Figure 1.
Location of the study area and distribution of field survey points.
Figure 1.
Location of the study area and distribution of field survey points.
Figure 2.
An explanation of convolutional neural networks (CNN) inputs.
Figure 2.
An explanation of convolutional neural networks (CNN) inputs.
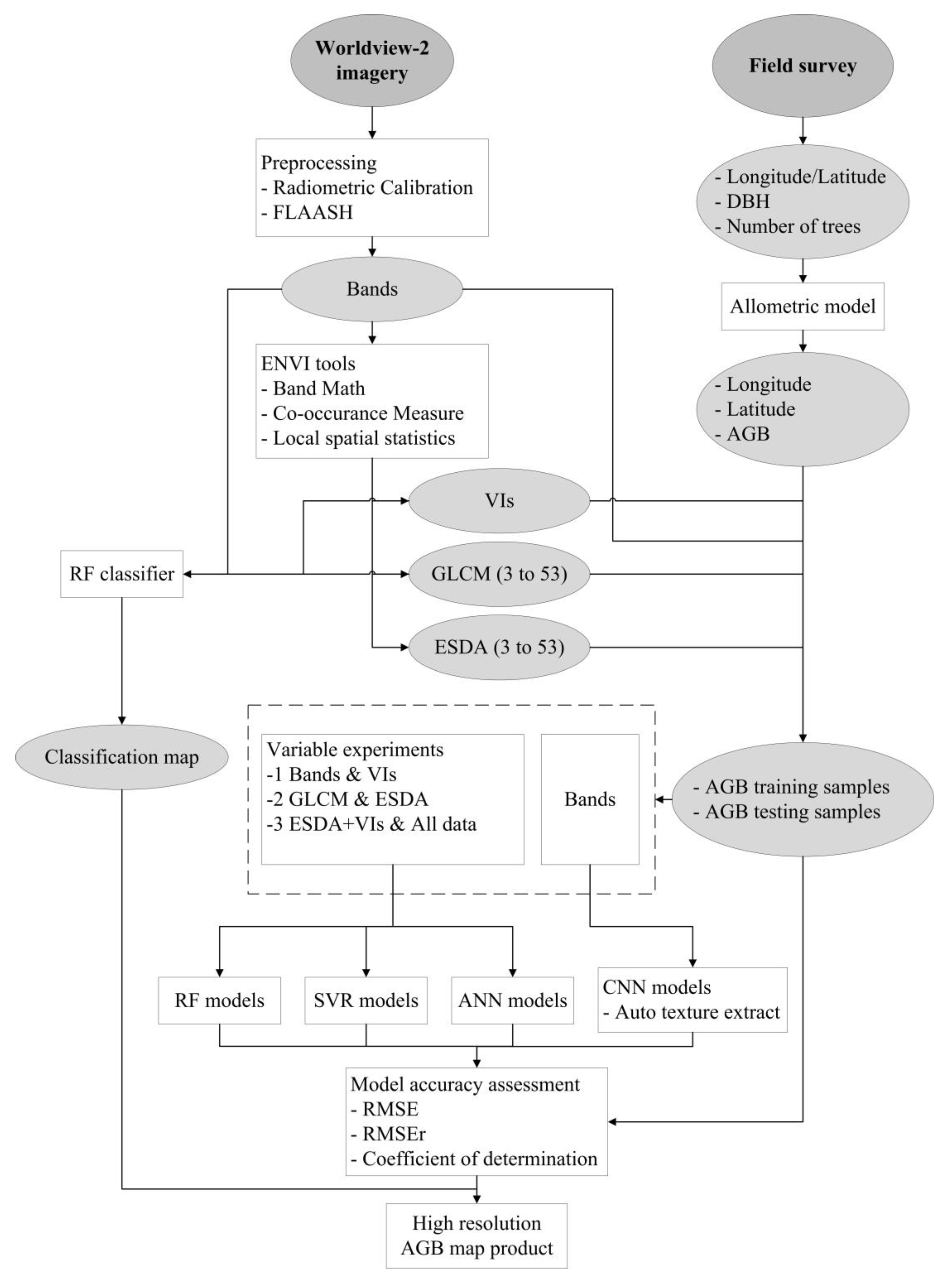
Figure 3.
