Improving K-Nearest Neighbor Approaches for Density-Based Pixel Clustering in Hyperspectral Remote Sensing Images
Abstract
1. Introduction
- Unsupervised: no labeled samples for training nor the actual number of clusters are available;
- Nonparametric: no information about the clusters’ characteristics (shape, size, density, dimensionality) is available;
- Easy parametrization: the method only relies on a small number of parameters that are intuitive;
- Deterministic: the clustering results do not depend on a random initialization step and are strictly reproducible.
2. Notation
3. Relation to Prior Works
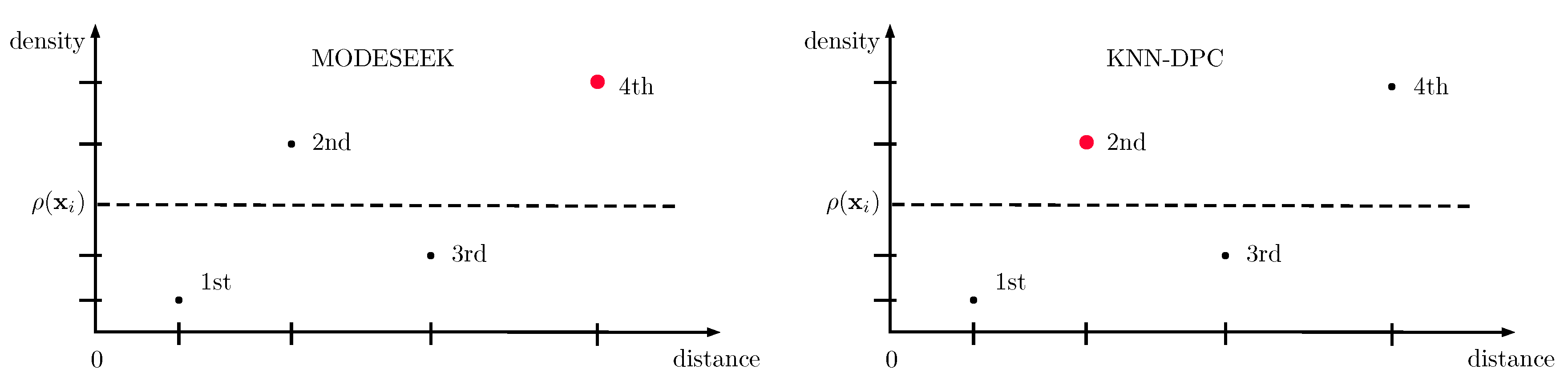
3.1. modeseek
