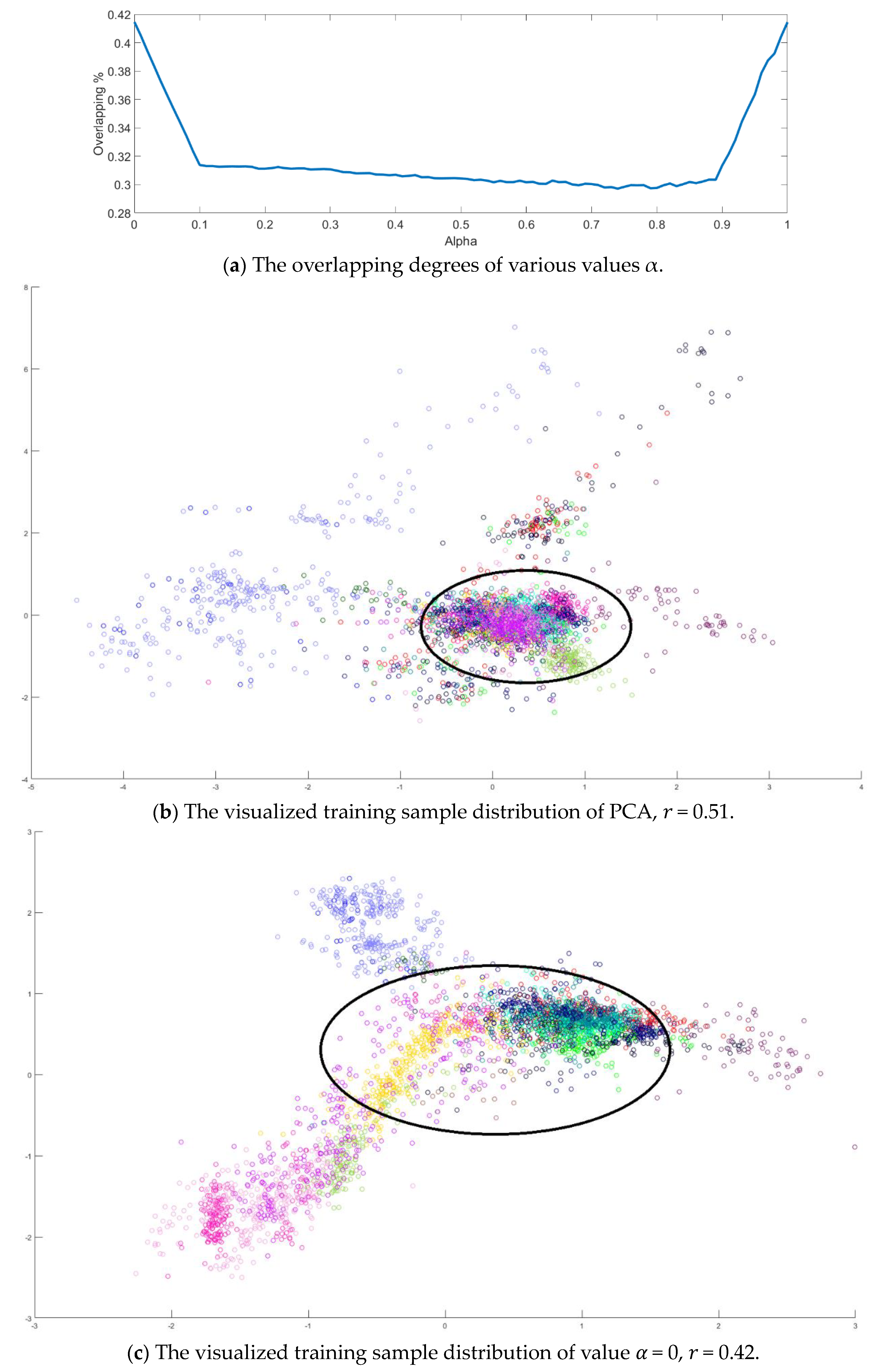
Next, an indicator is defined to determine the better value
in Equation (10) by measuring the overlapping degree among class samples. Consider a sample
in a specified class
, the Euclidean distances from every sample
to its corresponding class mean
are summed to represent the dispersion degree of samples as defined below:
It is defined to be the within-class distance for class
. On the other hand, the total Euclidean distance from every sample to the population mean
is also calculated as follows:
The dispersion index
is thus calculated and defined as the ratio between the summation of within-class distances and the population distance as follows.
