Improved Prototypical Network Model for Forest Species Classification in Complex Stand
Abstract
1. Introduction
- Improve the structure and parameter adjustment of a PrNet such that it is more suitable for the tree species classification of a small sample of airborne HIS.
- Analyze the effect of data dimensionality reduction on the classification accuracy and the network operation efficiency.
- Proposes a sample size suitable for PrNet to classify tree species of airborne HIS.
2. Materials and Methods
2.1. Study Area
2.2. Classification System and Survey of Field Sample Points
2.3. Acquisition and Preprocessing of Hyperspectral Images
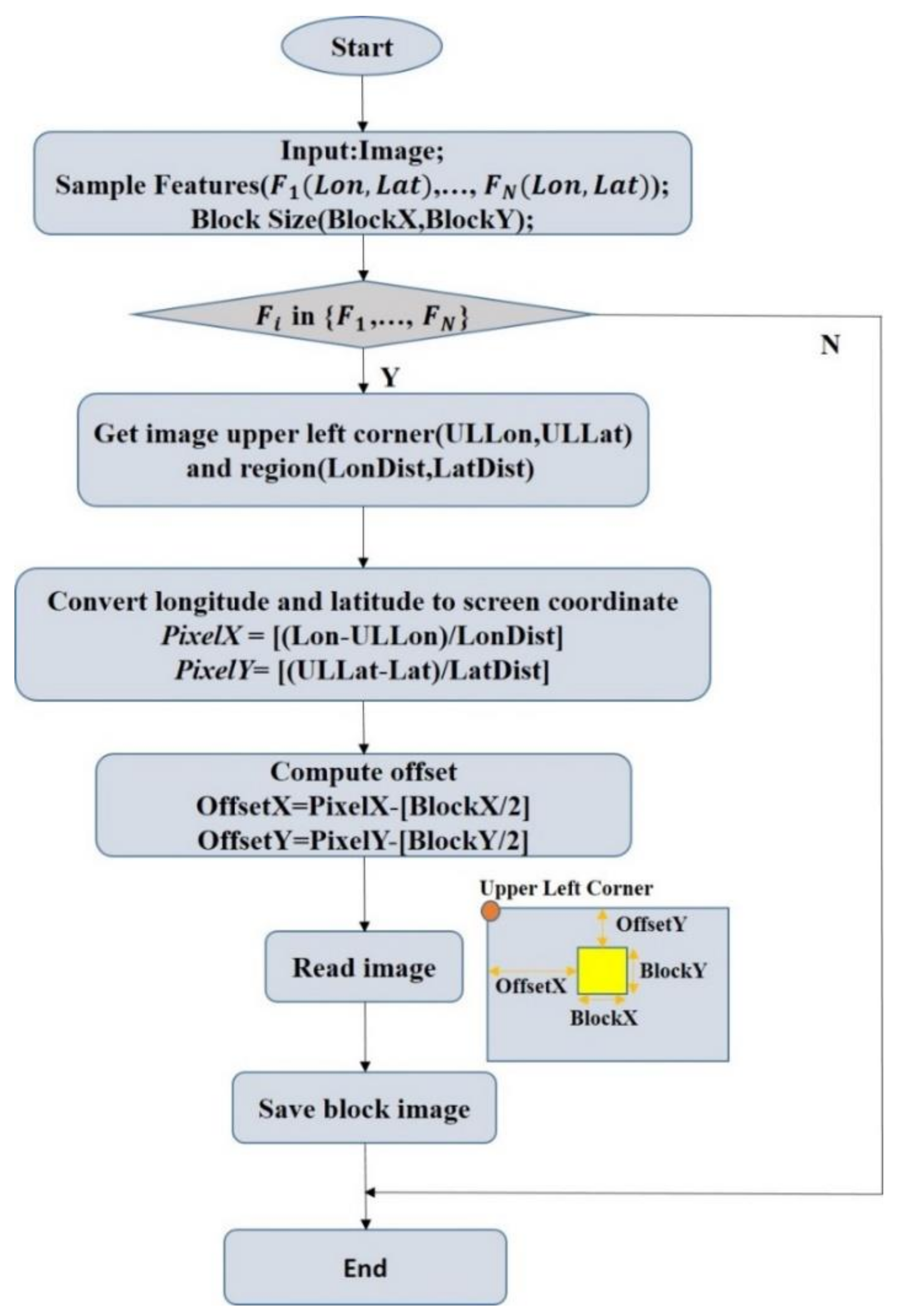
2.4. Sample Data Construction
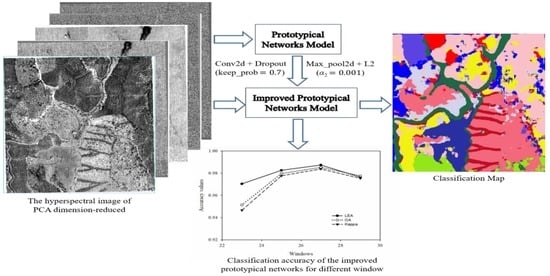
2.5. Prototypical Network Construction
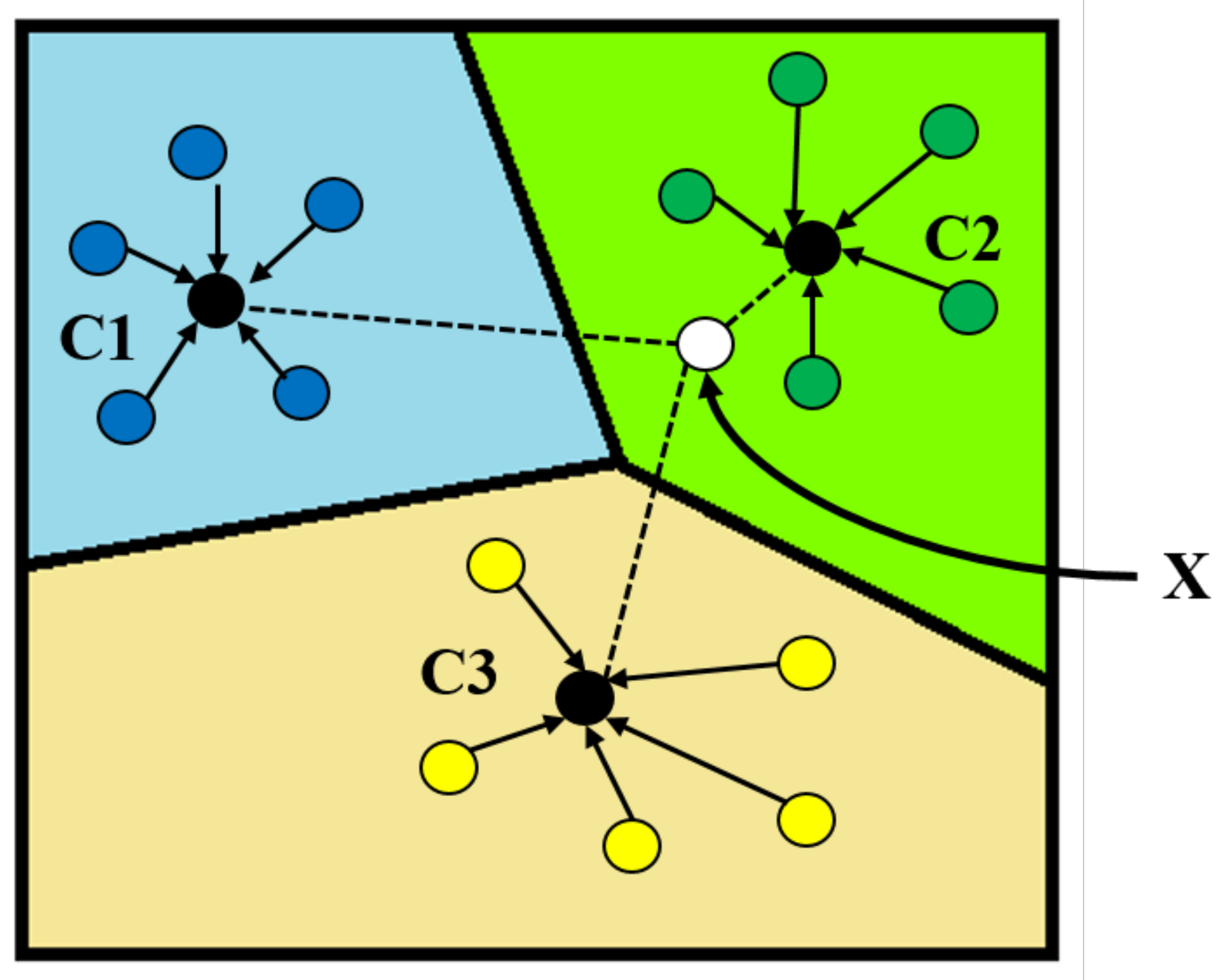
2.5.1. Prototypical Network Model
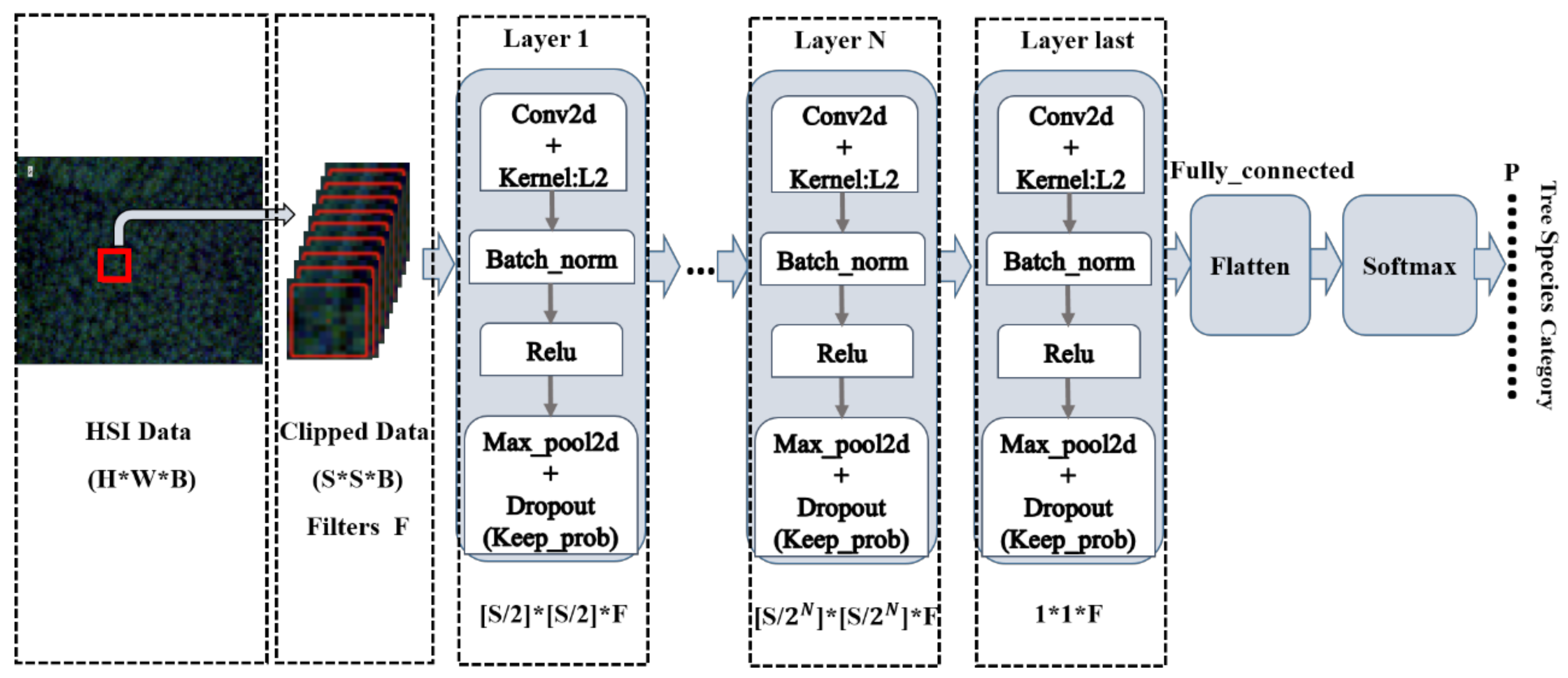
2.5.2. Prototypical Network Classification Algorithm and Improvement
2.5.3. Accuracy Verification
