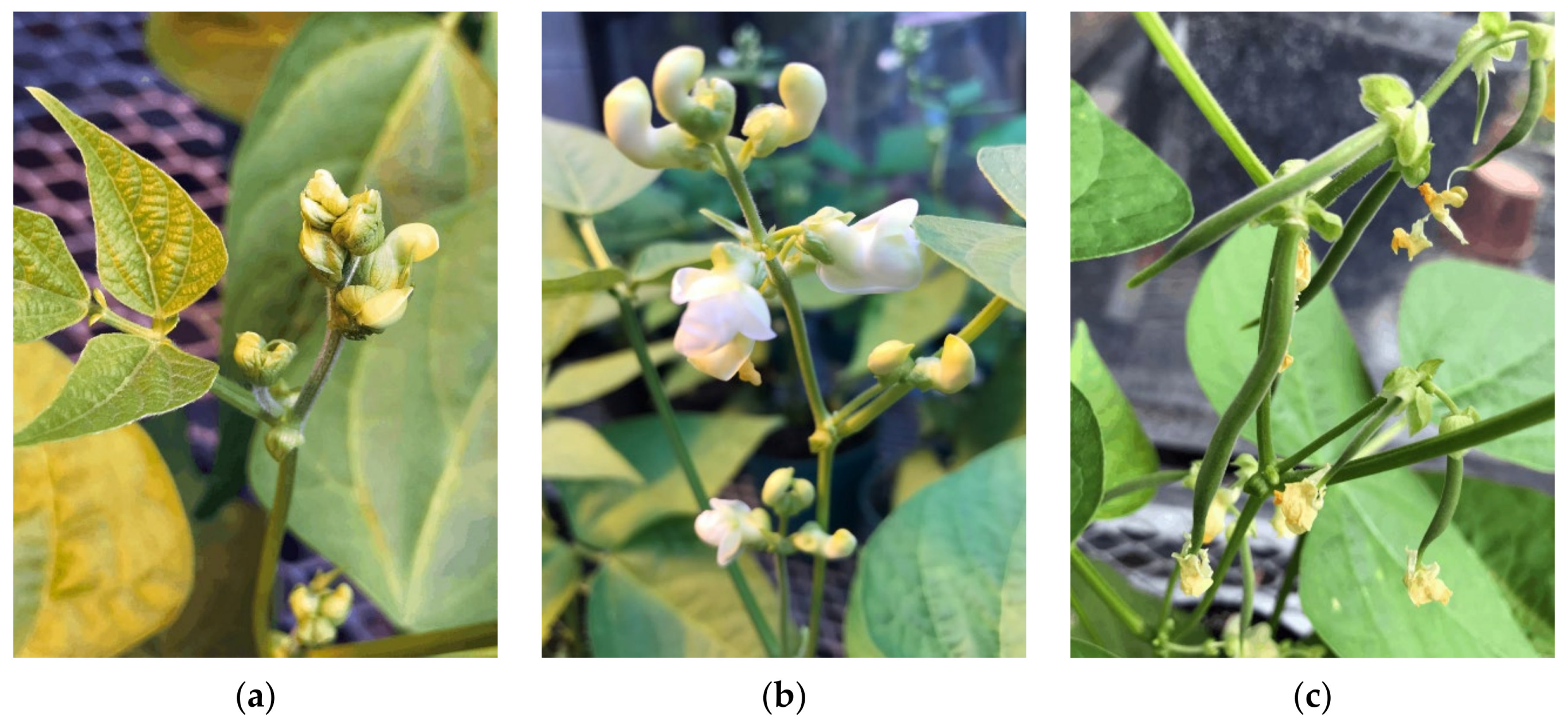
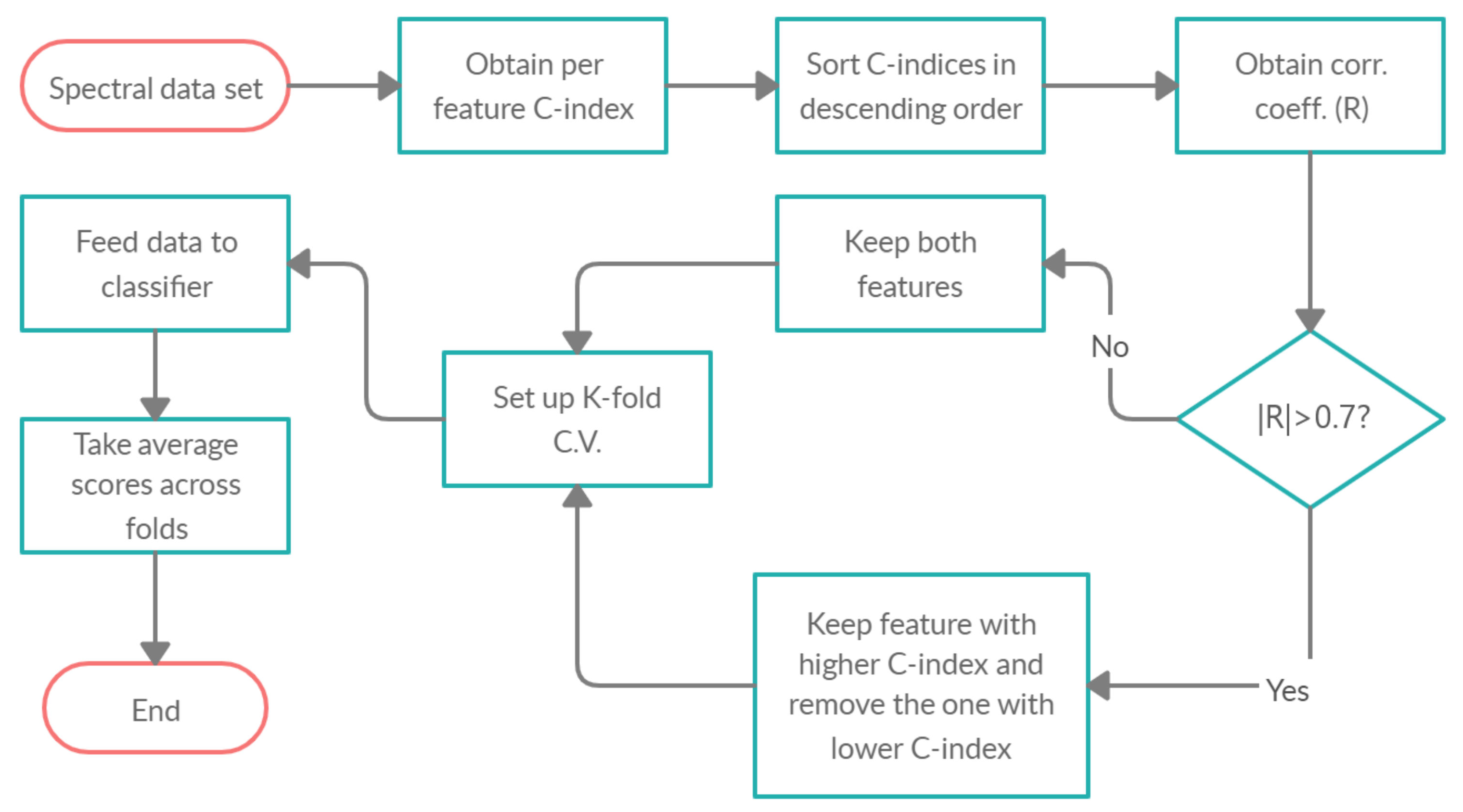
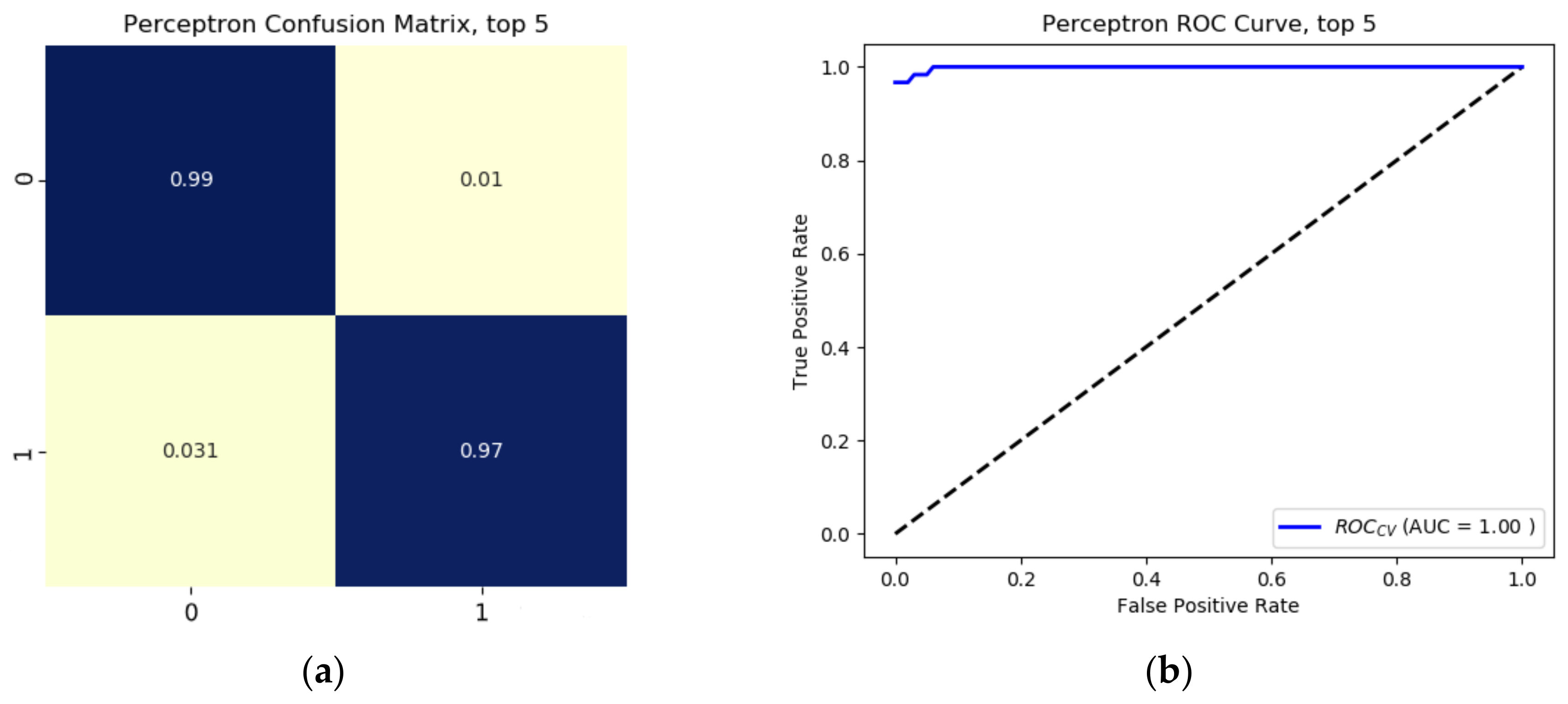
1. Introduction
Snap bean is one of the largest sources of broad-acre crop income in the United States, and is planted from California to New York [
1]. However, more than four million pounds of the harvested snap bean was disposed of unsold in 2017 [
1]. One of the contributing factors to overall “food loss” is our lack of understanding, developing, and adopting site-specific crop management, with one example being crop growth stage assessment, e.g., when and where to schedule the harvest, or apply pesticides and fertilizers. Precision agriculture, or site-specific management, has been shown to provide management inputs that could maximize output by optimizing inputs [
2]. This approach has been put into practice in recent decades via a variety of remote sensing systems [
3]. Remote sensing systems are non-destructive in nature, provide rapid data collection, and offer synoptic coverage, via ground-based (e.g., spectroradiometers), airborne (e.g., aircraft and UAS), or spaceborne (satellite) platforms [
4,
5]. Precision agriculture and remote sensing thus are closely tied and poised to take advantage of sensors, such as hyperspectral and structural systems, that exceed our visual assessment ability.