Flowchart of steps used in our study for comparison of different algorithms and variables for above-ground biomass (AGB) estimation. The meanings of the abbreviations are listed as follows: The Environment for Visualizing Images (ENVI), the Fast Line-of-Sight Atmospheric Analysis of Spectral Hypercubes (FLAASH), Diameters at Breast Height (DBH), Random Forest (RF), Support Vector Regression (SVR), Artificial Neural Network (ANN), Root Mean Square Error (RMSE), Relative Root Mean Square Error (RMSEr).
Figure 3.
Flowchart of steps used in our study for comparison of different algorithms and variables for above-ground biomass (AGB) estimation. The meanings of the abbreviations are listed as follows: The Environment for Visualizing Images (ENVI), the Fast Line-of-Sight Atmospheric Analysis of Spectral Hypercubes (FLAASH), Diameters at Breast Height (DBH), Random Forest (RF), Support Vector Regression (SVR), Artificial Neural Network (ANN), Root Mean Square Error (RMSE), Relative Root Mean Square Error (RMSEr).
Figure 4.
Confusion matrix of classification results and classification map.
Figure 4.
Confusion matrix of classification results and classification map.
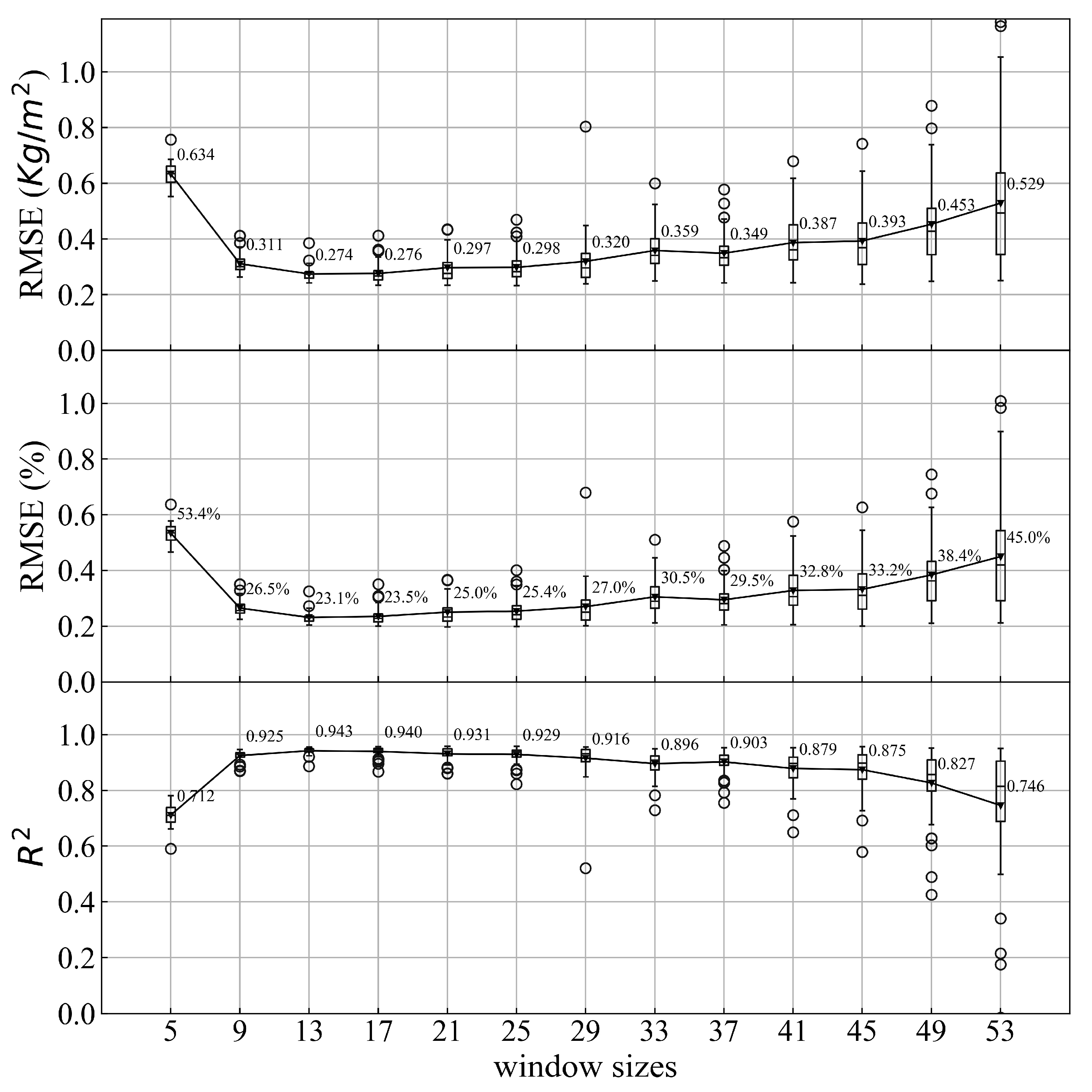
Figure 5.