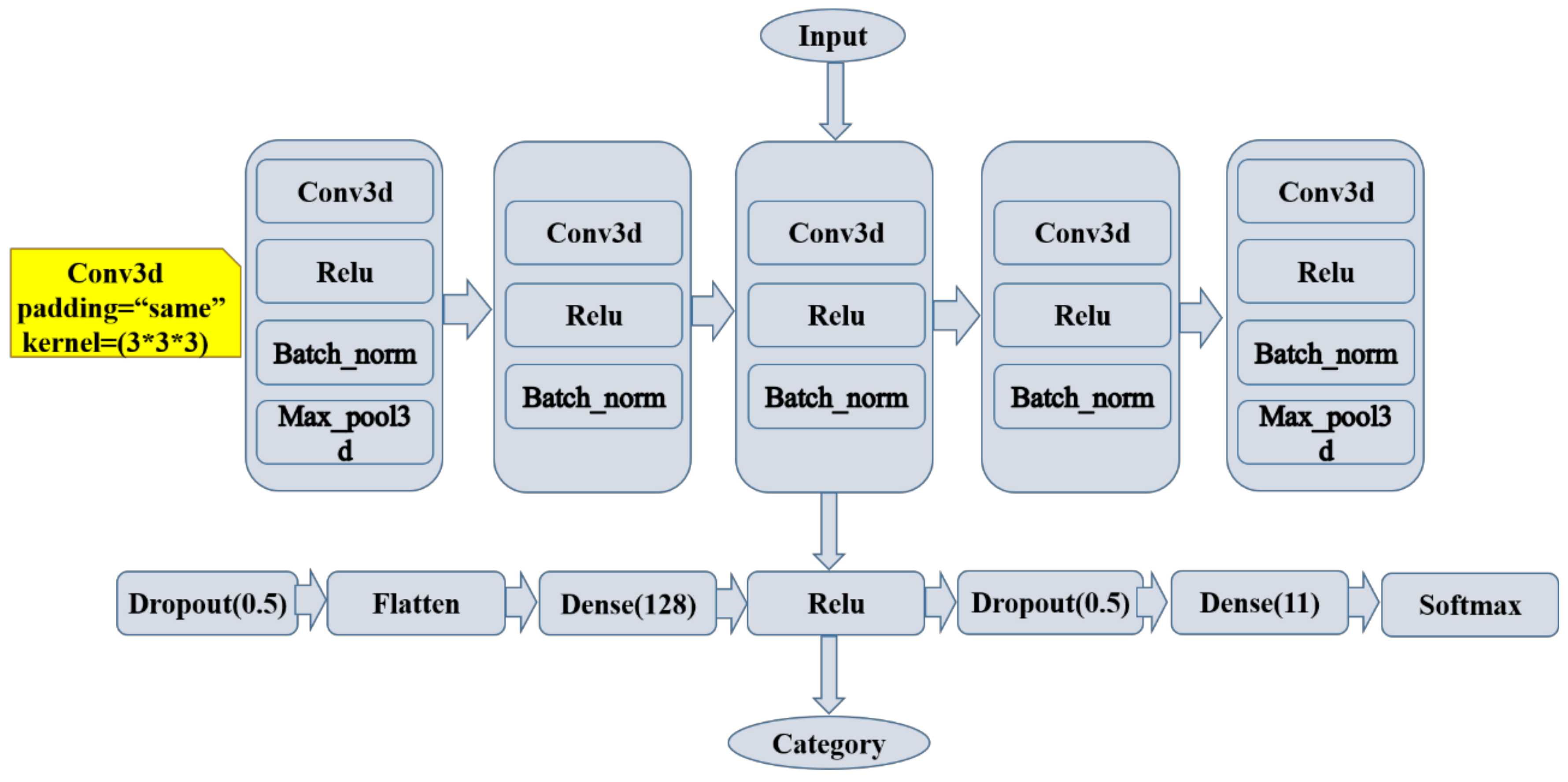
2.6. Three-Dimensional Convolutional Neural Network Construction
2.7. Experiments Design
- (1)
- Experiment A: Classification accuracy of the PrNet using different sample windows.
- (2)
- Experiment B: Classification accuracy of the IPrNet using different Keep_prob values.
- (3)
- Experiment C: Classification accuracy of the IPrNet using different data sources.
- (4)
- Experiment D: Classification accuracy of the 3D-CNN under the same conditions as Experiment C.