Most spectral imaging systems are categorized as either of the multispectral and hyperspectral (imaging spectroscopy) types, with the former defined as broader, non-contiguous bands and the latter as containing narrow, contiguous spectral coverage [
6]. The spectral range covered by these devices differ between applications, from visible-to-near-infrared (VNIR; 380–1000 nm) enabling assessments of (i) soil properties [
7,
8,
9,
10,
11], (ii) crop disease [
12,
13], (iii) crop yield [
14,
15,
16], (iv) crop ripeness [
15,
17,
18]; to shortwave- (SWIR; 1000–2500 nm), midwave-, and far-infrared (1–200 µm), with applications focused on chemical identification in the food industry [
19,
20,
21,
22] and soil analysis [
23,
24]. We will focus our discussion on the VNIR-SWIR, due to its pivotal role in crop analysis.
The electromagnetic energy incident on crops results in a unique spectral characterization, which is called a spectral signature (or spectral response), associated with either vegetation as general cover type, or the physiological state of a plant [
25]. As a plant grows, its biochemical composition is altered and in turn impacts the spectral response [
26]. This change in the spectral response of crops typically is more pronounced in the VNIR-SWIR region, in terms of spectral curve shape, amplitude, and absorption features [
27]. The change in crop spectral behavior in the VNIR-SWIR region is dominated by pigment levels, scattering due to intercellular leaf structure, water status, etc. [
28,
29,
30]. These variabilities in crop spectral behavior have led scholars to use spectral information to assess plant health, growth, and deviation from an “ideal” seasonal spectral evolution. We will focus on the applications related to growth stage assessment using two different approaches in this context, namely machine learning techniques and spectral indicators.
Machine learning-based crop growth stage assessment, among many other applications [
31,
32,
33,
34,
35], has gained traction with the advent of artificial intelligence approaches. Doktor et al. (2014) studied the use of random forests (RF) to extract the physiological status of summer barley crops using simulated signatures, as well as hyperspectral laboratory and field measurements in the VNIR-SWIR region [
36]. Their results show that random forests can explain much of the variability (R
2 = 0.80–0.89) in leaf area index (LAI) using spectral data. Yuan et al. (2017) also evaluated the use of machine learning techniques on VNIR hyperspectral data for LAI assessment [
37]. Their findings show that RF proved the most accurate when compared to other approaches, with a high coefficient of determination (R
2 = 0.74). Senthilnath et al. (2013), on the other hand, investigated crop stage classification using hyperspectral images, using dimensionality reduction via principal component analysis (PCA), followed by an unsupervised clustering method with the objective of distinguishing between three different growth stages of wheat [
38]. Findings from this study showed a high efficiency (ɳ = 81.5%), as well as an acceptable kappa coefficient (κ = 0.72).
Spectral indicators and vegetation indices (i.e., a mathematical transform or formulation based on two or more bands) have proven advantageous in terms of rapid assessment of crop spectra, with an extension to crop growth stage assessment [
26,
39]. Vegetation indices are classified as either of the broad- or narrow-band type [
40]. Narrow-band indices correspond to fine spectral resolution and are more sensitive to subtle changes in the spectrum, whereas broad-band indices take advantage of coarser spectral resolution. The peak in the first derivative of reflectance spectra between 680–750 nm, known as red edge peak, is a narrow-band spectral indicator of plant stress and age [
41,
42]. On the other hand, broadband indices, such as the ratio vegetation index (RVI; [
43]) and normalized vegetation index (NDVI; [
44]), highlight relative biomass, as well as plant vigor/health/leaf area [
43]. It also is important to briefly differentiate between plant maturity contexts, which focus either on the fruit-level or plant-level.