The results of CNN with different window sizes. RMSE, RMSEr, and R square (R2) are used to evaluate the results. The average results of the 50 repeats are shown with triangles and the values are written right beside.
Figure 5.
The results of CNN with different window sizes. RMSE, RMSEr, and R square (R2) are used to evaluate the results. The average results of the 50 repeats are shown with triangles and the values are written right beside.
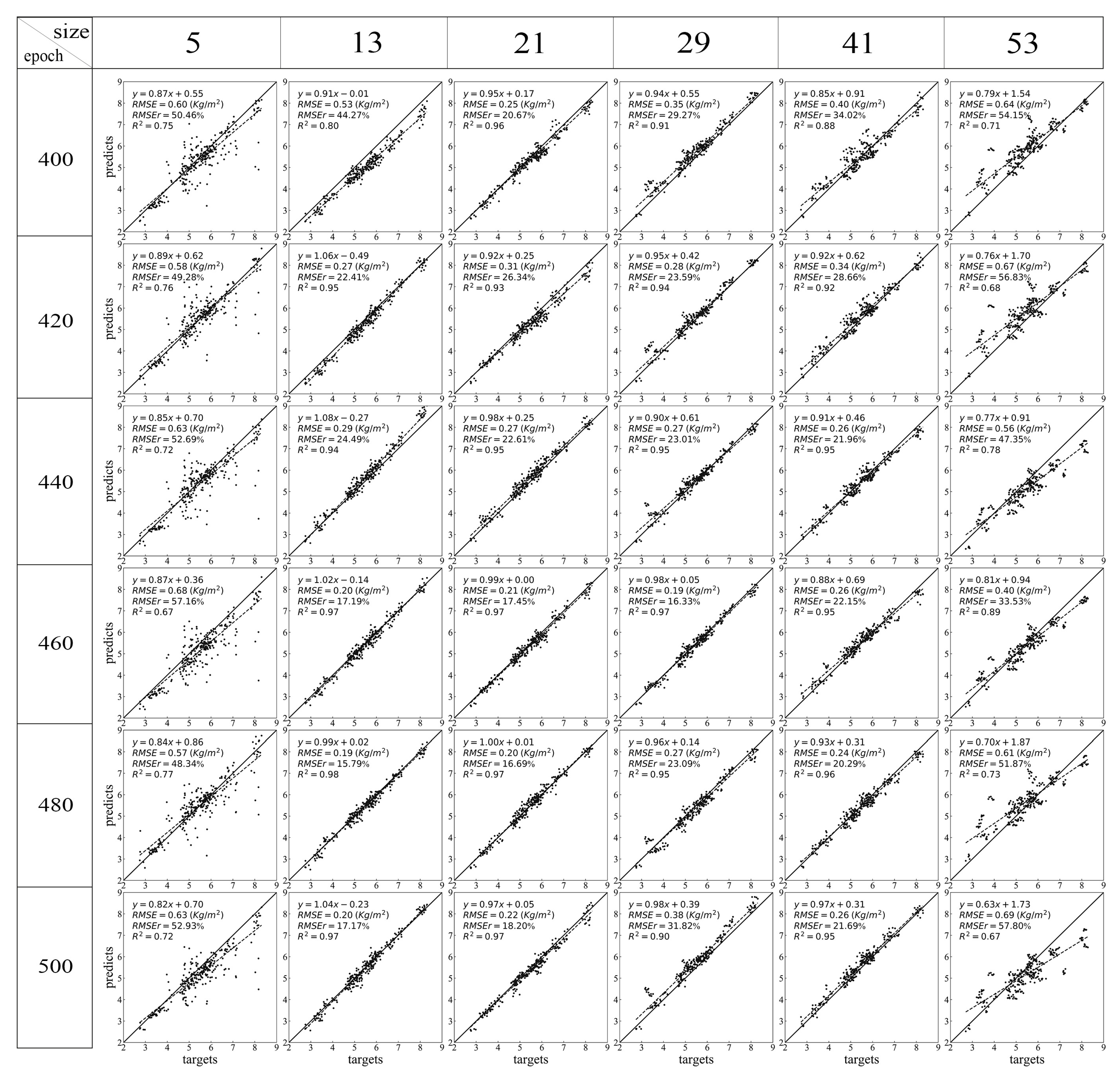
Figure 6.
Scatter plots of different epochs and window sizes of CNN. Different rows indicate the results from different training epochs and different columns indicate the results from inputs with different window sizes.
Figure 6.
Scatter plots of different epochs and window sizes of CNN. Different rows indicate the results from different training epochs and different columns indicate the results from inputs with different window sizes.
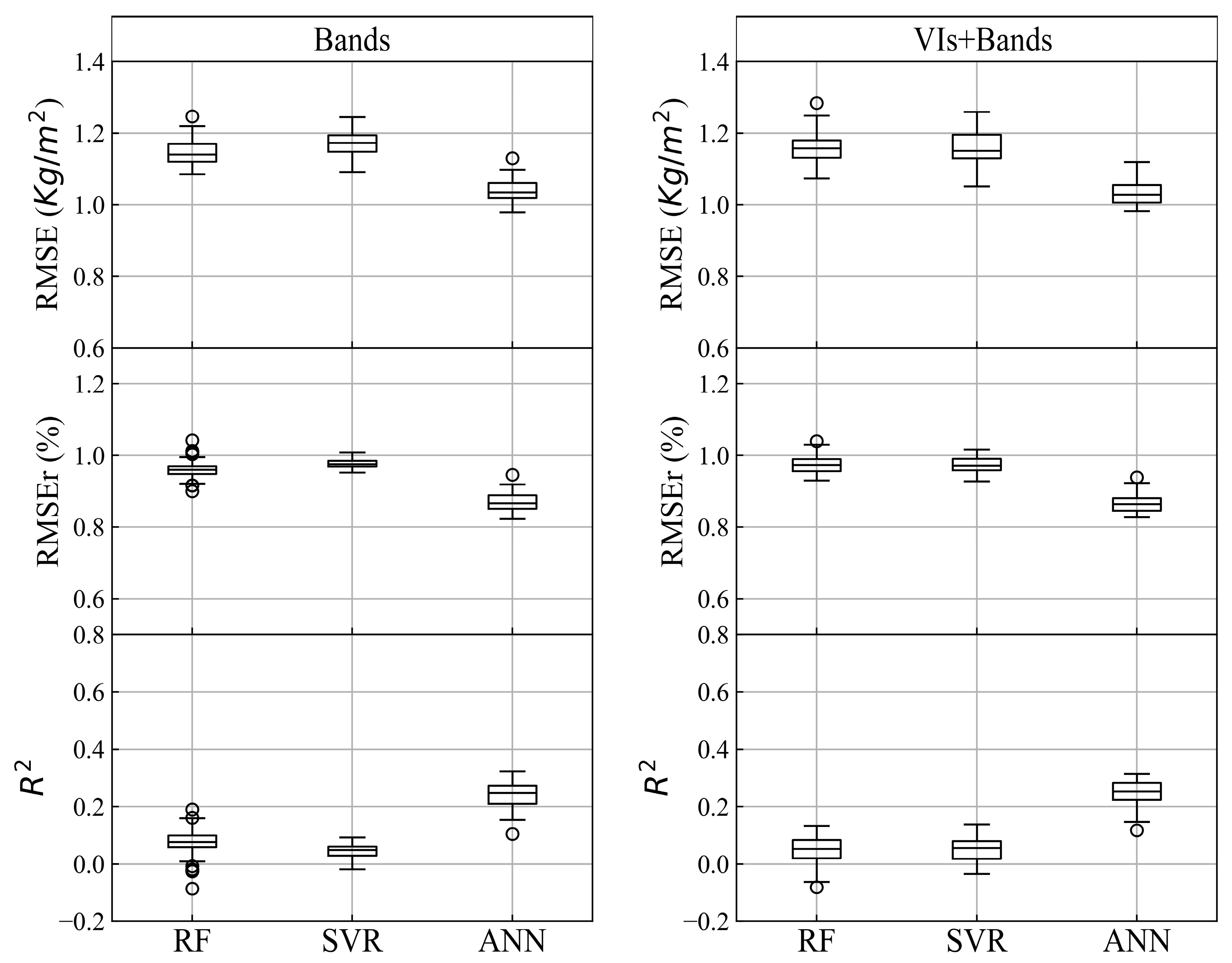
Figure 7.
The results (RMSE, RMSEr, R2) of bands and Vis + Bands using 3 Machine Learning (ML) algorithms with all the window sizes. (1) Figures of the left column are the results of three algorithms using bands. (2) Figures of the right column are the results of three algorithms using Vis + Bands.