3.2. Density Peaks Clustering—dpc
3.3. Graph Watershed Using Nearest Neighbors—gwenn
3.4. knnclust
3.5. Implementation Choices
4. Improvements of Two Nearest-Neighbor Density-Based Clustering Algorithms
4.1. Improvement of knn-dpc: m-knn-dpc
| Algorithm 1m-knn-dpc |
Input:
|
4.2. Improvement of knnclust-wm: m-knnclust-wm
| Algorithm 2m-knnclust-wm. |
Input:
|
4.3. Discussion
5. Improvements of Nearest-Neighbor Density-Based Methods for HSI Pixel Clustering
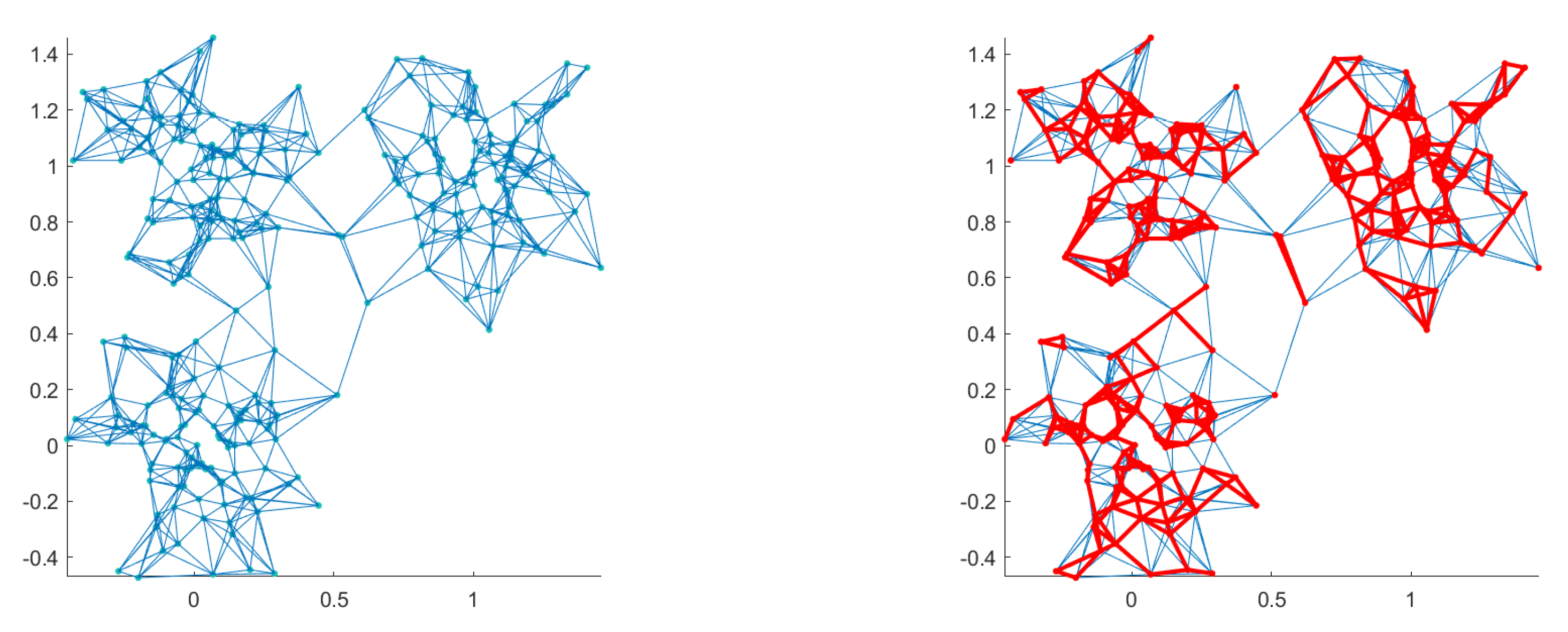
5.1. KNN Graph Regularization
5.2. Spatial Regularization
6. Clustering Experiments with the Baseline Methods on Synthetic Datasets
6.1. Cluster Validation Indices