Fruit maturity assessment often involves correlating internal characteristics (e.g., chlorophyll, firmness, soluble solid content, moisture content, etc.) with ripeness, using various techniques, such as colorimetry [
45,
46,
47,
48,
49], RGB imaging [
48,
49,
50,
51,
52], VNIR spectroscopy [
53,
54,
55,
56,
57] hyperspectral imaging [
58,
59,
60,
61,
62], and multispectral imaging [
60,
63,
64,
65,
66]. However, there is a general lack of research regarding plant- or leaf-level ripeness evaluation. A study by Johnson et al. (2004) addressed ripeness assessment of coffee fields via airborne multispectral imagery [
67]. The authors called for additional future work, given the inverse correlation of the introduced ripeness index with ground truth ripeness measurements. Furthermore, Merzlyak et al. (1999) assessed both fruit ripening and leaf-level senescence of multiple crops using visible to near-infrared (VNIR) spectroscopy [
68]. A reflectance index (plant senescence reflective index; PSRI) based on the 500 nm, 678 nm, and 700 nm bands, was identified as a quantitative measure that could explain both maturity measures (fruit ripening and leaf senescence). It is with studies such as these in mind, that we developed a detailed experimental setup to evaluate our ability to assess growth stages and optimal harvest scheduling, with snap bean as a proxy crop.
We hypothesize that growth stage and pod maturity assessment of snap bean for precision agriculture purposes can be addressed via hyperspectral systems, since growth and maturity are functions of spectral and biophysical features. The objectives of this research therefore were to: (i) evaluate the feasibility of snap bean growth stage assessment using a machine learning and spectral indices approach; (ii) assess snap bean pod maturity level via machine learning techniques, and (iii) determine spectral and biophysical attributes that correspond to accurate growth stage and pod maturity classification.
4. Discussion
The generated impact plots by PCA can be linked to the plants’ biophysical attributes (
Figure 6). The generated impact plot for PC-1 demonstrated changes in the 700–1800 nm range, especially for the 700–1250 nm range. This range is responsible for both water and structural absorption (e.g., cellulose) features [
101]; plant water intake, as well as structural characteristics, which change substantially with plant maturity. PC-2, on the other hand, showed changes in the visible and higher end of the SWIR region, denoting overall plant growth; the ratio between green reflectance and the red absorption trough, which increases as the plant matures, is tied to total visible light absorption by chlorophyll. PC-3 is mostly responsible for sugar, starch, and protein (both in the 800–1200 nm and ~1900 nm regions), cellulose and sugar absorption in the ~1490 nm region, as well as starch, protein, oil, and water absorption in the 800–1200 nm region [
101]. However, in an operational airborne data campaign setting, the 1400 nm and 1900 nm atmospheric water absorption features (low SNR) could impact the PC-3 outcome, thus diminishing the influence of these regions on the growth stage classifiers.
Our binary approach identified spectral wavelengths, at each growth stage, which proved to be discriminating. These spectral features, however, need to be scrutinized in terms of their remote sensing implications and extension to operational scenarios and sensors, and ultimately, they need to be linked to plant physiology. For example, the results from the most accurate model [CR-spec.] for binary classification of vegetative growth stage included chlorophyll, lignin, water, nitrogen, and starch-related chemical changes, as evidenced via spectral features:
The 610–620 nm range, identified as discriminating features for both continuum-removal approaches (see
Table 3) are strong indicators of chlorophyll absorption in the red spectral region, necessary for photosynthesis [
101]. Also, Card et al. (1988) found high correlation coefficients (R
2 = 0.93) between nitrogen and the 620 nm band, and between lignin and the 610 nm band (R
2 = 0.93) [
106].
The 700–710 nm wavelength region represents the well-known red edge spectral feature, and was identified by continuum-removal models. The red edge peak reflects plant growth, health/vigor, and maturity, as well as indicating sugar absorption [
42]. The red edge peak is an easy-to-detect spectral metric in first derivative spectra, since the slope of the red edge exhibits a common high value in comparison to other spectral metrics. However, it is notable that the continuum-removal approach proved useful in independently identifying the red edge peak.
The single wavelength at ~1390 nm in the SWIR region falls in the atmospheric water absorption feature, due to bending O–H bonds, as well as reflecting total nitrogen absorption [
101,
106].
The single wavelength at 1490 nm was deemed as being explanatory for cellulose and sugar absorption, via stretching of O–H bonds [
101].
The 1560–1570 nm region, resides in the vicinity of two regions, 1540 nm and 1580 nm, which have associations with starch and sugar, due to stretching O–H bonds [
101].
The 2000–2020 nm region, which also appeared in the raw and smoothed data sets, reflects starch absorption, causing deformation in O–H and C–O bonds [
101].
Finally, for the 2380–2390 nm region, we could not find any physiological link in the literature. This may require future efforts to concentrate on detecting ties between spectral signatures in the far-SWIR region and the biophysical/chemical changes due to plant growth.
Results from the continuum removal approach (
Table 4) show the budding maturity stage classification can be coupled with oil content in the plant, which was not important in differentiating vegetative growth, starch, protein, nitrogen, lignin, and water, with detected wavelengths as follows:
The best performing model for the flowering stage was identified as the continuum-removal method. The ability to capture this phenological change is of pivotal importance for plant maturity assessment, especially since it has bearing on disease risk (e.g., white mold caused by the fungus, Sclerotinia sclerotiorum; [
109]). This model identified spectral indicators indicative of oil (detected in budding classification), cellulose (not detected previously), starch, sugar, starch, and lignin, as below:
The 690–705 nm range, previously detected in the vegetative and budding stage, represents the red edge peak spectral metric.