Figure 7.
The results (RMSE, RMSEr, R2) of bands and Vis + Bands using 3 Machine Learning (ML) algorithms with all the window sizes. (1) Figures of the left column are the results of three algorithms using bands. (2) Figures of the right column are the results of three algorithms using Vis + Bands.
Figure 8.
The results (RMSE, RMSEr, R2) of GLCM + bands and ESDA + bands using 3 ML algorithms with all the window sizes. (1) Figures of the left column are the results of three algorithms using GLCM + band. (2) Figures of the right column are the results of three algorithms using ESDA + bands. Boxplots in red indicate RF; boxplots in blue indicate SVR; boxplots in green indicate ANN.
Figure 8.
The results (RMSE, RMSEr, R2) of GLCM + bands and ESDA + bands using 3 ML algorithms with all the window sizes. (1) Figures of the left column are the results of three algorithms using GLCM + band. (2) Figures of the right column are the results of three algorithms using ESDA + bands. Boxplots in red indicate RF; boxplots in blue indicate SVR; boxplots in green indicate ANN.
Figure 9.
The results (RMSE, RMSEr, R2) of all combined and ESDA + VIs using 3 ML algorithms with all the window sizes. (1) Figures of the left column are the results of three algorithms using All combined. (2) Figures of the right column are the results of three algorithms using ESDA + VIs. Boxplots in red indicate RF; boxplots in blue indicate SVR; boxplots in green indicate ANN.
Figure 9.
The results (RMSE, RMSEr, R2) of all combined and ESDA + VIs using 3 ML algorithms with all the window sizes. (1) Figures of the left column are the results of three algorithms using All combined. (2) Figures of the right column are the results of three algorithms using ESDA + VIs. Boxplots in red indicate RF; boxplots in blue indicate SVR; boxplots in green indicate ANN.
Figure 10.
AGB maps using the 4 algorithms (RF, SVR, ANN, CNN). The mean values and standard deviation of 10 repeats were shown. RF algorithm used GLCM with a window size of 37, SVR of 35, and ANN with 27. CNN used bands as inputs and used a window size of 13.
Figure 10.
AGB maps using the 4 algorithms (RF, SVR, ANN, CNN). The mean values and standard deviation of 10 repeats were shown. RF algorithm used GLCM with a window size of 37, SVR of 35, and ANN with 27. CNN used bands as inputs and used a window size of 13.