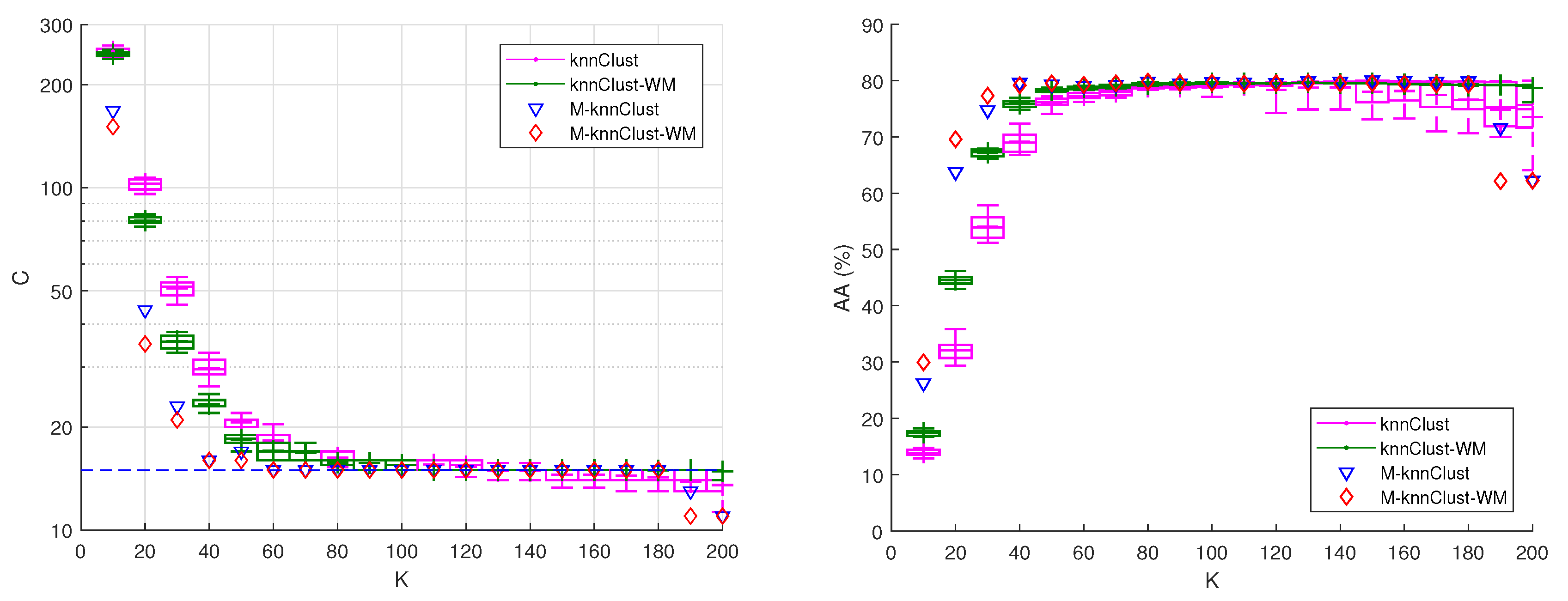
6.2. Comparison of knnclust Variants
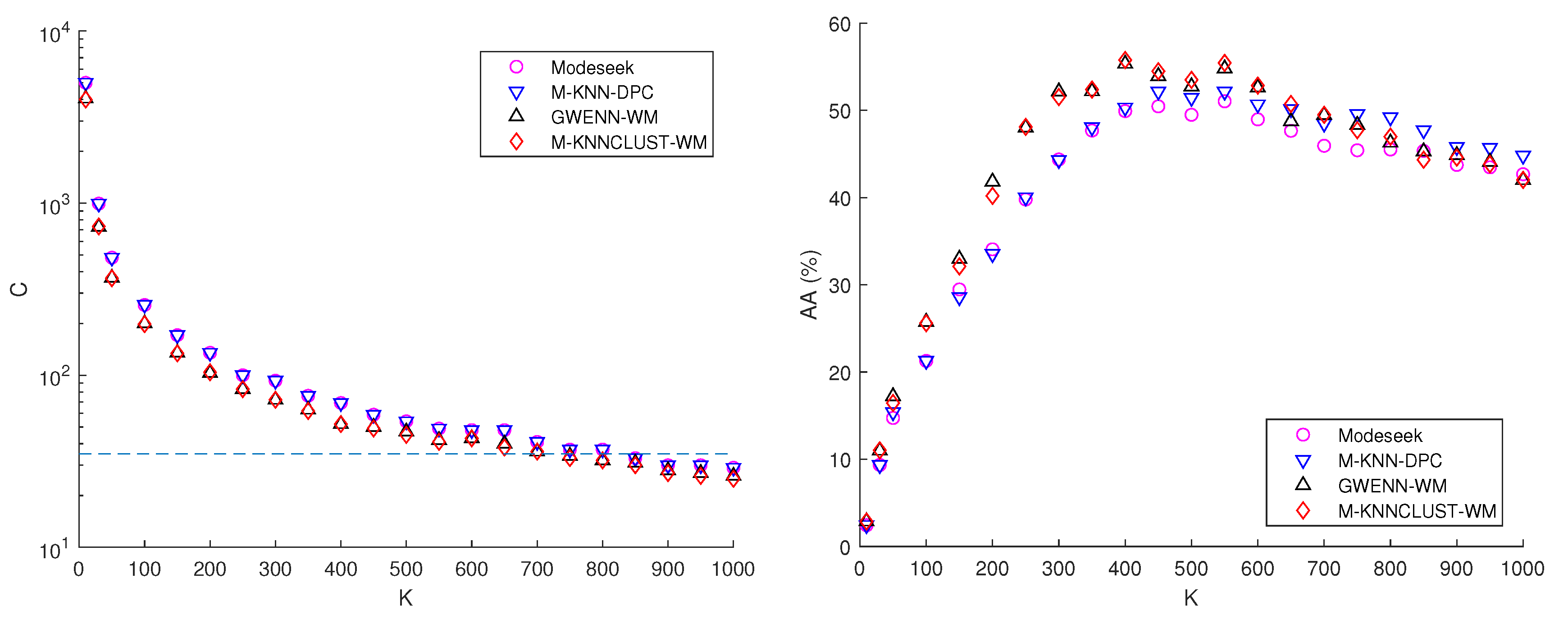
6.3. Comparison of the Baseline Clustering Methods
7. Application to Hyperspectral Remote Sensing Images
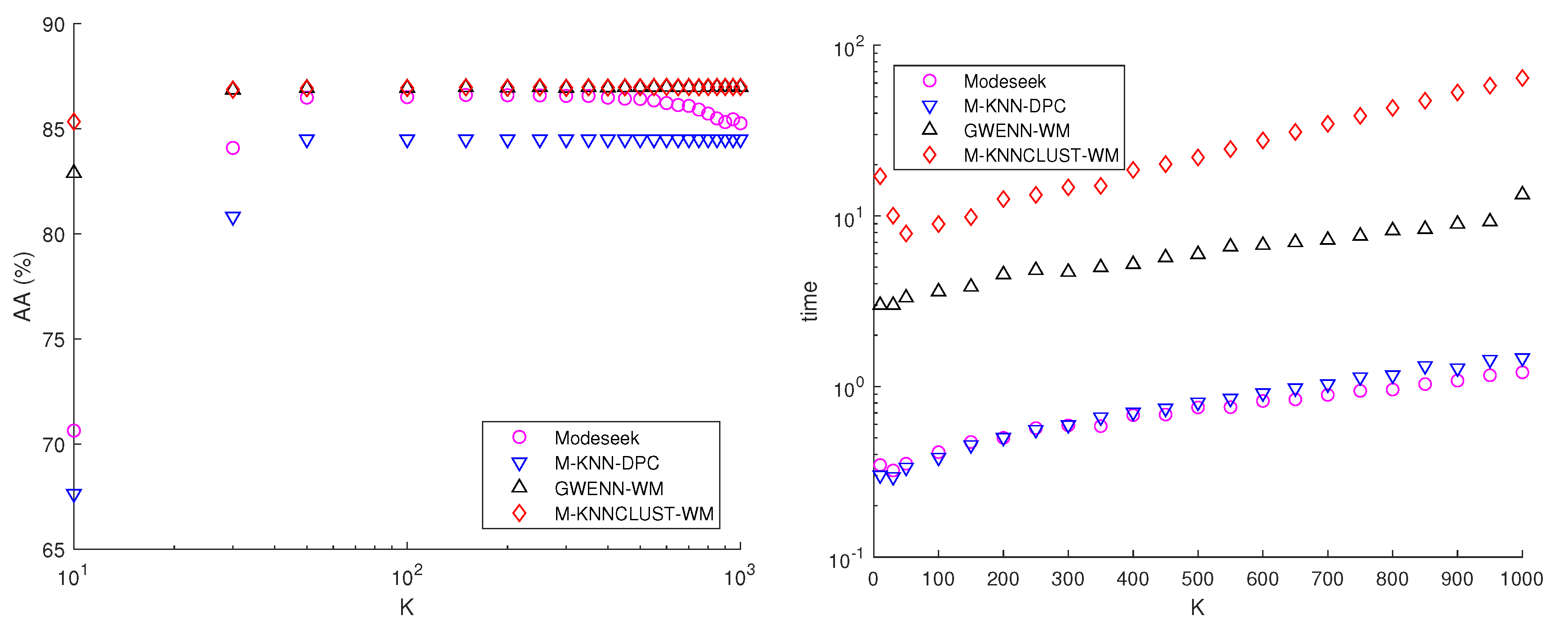
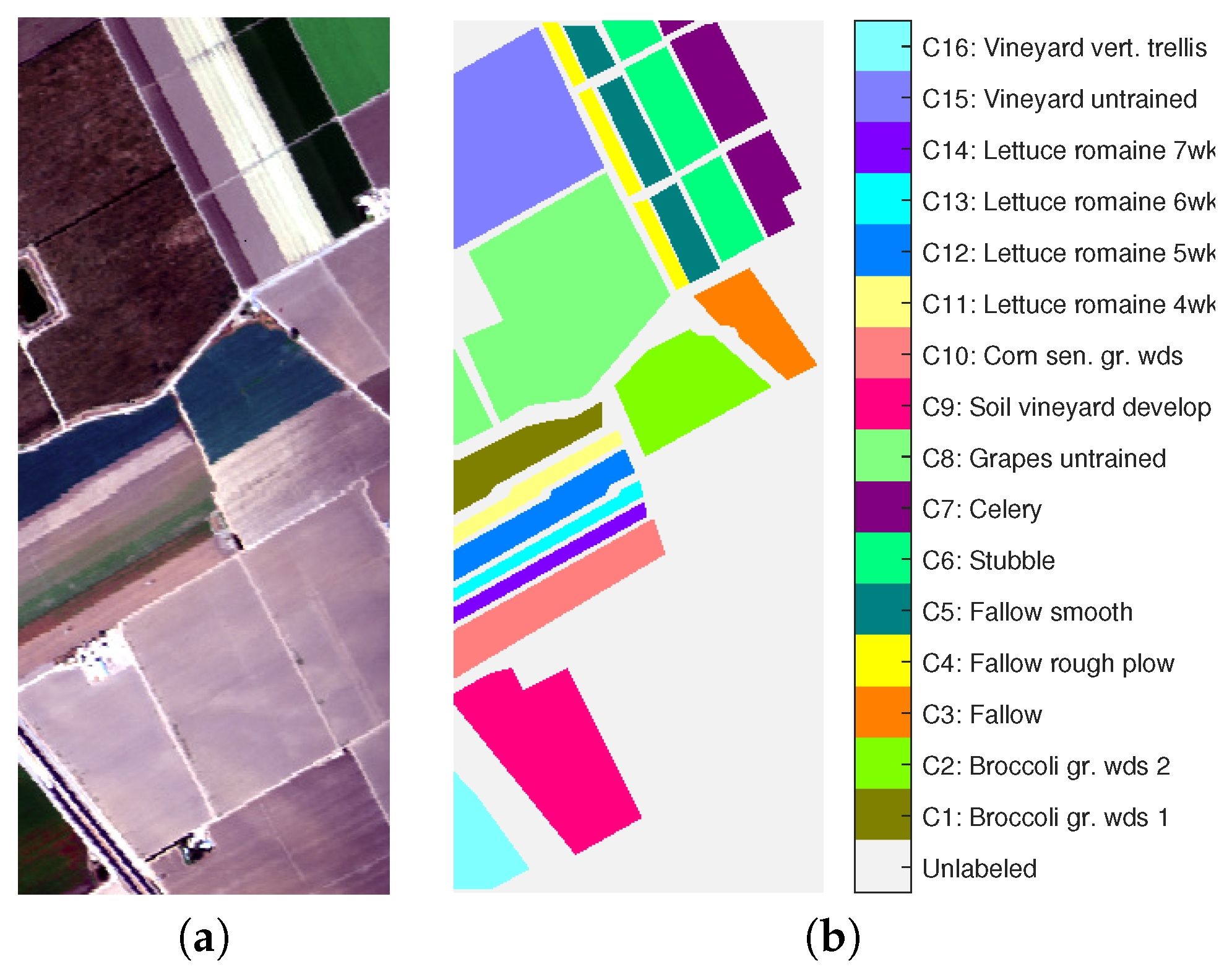
7.1. AVIRIS—Salinas Hyperspectral Image
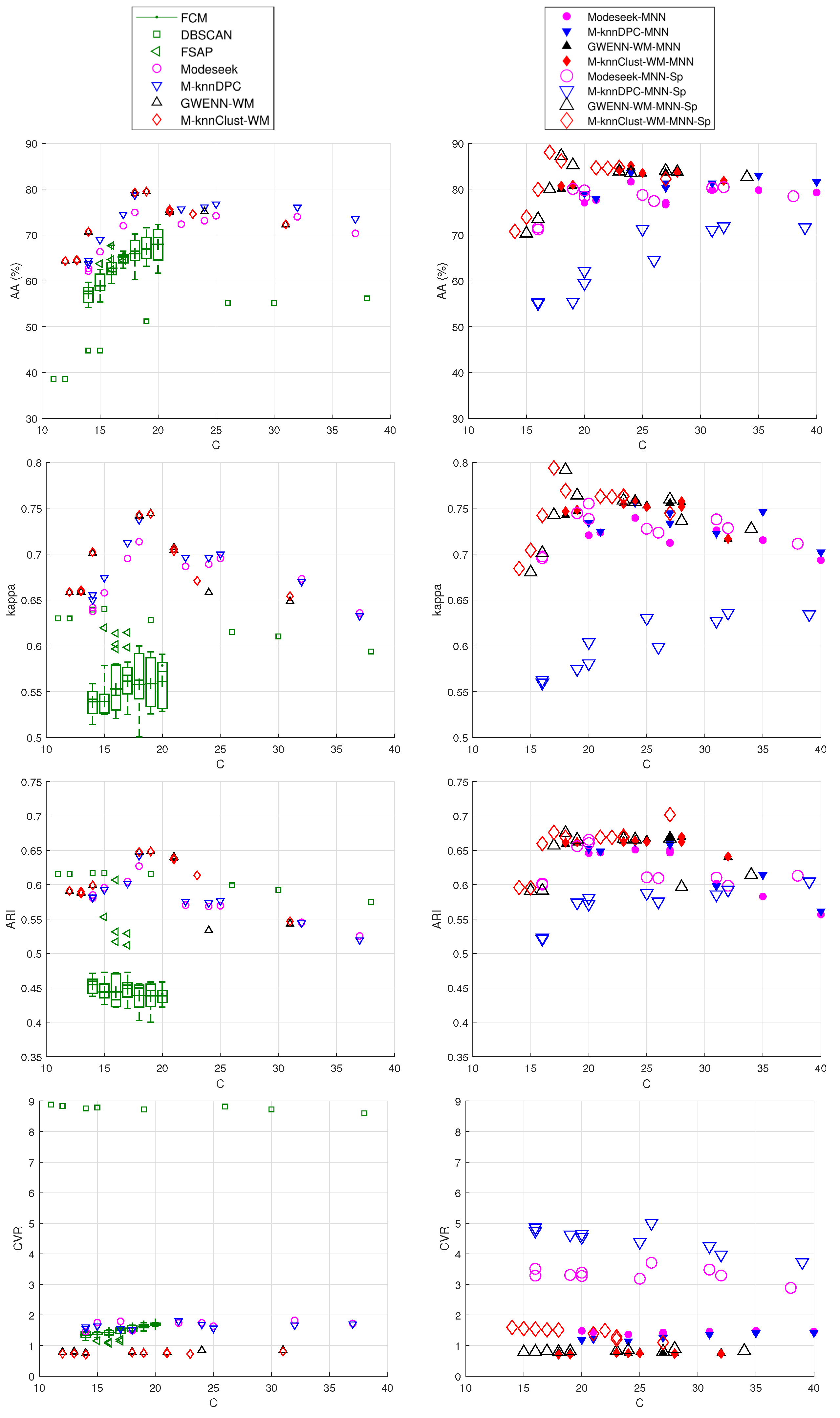
- On average, all the nearest-neighbor density-based methods perform better than dbscan, fsap and fcm whatever the number of clusters, found or imposed, and especially regarding the external performance indices. Exceptions to this observation are discussed below.