The wavelength at ~1740 nm is linked to lignin absorption [
108].
The ~2080 nm wavelength in the SWIR region is coupled to sugar and starch absorption, via stretching and deforming of O–H bonds [
101].
The wavelength region at ~2310 nm is attributed to oil absorption (previously found in budding stage), via bending in C–H bonds [
101].
A single wavelength was found at 2334 nm, in the vicinity of two absorption features due to starch and cellulose (not detected previously), and also linked to stretching of O–H/C–H bonds and being responsible for deformation in O–H/C–H/CH
2 bonds and molecules [
101].
Lastly, no relevant information was found for the wavelength at 1626 nm and the selection of the 2370–2390 nm region, which calls for future work. It is worth noting that the latter region has been given less attention in the literature due to the associated low SNR caused by atmospheric effects in aerial remote sensing systems.
Finally, pod formation, the final growth stage, is mainly dominated by chlorophyll absorption in the red portion of electromagnetic spectrum, coupled to water, starch, sugar, lignin, protein, nitrogen, and abundant cellulose absorption features (as identified by the most accurate model—continuum-removal). The corresponding wavelengths are:
The 610 nm wavelength, as previously detected in the vegetative classification, is linked to chlorophyll and lignin absorption [
106].
The 700–710 nm wavelength region, representing the red edge peak, proves to be crucial in all stages of growth (detected in all stages of growth; see
Table 3,
Table 4 and
Table 5).
The wavelength at ~1190 nm, in the proximity of water, cellulose, starch, lignin absorption ranges (~1200 nm), due to bending of O–H bonds.
The single wavelength at 1490 nm corresponds to cellulose and sugar absorption, with stretching in O–H bonds.
The wavelength at ~1820 nm (stretching of O–H/C–H bonds) is responsible for cellulose absorption.
The ~2350 nm wavelength, accountable for absorption of cellulose, protein, and nitrogen, causing CH
2 molecules to bend and deforming C–H bonds [
101]. Moreover, Card et al. (1988) identified that wavelength at 2350 nm correlates to plant’s lignin level [
106].
Finally, no information was found in the literature for the ~1230 nm, 2040 nm, and ~1380 nm wavelength regions, thus demanding future work. It also is worth noting that the selected ~2390 nm feature may be rendered useless due to its proximity to the longer SWIR region, which suffers from low SNR due to atmospheric considerations.
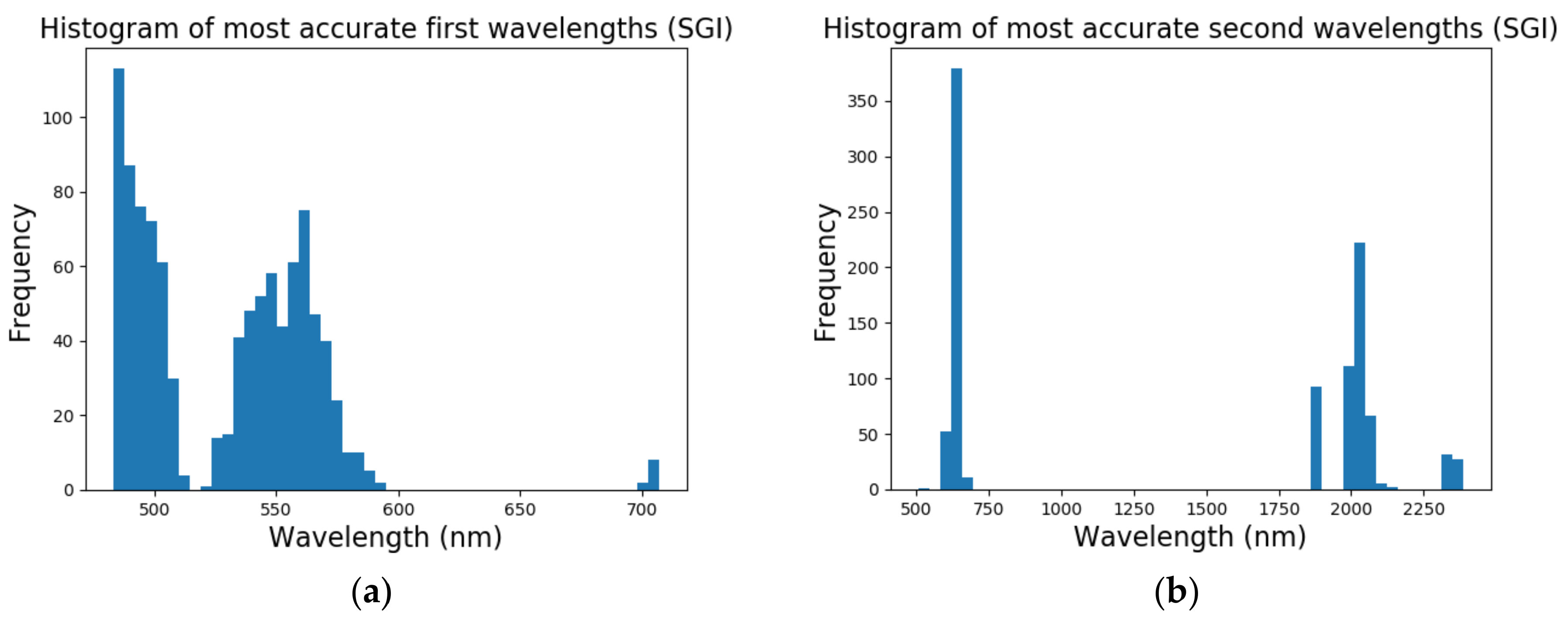
In short, the multi-class growth classification, using the proposed vegetation indices (ratio and normalized forms), identified two discriminative wavelengths, namely at 493 nm and 640 nm. Both wavelengths varied with chlorophyll absorption throughout the growth stages of the plant. Results from the pod maturity classification, on the other hand, showed that models that performed best (i.e., [CR-spec.] models) used detected wavelengths in the ~550 nm region (attributed to reflective green pigment and leaf senescence/yellowing), the 720–740 nm range (encompassing the red edge peak, between 2000-2070 nm (starch absorption, causing O–H and C–O deformation in the 2000 nm area), the 2040 nm spectral region (no relevant scientific link found), and protein and nitrogen absorption stretching and bending of N–H and N=H bands in the 2060 nm spectral region [
101].