3. Classification Process and Results
3.1. Classification Using the Prototypical Network
3.2. Classification Using the Improved Prototypical Network
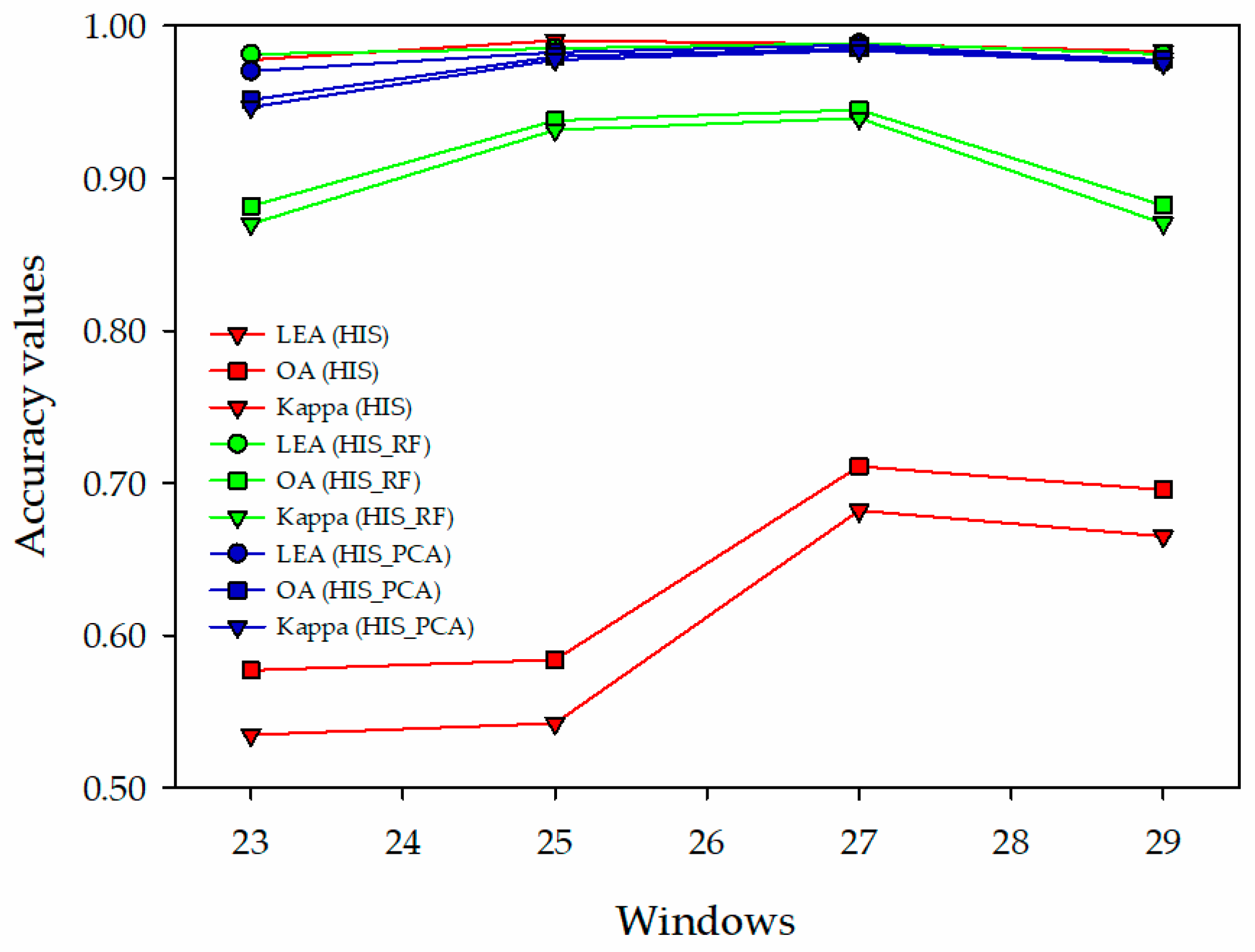
3.3. Classification Using the Improved Prototypical Network with Different Data Sources
3.4. Comparison to 3D-CNN
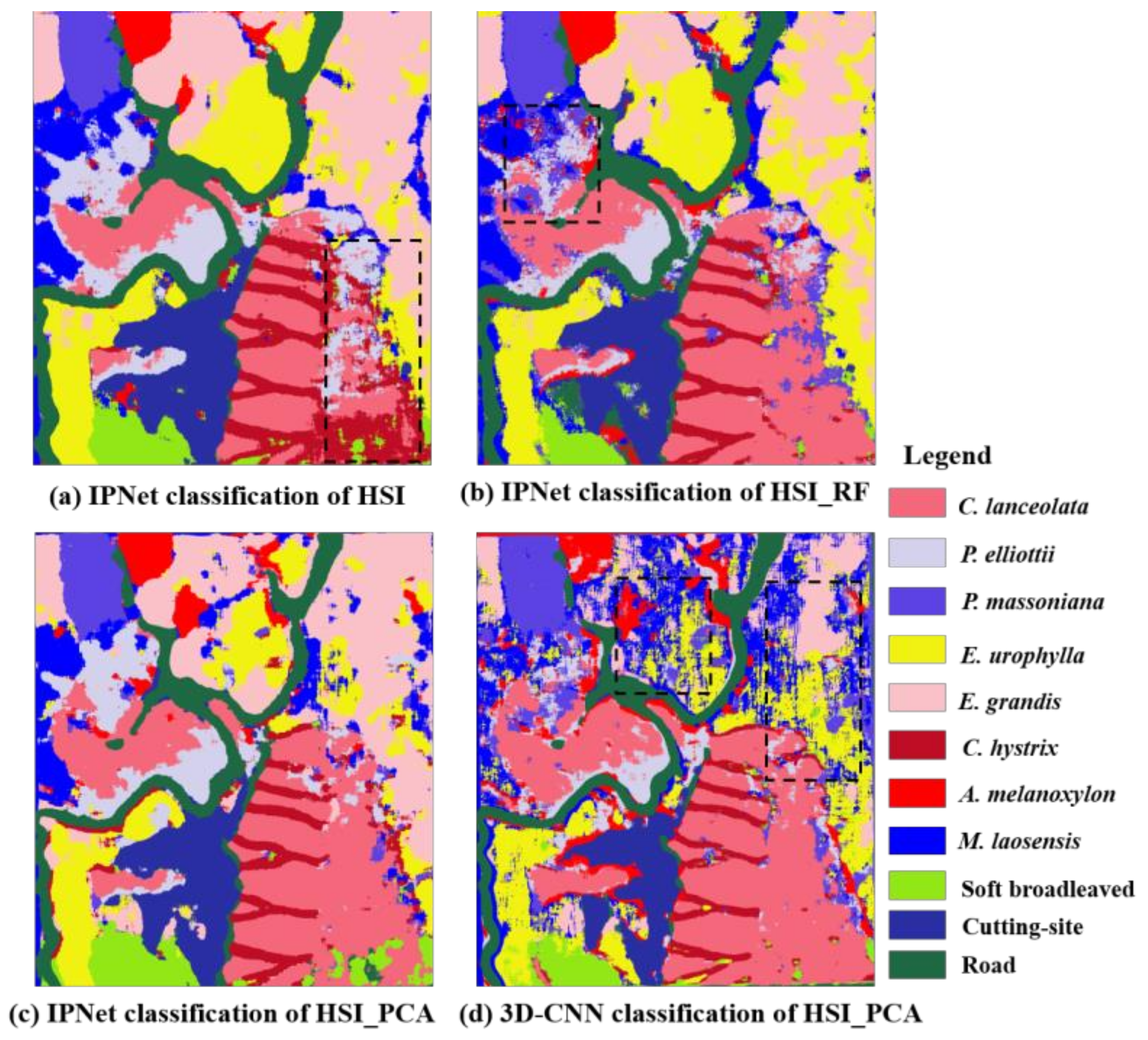
3.5. Classification Results Map
4. Discussion
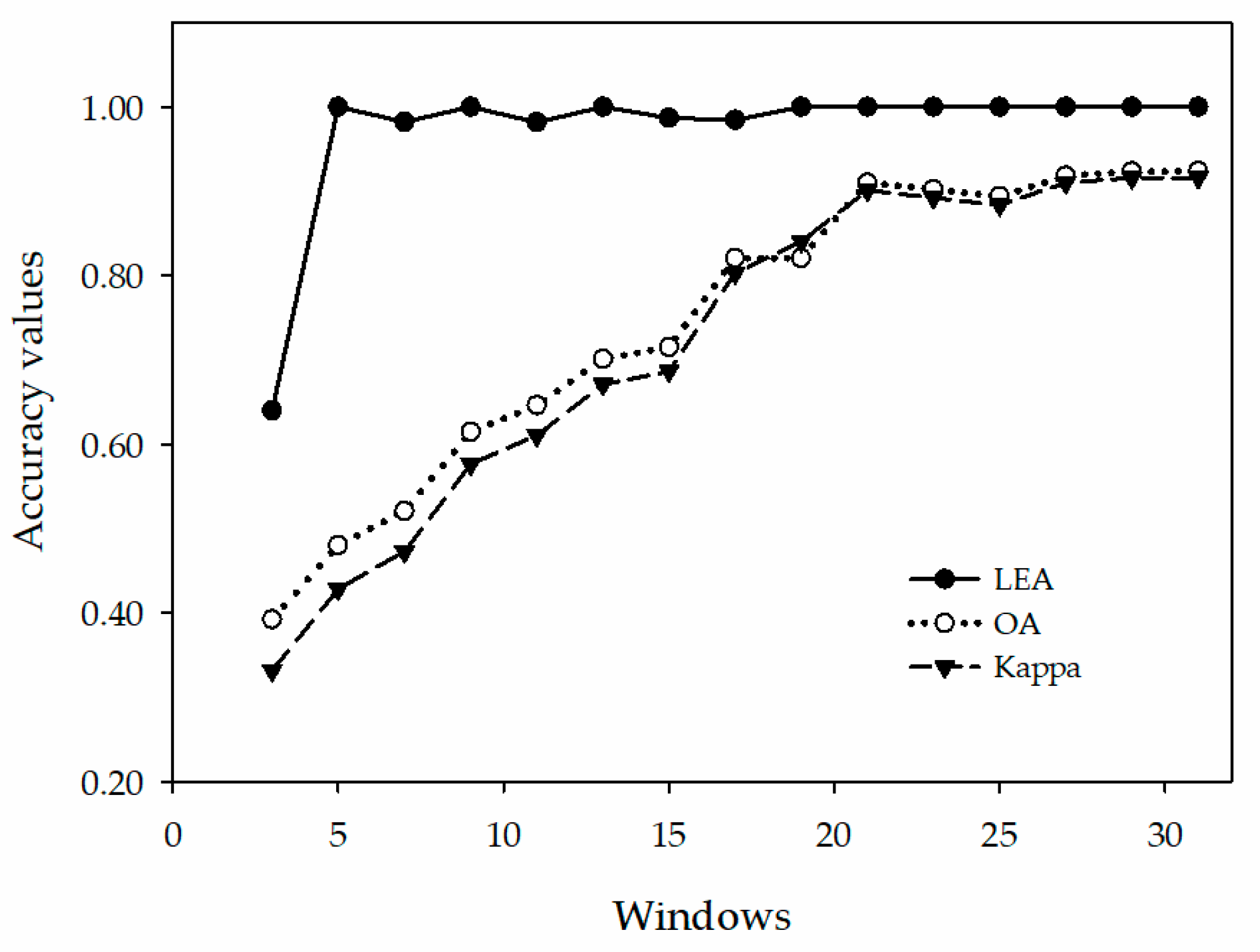
4.1. The Size of the Sample Windows on the Classification of the Prototypical Network
4.2. Different Data Sources on Classification of the Improved Prototypical Network
4.3. The Advantage of the Improved Prototypical Network
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, M.; Li, J.; Sheng, C.; Xu, J.; Wu, L. A review of wetland remote sensing. Sensors 2017, 17, 777. [Google Scholar] [CrossRef]
- Pu, R.; Gong, P.; Heald, R. In situ hyperspectral data analysis for nutrient estimation of giant sequoia. In Proceedings of the IEEE 1999 International Geoscience and Remote Sensing Symposium IGARSS’99, Hamburg, Germany, 28 June–2 July 1999; (Cat. No. 99CH36293). Volume 23, pp. 1827–1850. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote. Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Cao, J.; Leng, W.; Liu, K.; Liu, L.; He, Z.; Zhu, Y. Object-based mangrove species classification using unmanned aerial vehicle hyperspectral images and digital surface models. Remote. Sens. 2018, 10, 89. [Google Scholar] [CrossRef]
- Zhong, Y.; Cao, Q.; Zhao, J.; Ma, A.; Zhao, B.; Zhang, L. Optimal decision fusion for urban land-use/land-cover classification based on adaptive differential evolution using hyperspectral and LiDAR data. Remote. Sens. 2017, 9, 868. [Google Scholar] [CrossRef]
- Berhane, T.M.; Lane, C.R.; Wu, Q.; Autrey, B.C.; Anenkhonov, O.A.; Chepinoga, V.V.; Liu, H. Decision-tree, rule-based, and random forest classification of high-resolution multispectral imagery for wetland mapping and inventory. Remote. Sens. 2018, 10, 580. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Chen, C.; Jiang, F.; Yang, C.; Rho, S.; Shen, W.; Liu, S.; Liu, Z. Hyperspectral classification based on spectral–spatial convolutional neural networks. Eng. Appl. Artif. Intell. 2018, 68, 165–171. [Google Scholar] [CrossRef]
- Harsanyi, J.; Chang, C.-I. Hyperspectral image classification and dimensionality reduction: An orthogonal subspace projection approach. IEEE Trans. Geosci. Remote. Sens. 1994, 32, 779–785. [Google Scholar] [CrossRef]
- Sagan, V.; Asari, V.K.; Sagan, V. Progressively Expanded Neural Network (PEN Net) for hyperspectral image classification: A new neural network paradigm for remote sensing image analysis. ISPRS J. Photogramm. Remote. Sens. 2018, 146, 161–181. [Google Scholar] [CrossRef]
- Chang, C.-I.; Du, Q.; Sun, T.-L.; Althouse, M.L.G. A joint band prioritization and band-decorrelation approach to band selection for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 1999, 37, 2631–2641. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, T.; Wang, T.; Holzwarth, S.; Heiden, U.; Pinnel, N.; Zhu, X.; Heurich, M. Tree species classification using plant functional traits from LiDAR and hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 207–219. [Google Scholar] [CrossRef]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A review of supervised object-based land-cover image classification. ISPRS J. Photogramm. Remote. Sens. 2017, 130, 277–293. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, X. Object-based tree species classification using airborne hyperspectral images and LiDAR data. Forests 2019, 11, 32. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Learning multiscale and deep representations for classifying remotely sensed imagery. ISPRS J. Photogramm. Remote. Sens. 2016, 113, 155–165. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A comparison of selected classification algorithms for mapping bamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Benediktsson, J.A.; Swain, P.H.; Ersoy, O.K. Conjugate-gradient neural networks in classification of multisource and very-high-dimensional remote sensing data. Int. J. Remote. Sens. 1993, 14, 2883–2903. [Google Scholar] [CrossRef]
- Keshk, H.; Yin, X.-C. Classification of EgyptSat-1 images using deep learning methods. Int. J. Sensors, Wirel. Commun. Control. 2020, 10, 37–46. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.; Xu, X. MugNet: Deep learning for hyperspectral image classification using limited samples. ISPRS J. Photogramm. Remote. Sens. 2018, 145, 108–119. [Google Scholar] [CrossRef]
- Guo, Y.; Han, S.; Cao, H.; Zhang, Y.; Wang, Q. Guided filter based deep recurrent neural networks for hyperspectral image classification. Procedia Comput. Sci. 2018, 129, 219–223. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M. Urban tree species classification using a WorldView-2/3 and LiDAR data fusion approach and deep learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Cong, R.; Li, X.; Fu, H.; Lei, J. Joint spatial-spectral hyperspectral image classification based on convolutional neural network. Pattern Recognit. Lett. 2020, 130, 38–45. [Google Scholar] [CrossRef]
- Ashiquzzaman, A.; Tushar, A.K.; Islam, R.; Shon, D.; Im, K.; Park, J.-H.; Lim, D.-S.; Kim, J. Reduction of overfitting in diabetes prediction using deep learning neural network. In IT Convergence and Security 2017; Springer: Singapore, 2018; Volume 449, pp. 35–43. [Google Scholar] [CrossRef]
- Hinton, G.; Deng, L.; Yu, D.; Dahl, G.E.; Mohamed, A.-R.; Jaitly, N.; Senior, A.; Vanhoucke, V.; Nguyen, P.; Sainath, T.N.; et al. Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal. Process. Mag. 2012, 29, 82–97. [Google Scholar] [CrossRef]
- Tong, Q.X.; Xue, Y.Q.; Zhang, L.F. Progress in hyperspectral remote sensing science and technology in China over the past threedecades. IEEE J. Stars. 2014, 7, 70–91. [Google Scholar]
- Li, Y.; Li, Q.; Liu, Y.; Xie, W. A spatial-spectral SIFT for hyperspectral image matching and classification. Pattern Recognit. Lett. 2019, 127, 18–26. [Google Scholar] [CrossRef]
- Li, S.; Song, W.; Fang, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Deep learning for hyperspectral image classification: An overview. IEEE Trans. Geosci. Remote. Sens. 2019, 57, 6690–6709. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.; Xu, X. R-VCANet: A new deep-learning-based hyperspectral image classification method. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2017, 10, 1975–1986. [Google Scholar] [CrossRef]