Figure 11.
The mean value of 25 pixels of each plot. RF algorithm used GLCM with a window size of 37, SVR of 35, and ANN of 27. CNN used bands as inputs and used a window size of 13.
Figure 11.
The mean value of 25 pixels of each plot. RF algorithm used GLCM with a window size of 37, SVR of 35, and ANN of 27. CNN used bands as inputs and used a window size of 13.
Table 1.
Formulas of the selected variables.
Table 1.
Formulas of the selected variables.
| Feature Types | Feature Names | Details | References |
|---|
| bands | Green (G) | 450–510 nm | \ |
| | Blue (B) | 510–581 nm |
| | Red (R) | 630–690 nm |
| | Near-infrared red (NIR) | 770–895 nm |
| Vegetation Indices (VIs) | Difference Vegetation Index (DVI) | | |
| | Normalized Difference Vegetation Index (NDVI) | | [57] |
| | Normalized Difference Water Index (NDWI) | | [58] |
| | Ratio Vegetation Index (RVI) | | |
| Exploratory spatial data analysis (ESDA) | Moran’s I | c | [50] |
| | Geary’s C | | [50] |
| | G statistic | | [59] |
| Gray-level co-occurrence matrices (GLCM) | Mean (MEA) | | [54] |
| | Variance (VAR) | |
| | Homogeneity (HOM) | |
| | Contrast (CON) | |
| | Dissimilarity (DIS) | |
| | Entropy (ENT) | |
| | Augular Second Moment (ASM) | |
| | Correlation (COR) | |
| | Note: | |
|
Table 2.
Basic statistics of the field measured plant density, average DBH (D), and above ground biomass (AGB). Maximum value (Max), Minimum value (Min), mean value (Mean), Standard Deviation (SD) and Coefficient of Variation (CV) are listed.
Table 2.
Basic statistics of the field measured plant density, average DBH (D), and above ground biomass (AGB). Maximum value (Max), Minimum value (Min), mean value (Mean), Standard Deviation (SD) and Coefficient of Variation (CV) are listed.
| | Density (Plant/m2) | D (cm) | AGB (kg/m2) |
|---|
| Max | 5.160 | 4.671 | 8.175 |
| Min | 1.840 | 2.943 | 2.815 |
| Mean | 3.347 | 3.913 | 5.451 |
| SD | 0.755 | 0.372 | 1.200 |
| CV | 22.5% | 9.5% | 22% |
Table 3.
Classification results using GLCM and bands. The meanings of columns are as follows: CF: coniferous forest, FA: farmland, BR: broadleaf forest, TE: tea garden, BL: bare land, BU: building, BB: bamboo, RO: road, WA: water, CS: cutting site.
Table 3.
Classification results using GLCM and bands. The meanings of columns are as follows: CF: coniferous forest, FA: farmland, BR: broadleaf forest, TE: tea garden, BL: bare land, BU: building, BB: bamboo, RO: road, WA: water, CS: cutting site.
| | CF | FA | BR | TE | BL | BU | BB | RO | WA | CS |
|---|
| User Accuracy (UA) | 0.970 | 0.918 | 0.952 | 0.927 | 0.988 | 0.965 | 0.941 | 0.955 | 1.000 | 0.966 |
| Producer Accuracy (PA) | 0.946 | 0.882 | 0.962 | 0.950 | 0.934 | 0.953 | 0.960 | 0.988 | 1.000 | 1.000 |
| Overall Accuracy (OA) | 0.957 | | | | | | | | | |
| Kappa coefficient (kappa) | 0.952 | | | | | | | | | |
Table 4.
Parameter settings of random forest (RF) and support vector regression (SVR). For RF, the values of mtry and ntree for all the feature set are listed. For SVR, the values of C (cost of misclassification) and ε (the influence of a single sample).
Table 4.