5. Conclusions
We studied growth stage and pod maturity classification of snap bean, as a proxy broadacre crop, via an experimental greenhouse setup with a high spectral and temporal resolution. We approached growth stage assessment with binary (one-vs.-rest) and multi-class classification (vegetative index identifier), and pod maturity detection via multi-class classification. The reason we approached this study using both binary and multi-class fashion for growth stage classification, was to identify wavelengths that are not only discriminatory between four different levels of maturity, but to also generate results that could detect spectral wavelengths corresponding to each stage of snap-bean maturity, which is more conclusive and extensible to operational implementation.
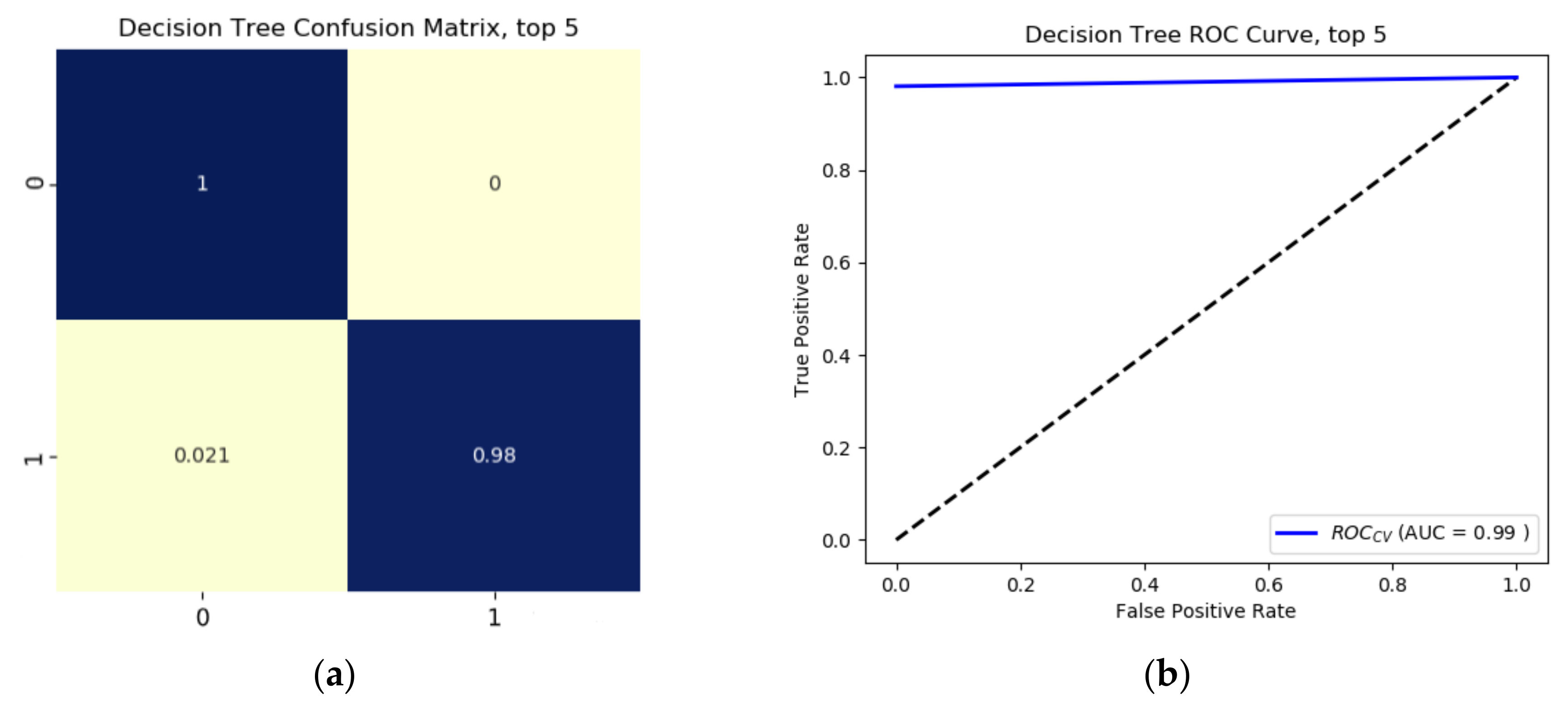
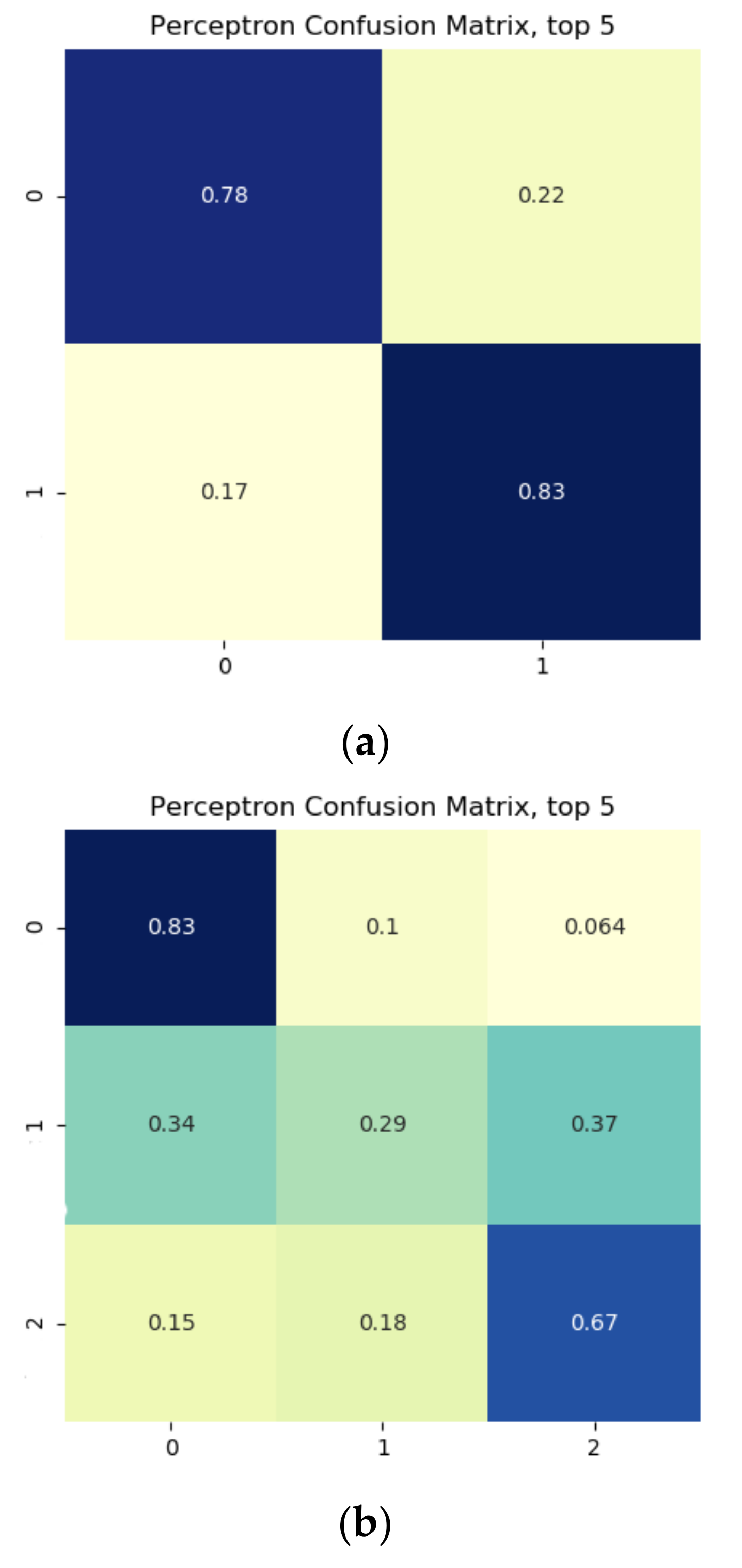
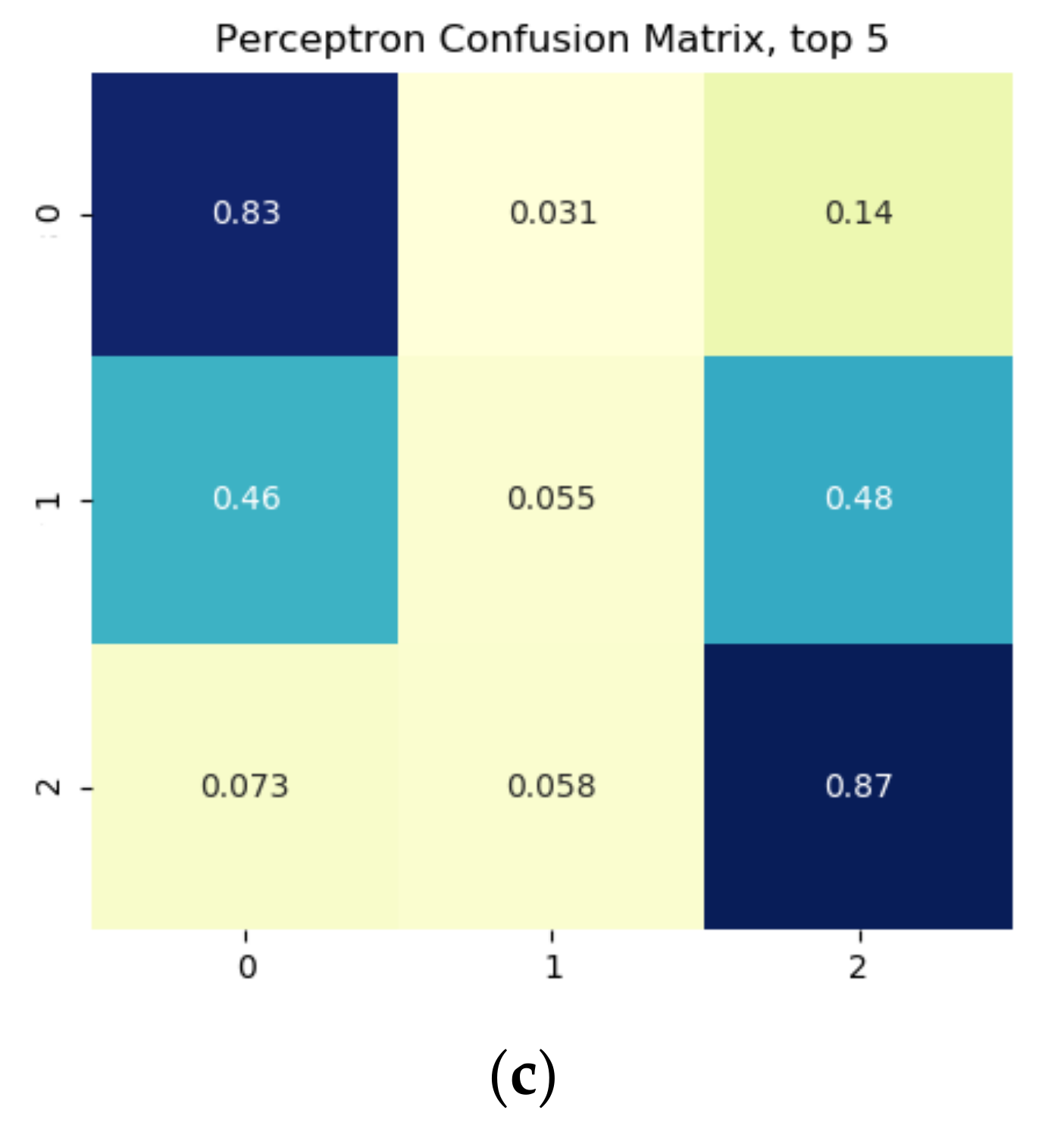
Results from the binary growth stage classification showed the continuum-removal technique was adept at detecting growth-related spectral features for all maturity stages, with accuracies ranging from 0.90–0.98. We determined that addition of biophysical features to spectral data for growth stage assessment enhanced the classification accuracy. Despite this improvement, it was found that the continuum-removal approach exhibited similarly promising results, with or without the