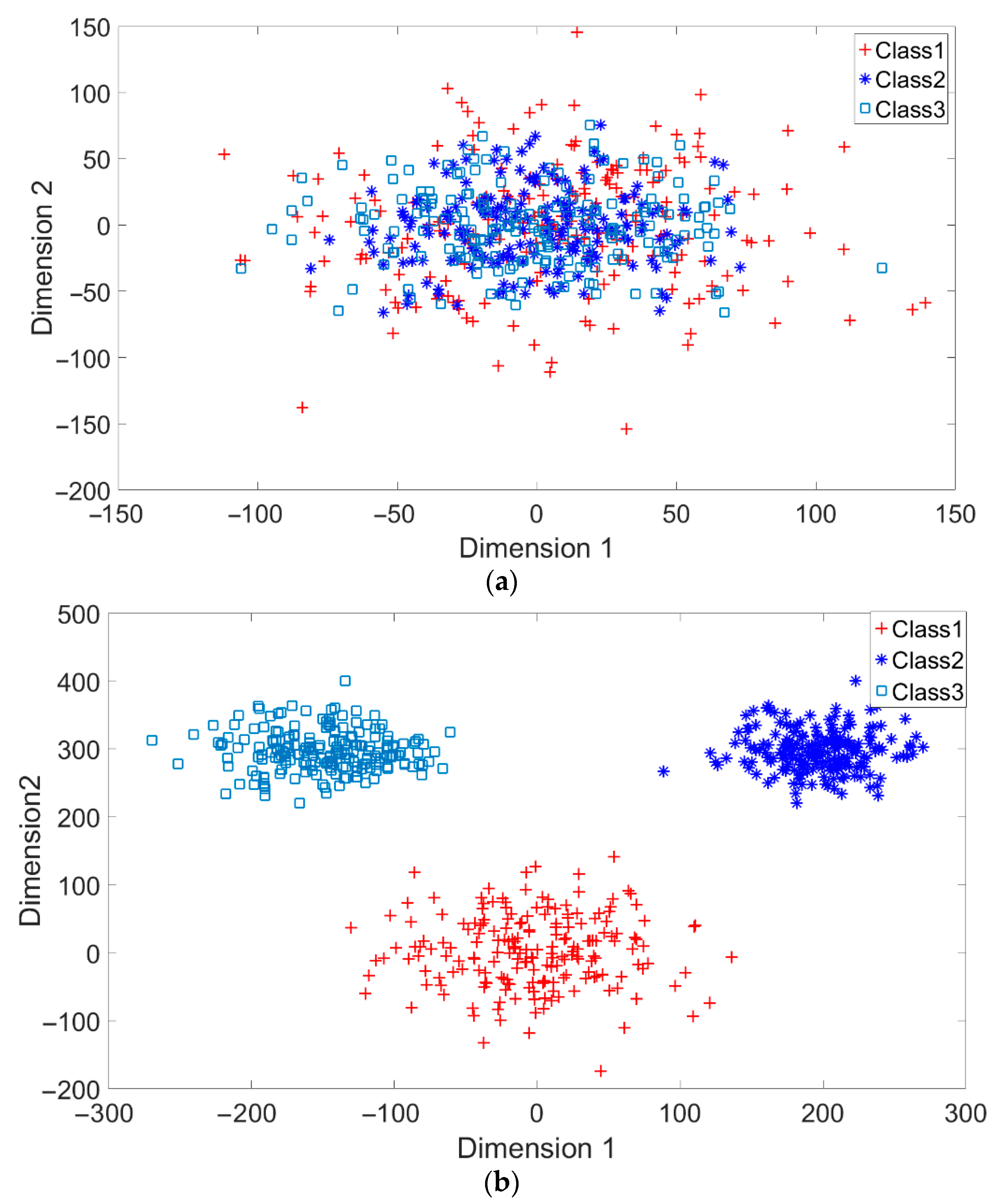
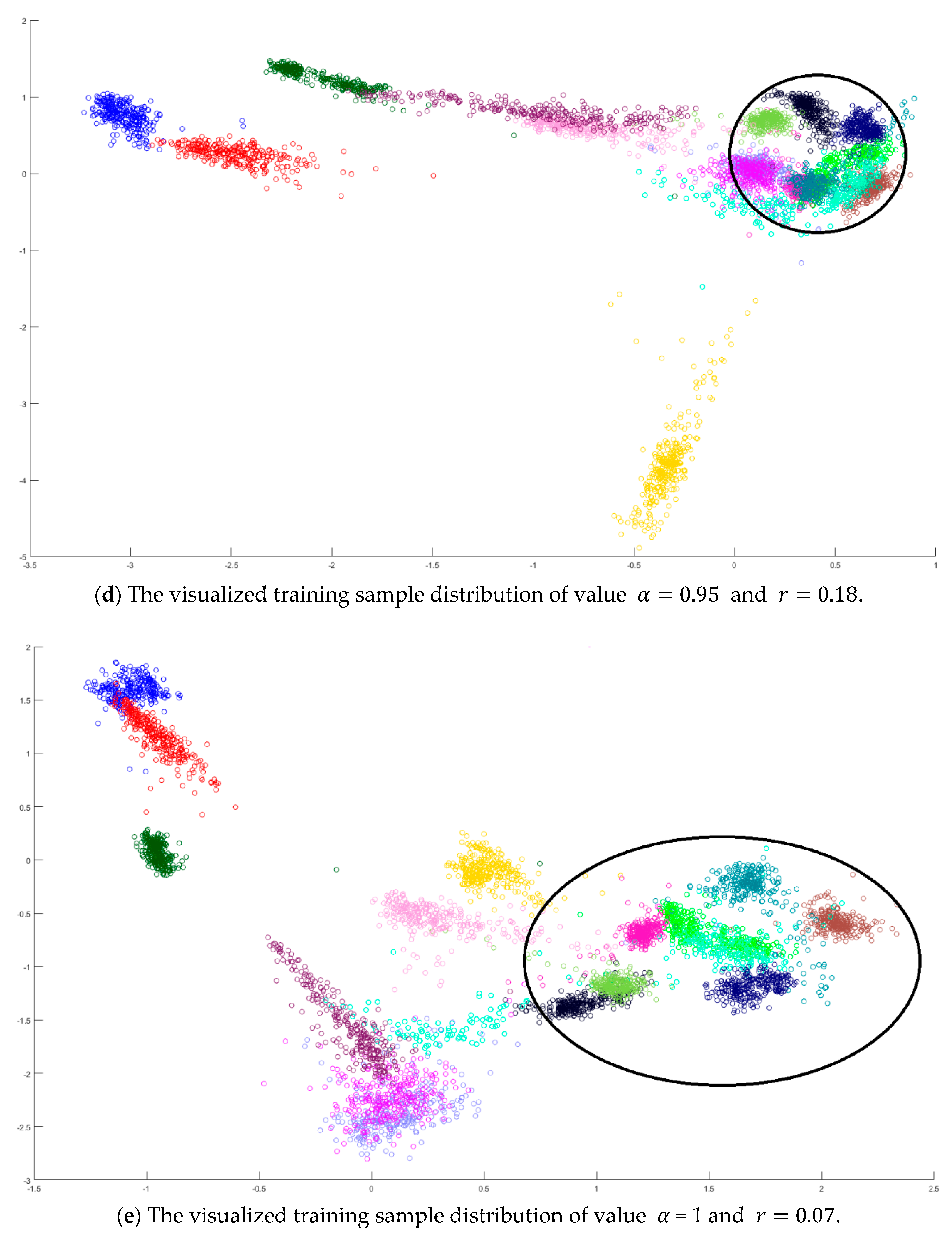
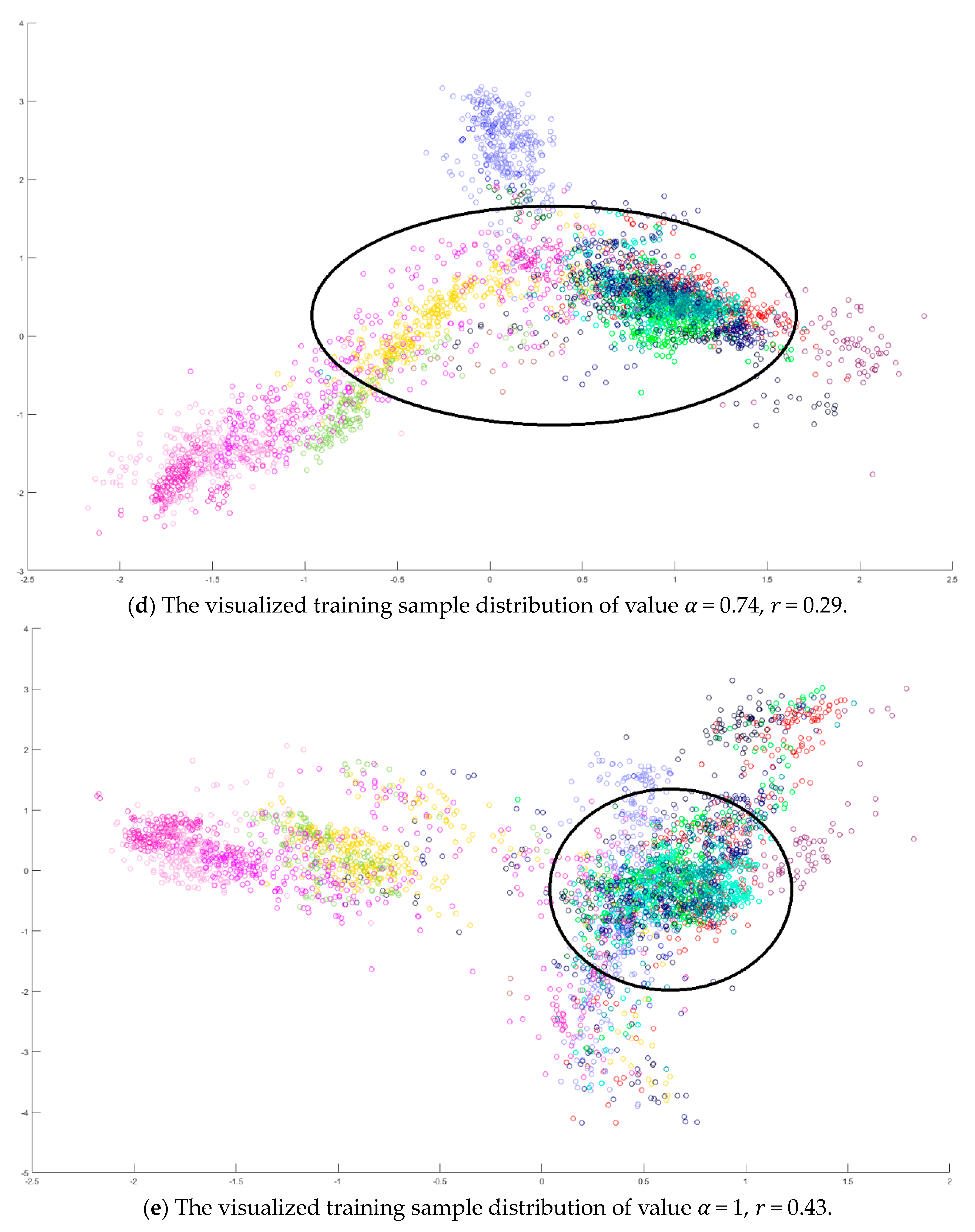
A toy example is given to present the dispersion degree of samples. Three class samples were randomly generated in the normal distribution forms
,
, and
as shown in
Figure 2a. All three class centers are located at point (0,0) and samples are distributed in various standard derivation values. The dispersion index
was calculated. Similarly, another toy example is given in
Figure 2b. Three classes were generated in the distributions
,
, and
. The class means located at three dispersive points that are far away from each other. The standard derivations are the same values as those in
Figure 2a. The dispersion index
is also calculated. From
Figure 2, the smaller dispersion index
, the larger dispersion between classes.