- Xu, Y.; Du, B.; Zhang, F.; Zhang, L. Hyperspectral image classification via a random patches network. ISPRS J. Photogramm. Remote. Sens. 2018, 142, 344–357. [Google Scholar] [CrossRef]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A Convolutional neural network classifier identifies tree species in mixed-conifer forest from hyperspectral imagery. Remote. Sens. 2019, 11, 2326. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Paoletti, M.; Haut, J.; Plaza, J.; Plaza, A. A new deep convolutional neural network for fast hyperspectral image classification. ISPRS J. Photogramm. Remote. Sens. 2018, 145, 120–147. [Google Scholar] [CrossRef]
- Audebert, N.; Le Saux, B.; Lefèvre, S. Deep learning for classification of hyperspectral data: A comparative review. IEEE Geosci. Remote. Sens. Mag. 2019, 7, 159–173. [Google Scholar] [CrossRef]
- Vilalta, R.; Drissi, Y. A perspective view and survey of meta-learning. Artif. Intell. Rev. 2002, 18, 77–95. [Google Scholar] [CrossRef]
- Bernhard, P.; Hilan, B.; Christophe, G.G. Meta-learning by landmarking various learning algorithms. In Proceedings of the 17th International Conference on Machine Learning, Stanford University, Stanford, CA, USA, 29 June–2 July 2000. [Google Scholar]
- Vinyals, O.; Blundell, C.; Lillicrap, T.; Kavukcuoglu, K.; Wierstra, D. Matching Networks for One Shot Learning. Proceedings of Advances in Neural Information Processing Systems (NIPS 2016), Barcelona, Spain, 5–10 December 2016; pp. 3630–3638. [Google Scholar]
- Fort, S. Gaussian Prototypical Networks for Few-Shot Learning on Omniglot. Proceedings of Bayesian Deep Learning NIPS 2017 Workshop, Long Beach, CA, USA, 9 December 2017. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R.S. Prototypical Networks for Few-shot Learning. Proceeding of Advances in Neural Information Processing Systems, NIPS 2017, Long Beach, CA, USA, 4–9 December 2017; pp. 4077–4087. [Google Scholar]
- Zhang, L.; Lu, G. Study on the biomass of chinese fir plantation in state-owned Gao Feng Forest Farm of Guangxi. For. Sci. Technol. Rep. 2019, 51, 43–47. [Google Scholar]
- Hao, J.; Yang, W.; Li, Y.; Hao, J. Atmospheric correction of multi-spectral imagery ASTER. Remote Sens. 2008, 1, 7–81. [Google Scholar]
- Zhang, N.; Zhang, X.; Yang, G.; Zhu, C.; Huo, L.; Feng, H. Assessment of defoliation during the Dendrolimus tabulaeformis Tsai et Liu disaster outbreak using UAV-based hyperspectral images. Remote. Sens. Environ. 2018, 217, 323–339. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, L.; Zhang, X. Three-dimensional convolutional neural network model for tree species classification using airborne hyperspectral images. Remote. Sens. Environ. 2020, 247, 111938. [Google Scholar] [CrossRef]
- Cao, K.; Zhang, X. An improved res-UNet model for tree species classification using airborne high-resolution images. Remote. Sens. 2020, 12, 1128. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Rezaee, M.; Mohammadimanesh, F.; Zhang, Y. Very deep convolutional neural networks for complex land cover mapping using multispectral remote sensing imagery. Remote. Sens. 2018, 10, 1119. [Google Scholar] [CrossRef]
- Yu, S.; Jia, S.; Xu, C. Convolutional neural networks for hyperspectral image classification. Neurocomputing 2017, 219, 88–98. [Google Scholar] [CrossRef]



| Hyperspectral: AISA Eagle II | |||
|---|---|---|---|
| Spectral resolution | 3.3 nm | Spatial resolution | 1 m |
| Angle of view | 37.7° | Spatial pixels | 1024 |