- Performing MNN graph regularization clearly helps with improving the results for all the methods, but at the cost of creating a higher number of clusters. These are consequences of graph pruning, which tends to create low-density clusters at the border of main clusters, while strengthening the density within the latter, thereby providing more robust results on average.
- The further inclusion of spatial context impacts differently the two subsets of density-based methods, i.e., on the one hand modeseek-mnn and m-knn-dpc-mnn, and on the other hand gwenn-wm-mnn and m-knnclust-mnn. On Figure 8, the performance indices are higher than those without spatial context for the latter subset, and they do not clearly improve or even degrade for the former subset, especially for m-knn-dpc-mnn-sp.
- The best overall kappa index () was obtained for m-knnclust-mnn-sp with , providing clusters, closely followed by for gwenn-wm-mnn-sp with same K, giving clusters. Notice that the running time between those two methods is very different, with 2338 s for m-knnclust-mnn-sp—mostly induced by the high number of labels at initialization—scaling down to 22 s for gwenn-wm-mnn-sp. These times exclude the computation of the original KNN graph, which is about 13 s for using MATLAB 2016 on a DELL 7810 Precision Tower (20-core, 32GB RAM), and the MNN procedure (78 s).
- The CVR internal clustering indices for the various methods are coherent with the results given by the external indices. It should be mentioned that CVR, contrarily to the other indices, is computed from the extensive cluster map and the original KNN graph, and therefore accounts for all the pixels, should they belong to the GT map or not. The best CVR values are obtained with the subset of gwenn-wm and m-knnclust methods and their MNN and spatial context variants. fcm provides higher CVR values, sometimes better than modeseek and m-knn-dpc.
- Among the state-of-the-art methods used for comparison, fsap provides the best results, at least visually, with a less noisy clustering map. However, there is some amount of confusion between the output clusters and the GT map which lowers the performance indices.
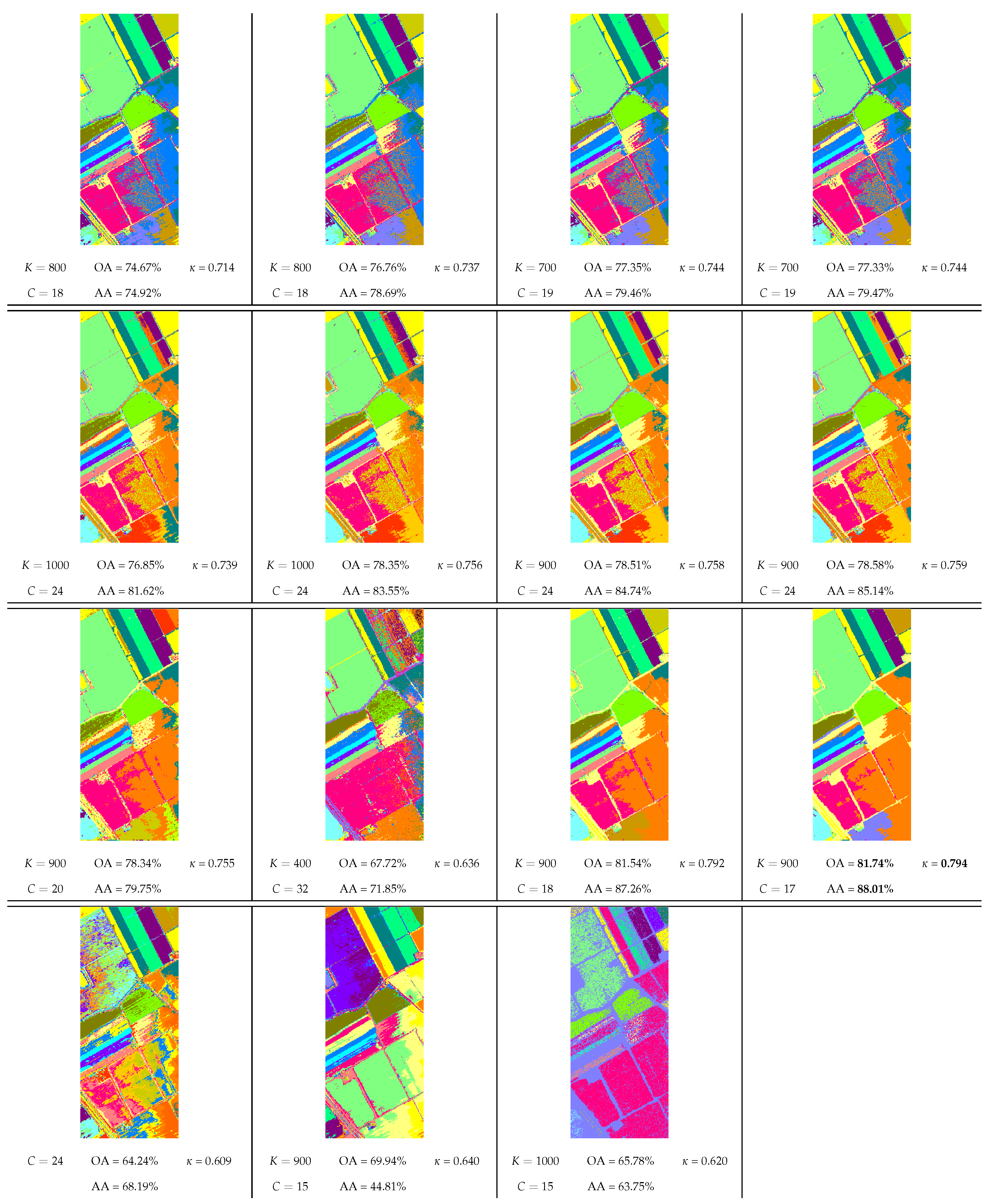
- The classes C8 (grapes untrained) and C15 (vineyard untrained) could not be retrieved by any method. Several published papers already show the difficulty to separate these two classes [65]. However, the class C7 (celery) was split into two clusters with visual coherence by all the density-based methods with MNN graph modification, which confirms the usefulness of this approach for detecting close clusters. This additional cluster disappears with the further use of spatial context which forces the links between those two clusters so as to merge in a single one.