Parameter settings of random forest (RF) and support vector regression (SVR). For RF, the values of mtry and ntree for all the feature set are listed. For SVR, the values of C (cost of misclassification) and ε (the influence of a single sample).
| Feature | RF | SVR | ANN |
|---|
| mtry | ntree | Denotation | C | ε | Denotation | Denotation |
|---|
| bands | 2 | 2000 | Bands_RF | 1000 | 0.1 | Bands_SVR | Bands_ANN |
| VIs | 3 | 2000 | VIs_RF | 10 | 0.1 | VIs_SVR | VIs_ANN |
| GLCM | 41 | 2000 | GLCM_RF_x | 0.8 | 0.01 | GLCM_SVR_x | GLCM_ANN_x |
| ESDA | 5 | 2000 | ESDA_RF_x | 15 | 0.01 | ESDA_SVR_x | ESDA_ANN_x |
| ESDA+VIs | 6 | 2000 | ESDA+VIs_RF_x | 0.8 | 0.1 | ESDA+VIs_SVR_x | ESDA+VIs_ANN_x |
| All combined | 48 | 2000 | All_RF_x | 0.8 | 0.1 | All_SVR_x | All_ANN_x |
Table 5.
The results (RMSE, RMSEr, R2) of bands and Vis + bands using 3 different algorithms. In this table, the mean values of 50 repeats are calculated. The highest result of each scenario is denoted in bold.
Table 5.
The results (RMSE, RMSEr, R2) of bands and Vis + bands using 3 different algorithms. In this table, the mean values of 50 repeats are calculated. The highest result of each scenario is denoted in bold.
| Algorithms | Bands | Bands+VIs |
|---|
| RMSE (Kg/m2) | RMSEr | R2 | RMSE (Kg/m2) | RMSEr | R2 |
|---|
| RF | 1.147 | 0.961 | 0.076 | 1.158 | 0.975 | 0.049 |
| SVR | 1.171 | 0.974 | 0.046 | 1.160 | 0.973 | 0.052 |
| ANN | 1.041 | 0.872 | 0.238 | 1.032 | 0.867 | 0.248 |
Table 6.
The results (RMSE, RMSEr, R2) of GLCM + bands and ESDA + bands using 3 different algorithms. In this table, the mean values of 50 repeats were calculated. Then the best result of all window sizes is reported as well as the window size. The highest result of each scenario is denoted in bold.
Table 6.
The results (RMSE, RMSEr, R2) of GLCM + bands and ESDA + bands using 3 different algorithms. In this table, the mean values of 50 repeats were calculated. Then the best result of all window sizes is reported as well as the window size. The highest result of each scenario is denoted in bold.
| Algorithms | GLCM | ESDA |
|---|
| RMSE (Kg/m2) | RMSEr | R2 | Window Size | RMSE (Kg/m2) | RMSEr | R2 | Window Size |
|---|
| RF | 0.169 | 0.142 | 0.979 | 37 | 0.610 | 0.514 | 0.735 | 51 |
| SVR | 0.203 | 0.170 | 0.970 | 35 | 0.749 | 0.500 | 0.591 | 47 |
| ANN | 0.460 | 0.387 | 0.831 | 27 | 0.708 | 0.594 | 0.646 | 41 |
Table 7.
The results (RMSE, RMSEr, R2) of all combined and ESDA + VIs using 3 different algorithms. In this table, the mean values of 50 repeats are calculated. Then the best result of all window sizes is reported as well as the window size. The highest result of each scenario is denoted in bold.
Table 7.
The results (RMSE, RMSEr, R2) of all combined and ESDA + VIs using 3 different algorithms. In this table, the mean values of 50 repeats are calculated. Then the best result of all window sizes is reported as well as the window size. The highest result of each scenario is denoted in bold.
| Algorithms | All Combined | ESDA + VIs |
|---|
| RMSE (Kg/m2) | RMSEr | R2 | Window Size | RMSE (Kg/m2) | RMSEr | R2 | Window Size |
|---|
| RF | 0.193 | 0.162 | 0.974 | 51 | 0.641 | 0.536 | 0.712 | 49 |
| SVR | 0.211 | 0.178 | 0.968 | 35 | 0.627 | 0.528 | 0.721 | 47 |
| ANN | 0.430 | 0.361 | 0.855 | 31 | 0.574 | 0.482 | 0.765 | 41 |