| Instantaneous angle of view | 0.646 mrad | Spectral sampling interval | 4.6 nm |
| Focal length | 18.5 mm | Bit depth | 12 bits |
| Layer | Output Shape (Height, Width, Depth, Numbers of Feature Map) | Parameters Number |
|---|---|---|
| Conv3d_1 | (27, 27, 5, 4) | 112 |
| Batch_norm_1 | (27, 27, 5, 4) | 16 |
| Max_pool3d_1 | (9, 9, 2, 4) | 0 |
| Conv3d_2 | (9, 9, 2, 8) | 872 |
| Batch_norm_2 | (9, 9, 2, 8) | 32 |
| Conv3d_3 | (9, 9, 2, 16) | 3472 |
| Batch_norm_3 | (9, 9, 2, 16) | 64 |
| Conv3d_4 | (9, 9, 2, 32) | 13856 |
| Batch_norm_4 | (9, 9, 2, 32) | 128 |
| Conv3d_5 | (9, 9, 2, 64) | 55360 |
| Batch_norm_5 | (9, 9, 2, 64) | 256 |
| Max_pool3d_2 | (3, 3, 1, 64) | 0 |
| Dropout_1 | (3, 3, 1, 64) | 0 |
| Flatten | (576) | 0 |
| Dense_1 | (128) | 73856 |
| Dropout_2 | (128) | 0 |
| Dense_2 | (11) | 1419 |
| Total parameters: 149,443 | ||
| Trainable parameters: 149,195 | ||
| Non-trainable parameters: 248 | ||
| Index | 3 × 3 | 5 × 5 | 7 × 7 | 9 × 9 | 11 × 11 | 13 × 13 | 15 × 15 | 17 × 17 | 19 × 19 | 21 × 21 | 23 × 23 | 25 × 25 | 27 × 27 | 29 × 29 | 31 × 31 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epochs/Iterations | 20/500 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 |
| LEA | 0.6402 | 1.0000 | 0.9822 | 1.0000 | 0.9818 | 1.0000 | 0.9867 | 0.9846 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| OA (%) | 39.22 | 48.02 | 52.04 | 61.46 | 64.61 | 70.10 | 71.52 | 82.08 | 85.50 | 91.01 | 90.22 | 89.42 | 91.85 | 92.35 | 92.40 |
| Kappa | 0.3315 | 0.4282 | 0.4723 | 0.5761 | 0.6107 | 0.6710 | 0.6868 | 0.8028 | 0.8404 | 0.9011 | 0.8924 | 0.8836 | 0.9103 | 0.9159 | 0.9163 |
| Training Times (S) | 68 | 43 | 57 | 122 | 159 | 247 | 314 | 447 | 626 | 732 | 872 | 1091 | 1252 | 1480 | 1708 |
| Index | 17 × 17 | 19 × 19 | 21 × 21 | 23 × 23 | ||||||||
| Dropout | 0.5 | 0.7 | 0.9 | 0.5 | 0.7 | 0.9 | 0.5 | 0.7 | 0.9 | 0.5 | 0.7 | 0.9 |
| Epochs/Iterations | 20/500 | 20/150 | 20/100 | 20/500 | 20/150 | 20/100 | 20/500 | 20/150 | 20/100 | 20/500 | 20/150 | 20/100 |
| LEA | 0.9084 | 0.9555 | 0.9829 | 0.9322 | 0.9840 | 0.9967 | 0.9358 | 0.9840 | 0.9958 | 0.9314 | 0.9816 | 0.9984 |
| OA (%) | 96.49 | 93.26 | 85.35 | 95.77 | 95.02 | 85.51 | 96.83 | 95.01 | 89.62 | 98.15 | 97.41 | 88.72 |
| Kappa | 0.9614 | 0.9259 | 0.8388 | 0.9535 | 0.9452 | 0.8406 | 0.9652 | 0.9451 | 0.8858 | 0.9796 | 0.9715 | 0.8759 |
| Training Times (S) | 2245 | 684 | 463 | 2961 | 887 | 593 | 3821 | 1132 | 753 | 4351 | 1299 | 874 |
| Index | 25 × 25 | 27 × 27 | 29 × 29 | 31 × 31 | ||||||||
| Dropout | 0.5 | 0.7 | 0.9 | 0.5 | 0.7 | 0.9 | 0.5 | 0.7 | 0.9 | 0.5 | 0.7 | 0.9 |
| Epochs/Iterations | 20/500 | 20/100 | 20/100 | 20/500 | 20/100 | 20/100 | 20/500 | 20/100 | 20/100 | 20/500 | 20/100 | 20/100 |
| LEA | 0.9504 | 0.9826 | 0.9976 | 0.9515 | 0.9874 | 1.0000 | 0.9506 | 0.9764 | 0.9980 | 0.9065 | 0.9795 | 0.9978 |
| OA (%) | 98.30 | 97.97 | 88.67 | 98.52 | 98.53 | 95.94 | 99.17 | 97.76 | 95.88 | 98.98 | 95.02 | 96.01 |
| Kappa | 0.9813 | 0.9777 | 0.8754 | 0.9837 | 0.9838 | 0.9553 | 0.9909 | 0.9754 | 0.9547 | 0.9888 | 0.9452 | 0.9561 |
| Training Times (S) | 5403 | 1122 | 1091 | 6458 | 1290 | 1275 | 7190 | 1449 | 1443 | 9565 | 1676 | 1669 |
| Index/Categories | 23 × 23 | 25 × 25 | 27 × 27 | 29 × 29 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSI | HSI_RF | HSI_PCA | HSI | HSI_RF | HSI_PCA | HSI | HSI_RF | HSI_PCA | HSI | HSI_RF | HSI_PCA | |