- The only exception regarding the good performances of the proposed methods is with the spatial variant of m-knn-dpc-mnn, which we recall only differs from modeseek by the way the NN of higher local density is selected (see Figure 1). Actually, including spatial neighbors in addition to "spectral" neighbors in this method is likely to drive the NN selection to the spectrally closest spatial neighbors, thereby separating compact clusters into subclusters while achieving the same final number of clusters as modeseek, as said above, hence dramatically reducing the clustering performance. In comparison, the NN selection rule set up in modeseek is more robust to some extent.
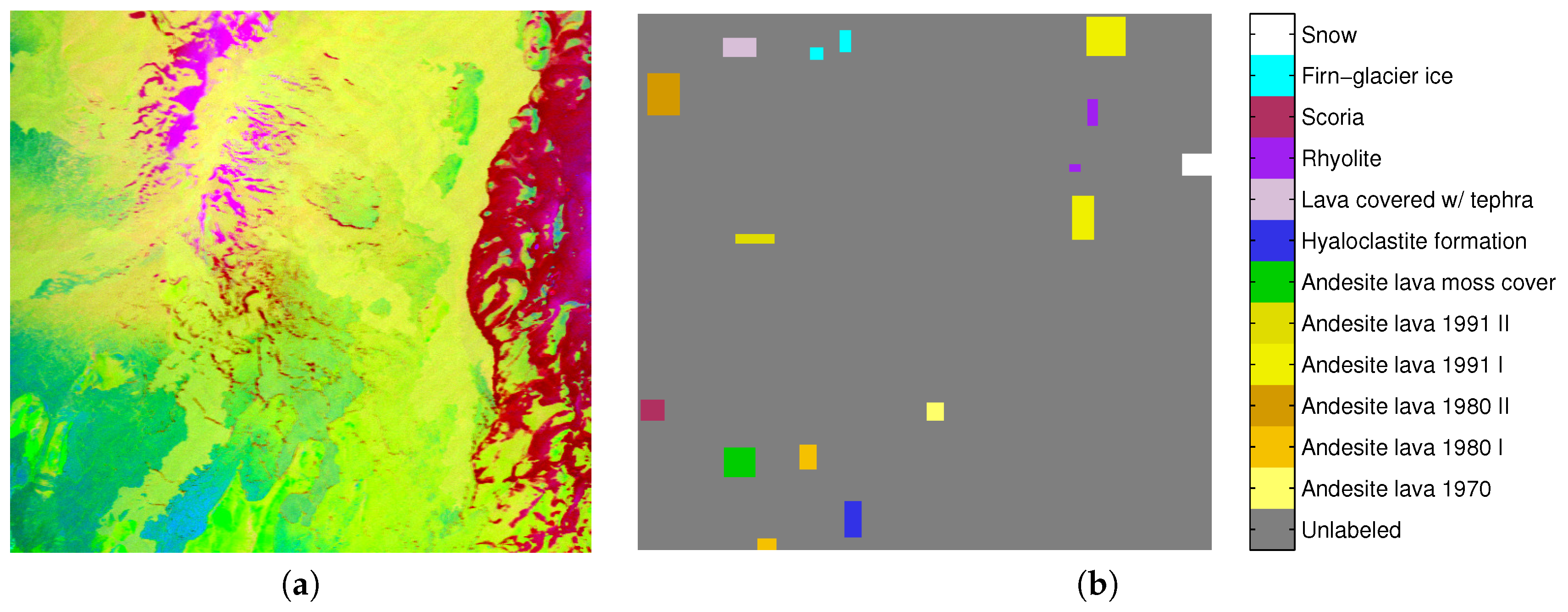
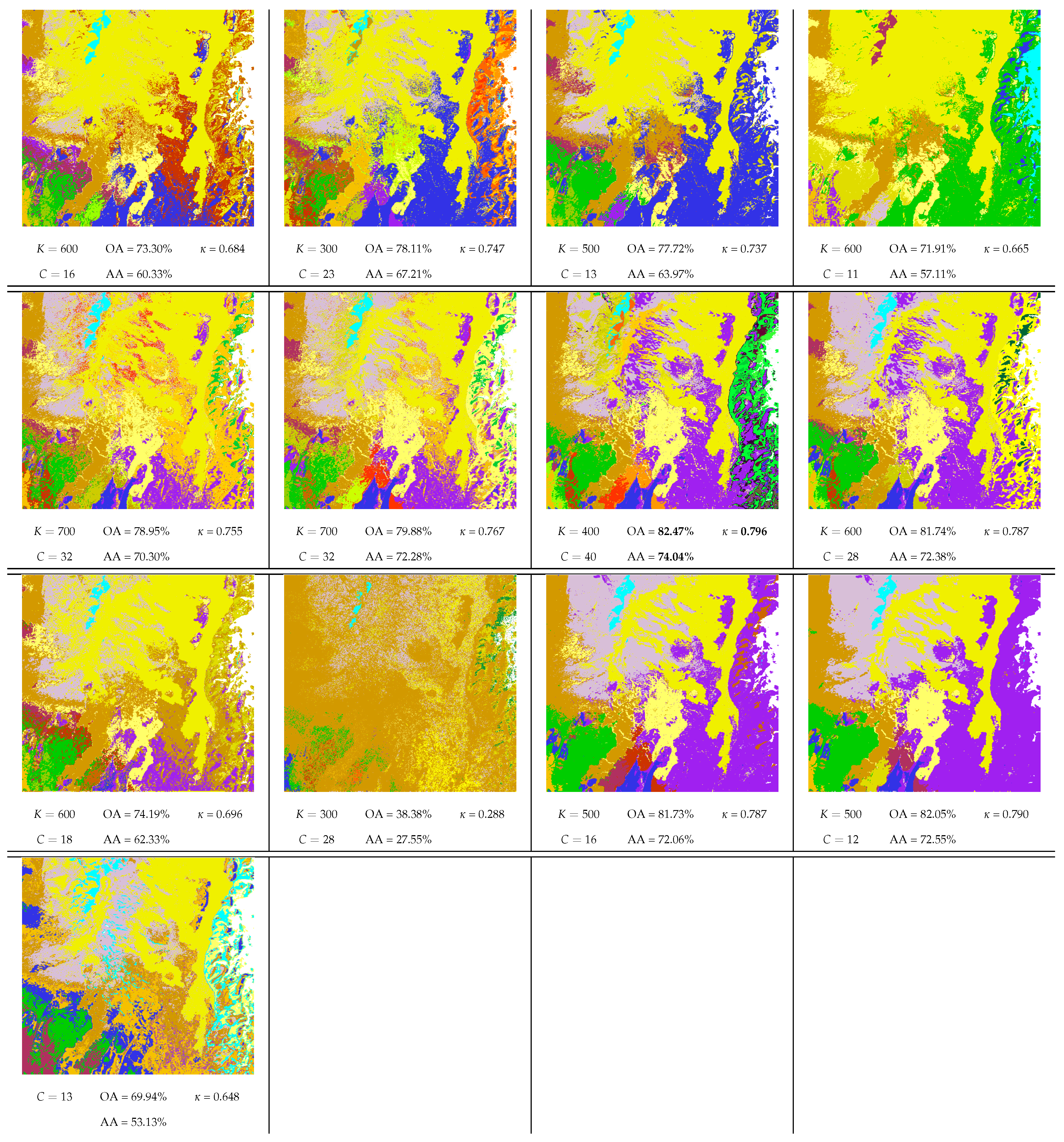
7.2. AVIRIS—Hekla Hyperspectral Image
7.3. Other Hyperspectral Images
- The DC Mall HSI (https://engineering.purdue.edu/~biehl/MultiSpec/hyperspectral.html) (1280 × 307 pixels, 191 spectral bands), acquired in 1995 over Washington, DC by the HYDICE instrument; the corresponding ground truth has 43368 pixels distributed in seven thematic classes [68];
- The Kennedy Space Center (KSC) HSI (https://www.ehu.es/ccwintco/index.php/Hyperspectral_Remote_Sensing_Scenes) (614 × 512 pixels, 176 spectral bands), acquired in 1996 over Titusville, Florida, by the AVIRIS sensor; the ground truth has 5211 pixels in 13 classes [53];
- The Massey University HSI (1579 × 1618 pixels, 339 spectral bands), acquired over Palmerston North, New Zealand, by the AisaFENIX sensor (Specim Ltd., Finland); the ground truth has 9564 pixels in 23 classes [53].
