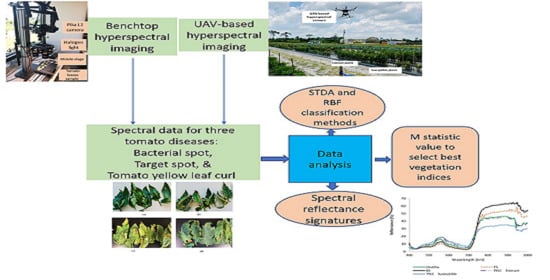
Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Tomato Yellow Leaf Curl (TYLC) Sample Collection
2.2. Target Spot and BS Sample Collection
2.2.1. Inoculation Methods
Tomato Yellow Leaf Curl Disease
Target Spot
Bacterial Spot
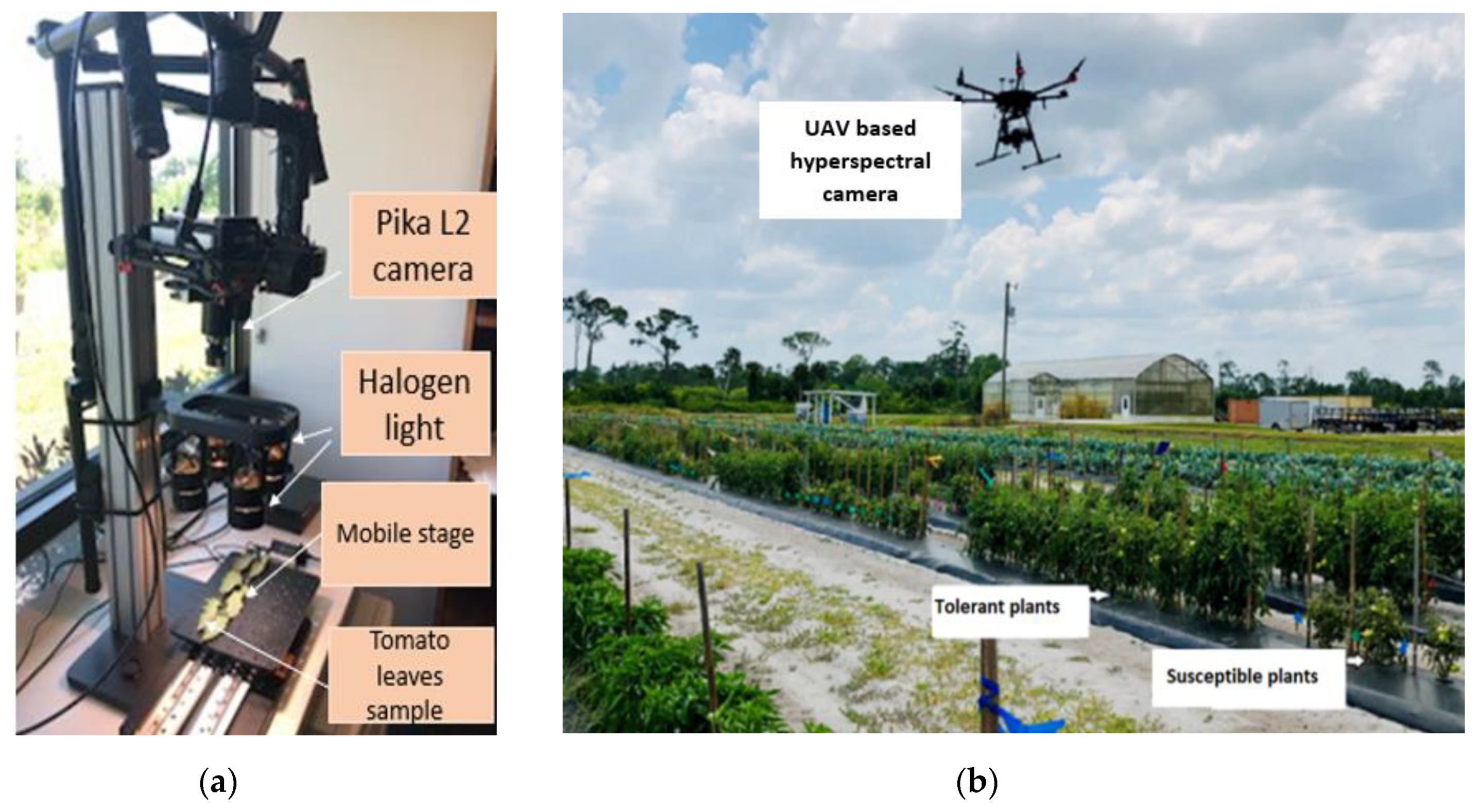
2.3. Laboratory Data Collection
2.4. UAV-Based Data Collection
2.5. Classification Methods
2.6. Vegetation Indices
Data Analysis for Selecting VIs
3. Results and Discussion
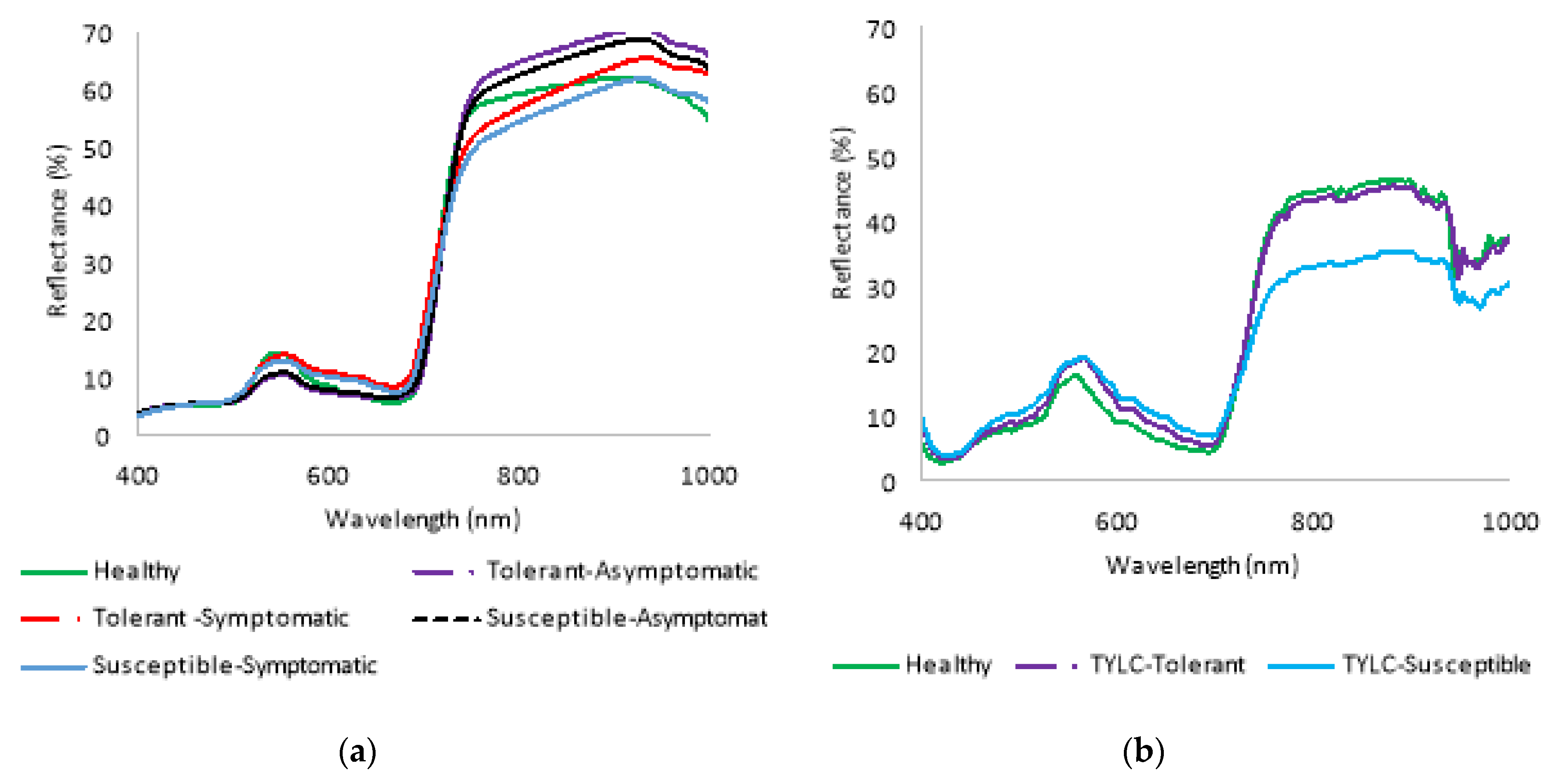