| Epochs/iterations | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 | 20/100 |
| LEA | 0.9776 | 0.9815 | 0.9704 | 0.9904 | 0.9851 | 0.9826 | 0.9880 | 0.9886 | 0.9874 | 0.9833 | 0.9815 | 0.9764 |
| OA (%) | 57.70 | 88.17 | 95.14 | 58.39 | 93.78 | 97.97 | 71.08 | 94.49 | 98.53 | 69.54 | 88.22 | 97.76 |
| Kappa | 0.5347 | 0.8699 | 0.9466 | 0.5423 | 0.9316 | 0.9777 | 0.6819 | 0.9394 | 0.9838 | 0.6650 | 0.8704 | 0.9754 |
| Training Times (S) | 1806 | 906 | 893 | 2057 | 1124 | 1122 | 2406 | 1308 | 1290 | 2774 | 1536 | 1449 |
| C. lanceolata | 0.5034 | 0.7624 | 0.9590 | 0.5306 | 0.9166 | 1.0000 | 0.4536 | 0.9186 | 1.0000 | 0.6470 | 0.9554 | 0.9976 |
| P. elliottii | 0.1852 | 0.6916 | 0.8210 | 0.3958 | 0.7810 | 0.8818 | 0.5558 | 0.7862 | 0.9374 | 0.5092 | 0.6754 | 0.9440 |
| P. massoniana | 0.4894 | 0.9342 | 0.9998 | 0.6598 | 0.9996 | 0.9970 | 0.8812 | 0.9962 | 1.0000 | 0.4814 | 0.6630 | 1.0000 |
| E. urophylla | 0.4980 | 0.7896 | 0.9258 | 0.5612 | 0.9776 | 0.9394 | 0.4604 | 0.8782 | 1.0000 | 0.8630 | 0.9554 | 0.9954 |
| E. grandis | 0.7660 | 0.9970 | 0.9238 | 0.9336 | 0.8920 | 0.9968 | 0.5574 | 0.9524 | 1.0000 | 0.9856 | 0.9464 | 0.9282 |
| C. hystrix | 0.5286 | 0.9590 | 0.9382 | 0.5912 | 0.8472 | 0.9552 | 0.9314 | 0.9330 | 0.9818 | 0.7570 | 0.9468 | 0.9922 |
| A. melanoxylon | 0.4792 | 0.8426 | 1.0000 | 0.5962 | 0.9962 | 0.9998 | 0.7338 | 0.9998 | 0.9992 | 0.4918 | 1.0000 | 1.0000 |
| M. laosensis | 0.3608 | 0.7292 | 0.9024 | 0.2752 | 0.9500 | 0.9568 | 0.4936 | 0.8870 | 0.9950 | 0.6176 | 0.6740 | 0.9696 |
| Soft broadleaf | 0.6344 | 0.9934 | 0.9958 | 0.7674 | 0.9558 | 0.9996 | 0.8376 | 0.9948 | 0.9602 | 0.7192 | 0.8874 | 1.0000 |
| Cutting site | 0.9450 | 1.0000 | 1.0000 | 0.3530 | 1.0000 | 1.0000 | 0.9610 | 1.0000 | 1.0000 | 0.8664 | 1.0000 | 1.0000 |
| Road | 0.9568 | 1.0000 | 1.0000 | 0.7592 | 1.0000 | 1.0000 | 0.9532 | 1.0000 | 1.0000 | 0.7114 | 1.0000 | 1.0000 |
| Index/Categories | IPrNet | 3D-CNN |
|---|---|---|
| LEA | 0.9874 | 0.8920 |
| OA (%) | 98.53 | 87.50 |
| Kappa | 0.9838 | 0.8625 |
| Training Times (S) | 1290 | 4986 |
| C. lanceolata | 1.0000 | 0.8214 |
| P. elliottii | 0.9374 | 0.8889 |
| P. massoniana | 1.0000 | 0.7586 |
| E. urophylla | 1.0000 | 0.7407 |
| E. grandis | 1.0000 | 0.8148 |
| C. hystrix | 0.9818 | 0.9167 |
| A. melanoxylon | 0.9992 | 0.8333 |
| M. laosensis | 0.9950 | 0.9375 |
| Soft broadleaf | 0.9602 | 1.0000 |
| Cutting site | 1.0000 | 1.0000 |
| Road | 1.0000 | 1.0000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, X.; Chen, L.; Zhang, X.; Chen, E. Improved Prototypical Network Model for Forest Species Classification in Complex Stand. Remote Sens. 2020, 12, 3839. https://doi.org/10.3390/rs12223839
Tian X, Chen L, Zhang X, Chen E. Improved Prototypical Network Model for Forest Species Classification in Complex Stand. Remote Sensing. 2020; 12(22):3839. https://doi.org/10.3390/rs12223839
Chicago/Turabian StyleTian, Xiaomin, Long Chen, Xiaoli Zhang, and Erxue Chen. 2020. "Improved Prototypical Network Model for Forest Species Classification in Complex Stand" Remote Sensing 12, no. 22: 3839. https://doi.org/10.3390/rs12223839
APA StyleTian, X., Chen, L., Zhang, X., & Chen, E. (2020). Improved Prototypical Network Model for Forest Species Classification in Complex Stand. Remote Sensing, 12(22), 3839. https://doi.org/10.3390/rs12223839