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A Survey of Clustering With Deep Learning: From the Perspective of Network Architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- MacQueen, J.B. Some Methods for Classification and Analysis of MultiVariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967; Volume 1, pp. 281–297. [Google Scholar]
- Bezdek, J. Pattern Recognition With Fuzzy Objective Function Algorithms; Plenum Press: New York, NY, USA, 1981. [Google Scholar]
- Ward, J. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Sneath, P.; Sokal, R. Numerical Taxonomy. The Principles and Practice of Numerical Classification; Freeman: London, UK, 1973. [Google Scholar]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD’96), Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Ankerst, M.; Breunig, M.M.; Kriegel, H.P.; Sander, J. OPTICS: Ordering Points To Identify the Clustering Structure. In Proceedings of the ACM SIGMOD International Conference on Management of Data (SIGMOD’99), Philadelphia, PA, USA, 31 May–3 June 1999; pp. 49–60. [Google Scholar]
- Fukunaga, K.; Hostetler, L.D. The estimation of the gradient of a density function, with applications in pattern recognition. IEEE Trans. Inf. Theory 1975, 21, 32–40. [Google Scholar] [CrossRef]
- Koontz, W.L.G.; Narendra, P.M.; Fukunaga, K. A Graph-Theoretic Approach to Nonparametric Cluster Analysis. IEEE Trans. Comput. 1976, 25, 936–944. [Google Scholar] [CrossRef]
- Duin, R.P.W.; Fred, A.L.N.; Loog, M.; Pekalska, E. Mode Seeking Clustering by KNN and Mean Shift Evaluated. In Proceedings of the SSPR/SPR, Hiroshima, Japan, 7–9 November 2012; Lecture Notes in Computer Science. Springer: Berlin/Heidelberg, Germany, 2012; Volume 7626, pp. 51–59. [Google Scholar]
- Rodriguez, A.; Laio, A. Clustering by fast search and find of density peaks. Science 2014, 344, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Series B. 1977, 39, 1–38. [Google Scholar]
- Celeux, G.; Govaert, G. A classification EM algorithm for clustering and two stochastic versions. Comput. Stat. Data Anal. 1992, 14, 315–332. [Google Scholar] [CrossRef]
- Rasmussen, C.E. The Infinite Gaussian Mixture Model. In Proceedings of the 12th International Conference on Neural Information Processing Systems (NIPS’99), 29 November–4 Decemeber 1999; Solla, S.A., Leen, T.K., Müller, K.R., Eds.; MIT Press: Cambridge, MA, USA, 2000; pp. 554–560. [Google Scholar]
- Shi, J.; Malik, J. Normalized Cuts and Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 888–905. [Google Scholar]
- Schölkopf, B.; Smola, A.; Müller, K.R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998, 10, 1299–1319. [Google Scholar] [CrossRef]
- Frey, B.J.; Dueck, D. Clustering by Passing Messages Between Data Points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef]
- Sugiyama, M.; Niu, G.; Yamada, M.; Kimura, M.; Hachiya, H. Information-Maximization Clustering Based on Squared-Loss Mutual Information. Neural Comput. 2014, 26, 84–131. [Google Scholar] [CrossRef] [PubMed]
- Hocking, T.; Vert, J.P.; Bach, F.R.; Joulin, A. Clusterpath: An Algorithm for Clustering using Convex Fusion Penalties. In Proceedings of the 28th International Conference on International Conference on Machine Learning (ICML’11), Bellevue, WA, USA, 28 June–2 July 2011; pp. 745–752. [Google Scholar]
- Elhamifar, E.; Vidal, R. Sparse Subspace Clustering: Algorithm, Theory, and Applications. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2765–2781. [Google Scholar] [CrossRef] [PubMed]
- Chazal, F.; Guibas, L.J.; Oudot, S.Y.; Skraba, P. Persistence-Based Clustering in Riemannian Manifolds. J. ACM 2013, 60, 38. [Google Scholar] [CrossRef]
- Masulli, F.; Rovetta, S. Clustering High-Dimensional Data. In Proceedings of the 1st International Workshop on Clustering High-Dimensional Data (Revised Selected Papers); Volume LNCS-7627, New York, NY, USA, 2015; Volume Springer, pp. 1–13. [Google Scholar]
- Xu, R.; Wunsch, D. Survey of clustering algorithms. IEEE Trans. Neural Netw. 2005, 16, 645–678. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.N.; Wehrens, R.; Buydens, L.M.C. KNN-kernel density-based clustering for high-dimensional multivariate data. Comput. Stat. Data Anal. 2006, 51, 513–525. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K. Unsupervised Nearest Neighbors Clustering with Application to Hyperspectral Images. IEEE J. Sel. Top. Signal Process. 2015, 9, 1105–1116. [Google Scholar] [CrossRef]
- Xie, J.; Gao, H.; Xie, W.; Liu, X.; Grant, P.W. Robust clustering by detecting density peaks and assigning points based on fuzzy weighted K-nearest neighbors. Inf. Sci. 2016, 354, 19–40. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, J.; Zhang, C.; Hua, X.S. Finding image exemplars using fast sparse affinity propagation. In ACM Multimedia; El-Saddik, A., Vuong, S., Griwodz, C., Bimbo, A.D., Candan, K.S., Jaimes, A., Eds.; ACM: New York, NY, USA, 2008; pp. 639–642. [Google Scholar]
- Hinneburg, A.; Aggarwal, C.C.; Keim, D.A. What Is the Nearest Neighbor in High Dimensional Spaces? In Proceedings of the 26th VLDB Conference, Cairo, Egypt, 10–14 September 2000; Abbadi, A.E., Brodie, M.L., Chakravarthy, S., Dayal, U., Kamel, N., Schlageter, G., Whang, K.Y., Eds.; Morgan Kaufmann: San Fransisco, CA, USA, 2000; pp. 506–515. [Google Scholar]
- Radovanovic, M.; Nanopoulos, A.; Ivanovic, M. Hubs in Space: Popular Nearest Neighbors in High-Dimensional Data. J. Mach. Learn. Res. 2010, 11, 2487–2531. [Google Scholar]
- Arya, S.; Mount, D.; Netanyahu, N.; Silverman, R.; Wu, A. An Optimal Algorithm for Approximate Nearest Neighbor Searching Fixed Dimensions. J. ACM 1998, 45, 891–923. [Google Scholar] [CrossRef]