addition of biophysical attributes. Findings for the multi-class growth stage classification approach showed that two distinguishing wavelengths at 493 nm and 640 nm yielded the best separation between the four growth classes. Accuracies of 79% for the ratio index (i.e., snap-bean growth index [SGI]) and 78% for the normalized difference form (i.e., normalized difference snap-bean growth index [NDSI]) were observed. The selected binary classification algorithm was effective at determining plant growth stage, relying on both VIS-NIR and SWIR regions for accurate outcomes. However, the multi-class classification and the proposed indices also yielded solid classification results, even if with marginally lower accuracies, with the distinct benefit of only using two wavelengths in the visible region.
Findings from the pod maturity assessment, coupled to harvest scheduling, proved that distinction between not ready-to-harvest and ready-to-harvest sieve sizes (immature + mid-mature vs. mature + over-mature; i.e., industry sieve sizes 1–4 vs. 5–6) is feasible with solid accuracy measures (~78% with only two selected narrow-band wavelengths), while the algorithm failed to differentiate immature vs. mid-mature and mature vs. over-mature due to the similarity of spectral data between these pod maturity levels.
Specific limitations of this study include the relatively small sample size, our focus on only one snap bean cultivar (i.e., cv. Huntington), and the absence of mixed-pixel effects and atmospheric noise by collecting data under controlled laboratory conditions. All of these factors can be evaluated in future studies, e.g., an extension to unmanned aerial systems (UAS) and subsequent transitioning to other airborne remote sensing platforms would introduce both mixed-pixel and atmospheric noise and enable sampling of thousands of individual plants. In terms of the advantages of this approach, we noted the robustness of the algorithms to newly introduced data, while the growth stage classification results exhibited high classification accuracy via the SWIR region, and solid performance using only two bands in the visible spectral domain. We are especially intrigued by this last finding, i.e., the potential of using only two wavelengths in the “cheap” silicon detector range to develop cost-effective, operational platforms to accurately assess snap bean growth stages. An acceptable classification performance for pod maturity to differentiate between not ready-to-harvest and ready-to-harvest, with identified wavelengths residing in the silicon and SWIR spectral regions, may prove to be advantageous to operational implementation, as long as a practical and affordable solution is available to merge the two detectors into one multispectral sensor suite.