- Fränti, P.; Virmajoki, O.; Hautamäki, V. Fast Agglomerative Clustering Using a k-Nearest Neighbor Graph. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 1875–1881. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K. A new k-nearest neighbor density-based clustering method and its application to hyperspectral images. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Beijing, China, 10–15 July 2016; pp. 6161–6164. [Google Scholar]
- Goodenough, D.G.; Chen, H.; Richardson, A.; Cloude, S.; Hong, W.; Li, Y. Mapping fire scars using Radarsat-2 polarimetric SAR data. Can. J. Remote Sens. 2011, 37, 500–509. [Google Scholar] [CrossRef]
- Du, M.; Ding, S.; Jia, H. Study on density peaks clustering based on k-nearest neighbors and principal component analysis. Knowl.-Based Syst. 2016, 99, 135–145. [Google Scholar] [CrossRef]
- Yaohui, L.; Ma, Z.; Fang, Y. Adaptive density peak clustering based on K-nearest neighbors with aggregating strategy. Knowl.-Based Syst. 2017, 133, 208–220. [Google Scholar] [CrossRef]
- Wang, G.; Song, Q. Automatic Clustering via Outward Statistical Testing on Density Metrics. IEEE Trans. Knowl. Data Eng. 2016, 28, 1971–1985. [Google Scholar] [CrossRef]
- Liu, R.; Wang, H.; Yu, X. Shared-nearest-neighbor-based clustering by fast search and find of density peaks. Inf. Sci. 2018, 450, 200–226. [Google Scholar] [CrossRef]
- Vincent, L.; Soille, P. Watersheds in Digital Spaces: An Efficient Algorithm Based on Immersion Simulations. IEEE Trans. PAMI 1991, 13, 583–598. [Google Scholar] [CrossRef]
- Desquesnes, X.; Elmoataz, A.; Lézoray, O. Eikonal Equation Adaptation on Weighted Graphs: Fast Geometric Diffusion Process for Local and Non-local Image and Data Processing. J. Math. Imaging Vis. 2013, 46, 238–257. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K. Nearest-neighbor density-based clustering methods for large hyperspectral images. In Proceedings of the SPIE Image and Signal Processing for Remote Sensing XXIII, Warsaw, Poland, 11–14 September 2017; Volume 10427. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K. Application of unsupervised nearest-neighbor density-based approaches to sequential dimensionality reduction and clustering of hyperspectral images. In Proceedings of the SPIE Image and Signal Processing for Remote Sensing XXIV, Berlin, Germany, 10–13 September 2018; Volume 10789. [Google Scholar]
- Aggarwal, C.C.; Hinneburg, A.; Keim, D.A. On the surprising behavior of distance metrics in high dimensional space. In Lecture Notes in Computer Science; Springer: Berlin, Germany, 2001; pp. 420–434. [Google Scholar]
- Mahalanobis, P.C. On the generalized distance in statistics. Proc. Natl. Inst. Sci. (Calcutta) 1936, 2, 49–55. [Google Scholar]
- Xu, X.; Ju, Y.; Liang, Y.; He, P. Manifold Density Peaks Clustering Algorithm. In Proceedings of the Third International Conference on Advanced Cloud and Big Data, Yangzhou, China, 30 October–1 November 2015; IEEE Computer Society: Piscataway, NJ, USA, 2015; pp. 311–318. [Google Scholar] [CrossRef]
- Du, M.; Ding, S.; Xu, X.; Xue, Y. Density peaks clustering using geodesic distances. Int. J. Mach. Learn. Cybern. 2018, 9, 1335–1349. [Google Scholar] [CrossRef]
- Schölkopf, B. The Kernel Trick for Distances. In Proceedings of the 13th International Conference on Neural Information Processing Systems (NIPS’00); Leen, T.K., Dietterich, T.G., Tresp, V., Eds.; MIT Press: Cambridge, MA, USA, 2000; pp. 283–289. [Google Scholar]
- Scott, D.W.; Sain, S.R. Multi-dimensional density estimation. In Data Mining and Data Visualization; Handbook of Statistics: Amsterdam, The Netherlands, 2005; Volume 24. [Google Scholar]
- Sieranoja, S.; Fränti, P. Fast and general density peaks clustering. Pattern Recognit. Lett. 2019, 128, 551–558. [Google Scholar] [CrossRef]
- Geng, Y.A.; Li, Q.; Zheng, R.; Zhuang, F.; He, R.; Xiong, N. RECOME: A new density-based clustering algorithm using relative KNN kernel density. Inf. Sci. 2018, 436–437, 13–30. [Google Scholar] [CrossRef]
- Le Moan, S.; Cariou, C. Parameter-Free Density Estimation for Hyperspectral Image Clustering. In Proceedings of the International Conference on Image and Vision Computing New Zealand (IVCNZ), Auckland, New Zealand, 19–21 November 2018. [Google Scholar]
- Liang, Z.; Chen, P. Delta-density based clustering with a divide-and-conquer strategy: 3DC clustering. Pattern Recognit. Lett. 2016, 73, 52–59. [Google Scholar] [CrossRef]
- Stevens, J.R.; Resmini, R.G.; Messinger, D.W. Spectral-Density-Based Graph Construction Techniques for Hyperspectral Image Analysis. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5966–5983. [Google Scholar] [CrossRef]
- Le Moan, S.; Cariou, C. Minimax Bridgeness-Based Clustering for Hyperspectral Data. Remote Sens. 2020, 12, 1162. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K.; Le Moan, S. Improved Nearest Neighbor Density-Based Clustering Techniques with Application to Hyperspectral Images. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 4127–4131. [Google Scholar]
- Zou, X.; Zhu, Q. Adaptive Neighborhood Graph for LTSA Learning Algorithm without Free-Parameter. Int. J. Comput. Appl. 2011, 19, 28–33. [Google Scholar]
- Kuhn, H. The Hungarian method for the assignment problem. Nav. Res. Logist. Q. 1955, 2, 83–97. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J. Spectral-spatial classification of hyperspectral imagery based on partitional clustering techniques. IEEE Trans. Geosci. Remote Sens. 2009, 47, 2973–2987. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Manning, C.D.; Raghavan, P.; Schütze, H. An Introduction to Information Retrieval; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Fränti, P.; Rezaei, M.; Zhao, Q. Centroid index: Cluster level similarity measure. Pattern Recognit. 2014, 47, 3034–3045. [Google Scholar] [CrossRef]
- Ver Steeg, G.; Galstyan, A.; Sha, F.; DeDeo, S. Demystifying Information-Theoretic Clustering. In Proceedings of the 31st International Conference on Machine Learning, Beijing, China, 22–24 June 2014. [Google Scholar]
- Fränti, P.; Sieranoja, S. How much can k-means be improved by using better initialization and repeats? Pattern Recognit. 2019, 93, 95–112. [Google Scholar] [CrossRef]
- Gualtieri, J.A.; Chettri, S.R.; Cromp, R.F.; Johnson, L.F. Support Vector Machine Classifiers as Applied to AVIRIS Data. In Proceedings of the Summaries of the Eighth JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 9–11 February 1999; JPL-NASA: Pasadena, CA, USA, 1999; pp. 1–10. [Google Scholar]
- Huang, S.; Zhang, H.; Du, Q.; Pizurica, A. Sketch-Based Subspace Clustering of Hyperspectral Images. Remote Sens. 2020, 12, 775. [Google Scholar] [CrossRef]
- Waske, B.; Benediktsson, J.A.; Árnason, K.; Sveinsson, J.R. Mapping of hyperspectral AVIRIS data using machine-learning algorithms. Can. J. Remote Sens. 2009, 35, S106–S116. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Graig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Landgrebe, D. Hyperspectral image data analysis. IEEE Signal Process. Mag. 2002, 19, 17–28. [Google Scholar] [CrossRef]


| Density Estimate | References | |
|---|---|---|
| Non-parametric model | [32,48] | |
| [41] | ||
| [34] | ||
| Parametric model | [49] | |
| [50] |
| modeseek [10] | m-knn-dpc | m-knnclust-wm | gwenn-wm | |
|---|---|---|---|---|
| Iterative | yes | yes | yes | no |
| Speed | high | high | low | average |
| Initial labeling | none | none | one label per sample | none |
| Local labeling | label of NN with | label of closest NN with | weighted mode of | weighted mode of |
| decision rule | highest density | higher density | NNs’ labels | NNs’ labels |
| Stopping rule | upon convergence | upon convergence | upon convergence | not applicable |
| Optimal K, #Clusters and Criteria → | K | C | AA | OA | Purity | NMI | CVR | ||
|---|---|---|---|---|---|---|---|---|---|
| DC Mall : N = 43,368, , = 7 | |||||||||
| modeseek | 2200 | 5 | 61.72 | 82.22 | 0.777 | 0.874 | 0.721 | 1.431 | |
| Constant | m-knn-dpc | 2200 | 5 | 63.57 | 84.43 | 0.805 | 0.898 | 0.753 | 0.874 |
| K | gwenn-wm | 3000 | 5 | 62.53 | 82.97 | 0.787 | 0.883 | 0.743 | 0.716 |
| m-knnclust-wm | 3000 | 5 | 62.96 | 83.63 | 0.795 | 0.890 | 0.745 | 0.706 | |
| MNN | modeseek-mnn | 3000 | 6 | 73.07 | 84.45 | 0.809 | 0.873 | 0.756 | 0.654 |
| m-knn-dpc-mnn | 3000 | 6 | 72.86 | 84.35 | 0.808 | 0.872 | 0.756 | 0.657 | |
| gwenn-wm-mnn | 3000 | 6 | 73.05 | 84.81 | 0.813 | 0.877 | 0.762 | 0.521 | |
| m-knnclust-wm-mnn | 3000 | 6 | 73.04 | 84.82 | 0.813 | 0.877 | 0.763 | 0.521 | |
| fcm | - | 6 | 71.06 | 80.01 | 0.754 | 0.827 | 0.717 | 1.179 | |
| dbscan | 320 | 5 | 37.86 | 76.63 | 0.608 | 0.778 | 0.591 | 2.116 | |
| KSC : N = 5211, , = 13 | |||||||||
| modeseek | 120 | 9 | 42.44 | 52.58 | 0.478 | 0.685 | 0.592 | 1.807 | |
| Constant | m-knn-dpc | 120 | 9 | 45.18 | 57.15 | 0.526 | 0.730 | 0.641 | 1.373 |
| K | gwenn-wm | 110 | 9 | 44.15 | 56.75 | 0.521 | 0.730 | 0.633 | 0.731 |
| m-knnclust-wm | 110 | 9 | 44.26 | 56.88 | 0.523 | 0.730 | 0.634 | 0.713 | |
| MNN | modeseek-mnn | 150 | 10 | 43.92 | 57.59 | 0.531 | 0.718 | 0.628 | 2.794 |
| m-knn-dpc-mnn | 150 | 10 | 46.54 | 60.45 | 0.563 | 0.751 | 0.672 | 2.241 | |
| gwenn-wm-mnn | 140 | 9 | 43.86 | 57.34 | 0.526 | 0.752 | 0.646 | 1.617 | |
| m-knnclust-wm-mnn | 130 | 9 | 44.04 | 57.49 | 0.528 | 0.752 | 0.647 | 1.337 | |
| fcm | - | 11 | 40.60 | 52.29 | 0.471 | 0.604 | 0.560 | 5.383 | |
| dbscan | 70 | 9 | 47.68 | 68.23 | 0.530 | 0.692 | 0.795 | 104.0 | |
| Massey University : N = 9564, , = 23 | |||||||||
| modeseek | 130 | 14 | 38.33 | 51.49 | 0.460 | 0.725 | 0.625 | 2.860 | |
| Constant | m-knn-dpc | 130 | 14 | 39.19 | 53.32 | 0.479 | 0.746 | 0.672 | 2.060 |
| K | gwenn-wm | 110 | 16 | 41.09 | 52.97 | 0.480 | 0.702 | 0.664 | 0.990 |
| m-knnclust-wm | 110 | 16 | 41.08 | 52.89 | 0.480 | 0.703 | 0.668 | 0.949 | |
| MNN | modeseek-mnn | 130 | 22 | 45.29 | 55.73 | 0.507 | 0.715 | 0.669 | 2.380 |
| m-knn-dpc-mnn | 130 | 22 | 50.72 | 60.78 | 0.564 | 0.747 | 0.729 | 1.639 | |
| gwenn-wm-mnn | 110 | 27 | 55.39 | 59.06 | 0.552 | 0.684 | 0.738 | 0.961 | |
| m-knnclust-wm-mnn | 110 | 28 | 55.27 | 57.69 | 0.544 | 0.621 | 0.727 | 1.073 | |
| fcm | - | 21 | 39.41 | 40.73 | 0.371 | 0.471 | 0.603 | 8.859 | |
| dbscan | 450 | 4 | 6.28 | 75.07 | 0.589 | 0.751 | 0.753 | 87.11 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cariou, C.; Le Moan, S.; Chehdi, K. Improving K-Nearest Neighbor Approaches for Density-Based Pixel Clustering in Hyperspectral Remote Sensing Images. Remote Sens. 2020, 12, 3745. https://doi.org/10.3390/rs12223745
Cariou C, Le Moan S, Chehdi K. Improving K-Nearest Neighbor Approaches for Density-Based Pixel Clustering in Hyperspectral Remote Sensing Images. Remote Sensing. 2020; 12(22):3745. https://doi.org/10.3390/rs12223745
Chicago/Turabian StyleCariou, Claude, Steven Le Moan, and Kacem Chehdi. 2020. "Improving K-Nearest Neighbor Approaches for Density-Based Pixel Clustering in Hyperspectral Remote Sensing Images" Remote Sensing 12, no. 22: 3745. https://doi.org/10.3390/rs12223745
APA StyleCariou, C., Le Moan, S., & Chehdi, K. (2020). Improving K-Nearest Neighbor Approaches for Density-Based Pixel Clustering in Hyperspectral Remote Sensing Images. Remote Sensing, 12(22), 3745. https://doi.org/10.3390/rs12223745