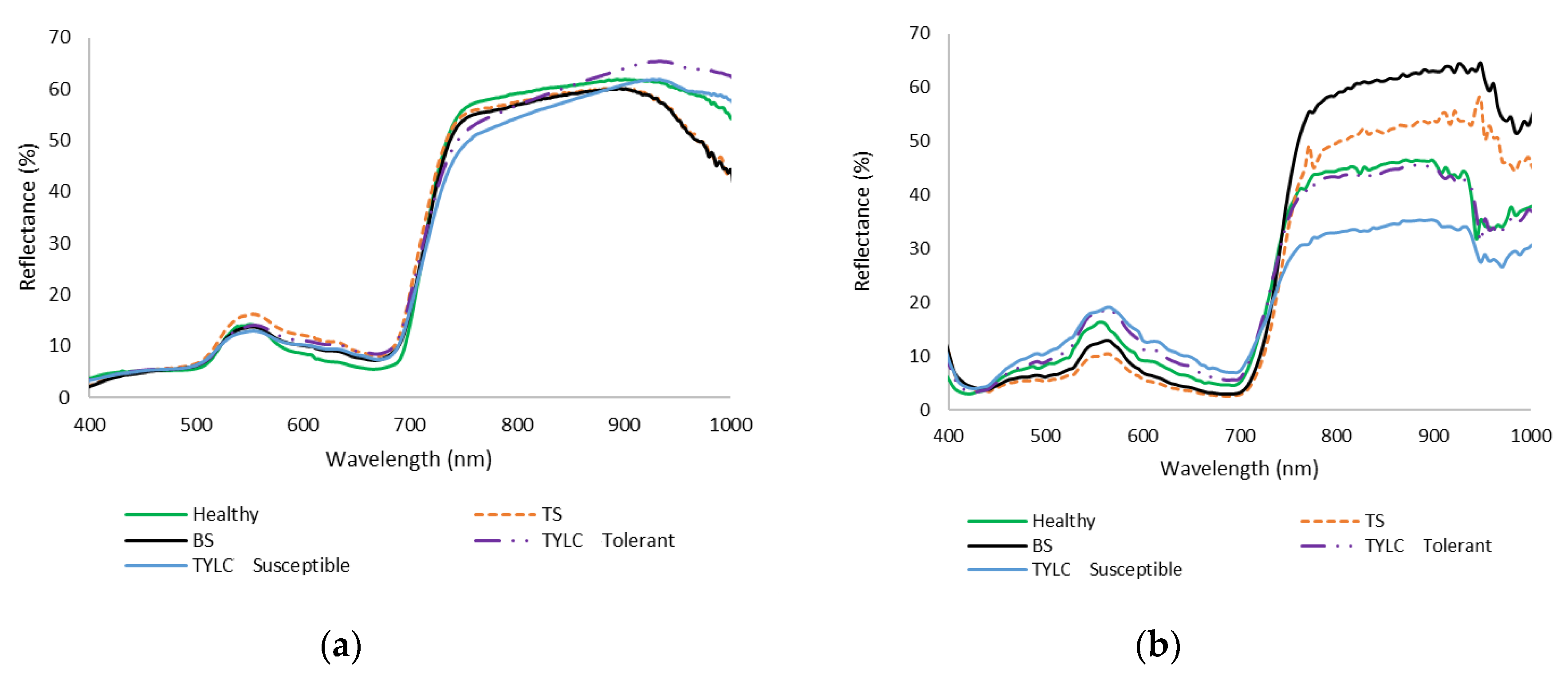
3.1. Spectral Reflectance Analysis
3.1.1. Spectral Reflectance of TYLC, BS, and TS Diseases: Laboratory-Based Analysis
3.1.2. Spectral Reflectance of TYLC, BS, and TS Diseases: Field (UAV)-Based Analysis
3.2. Classification Results
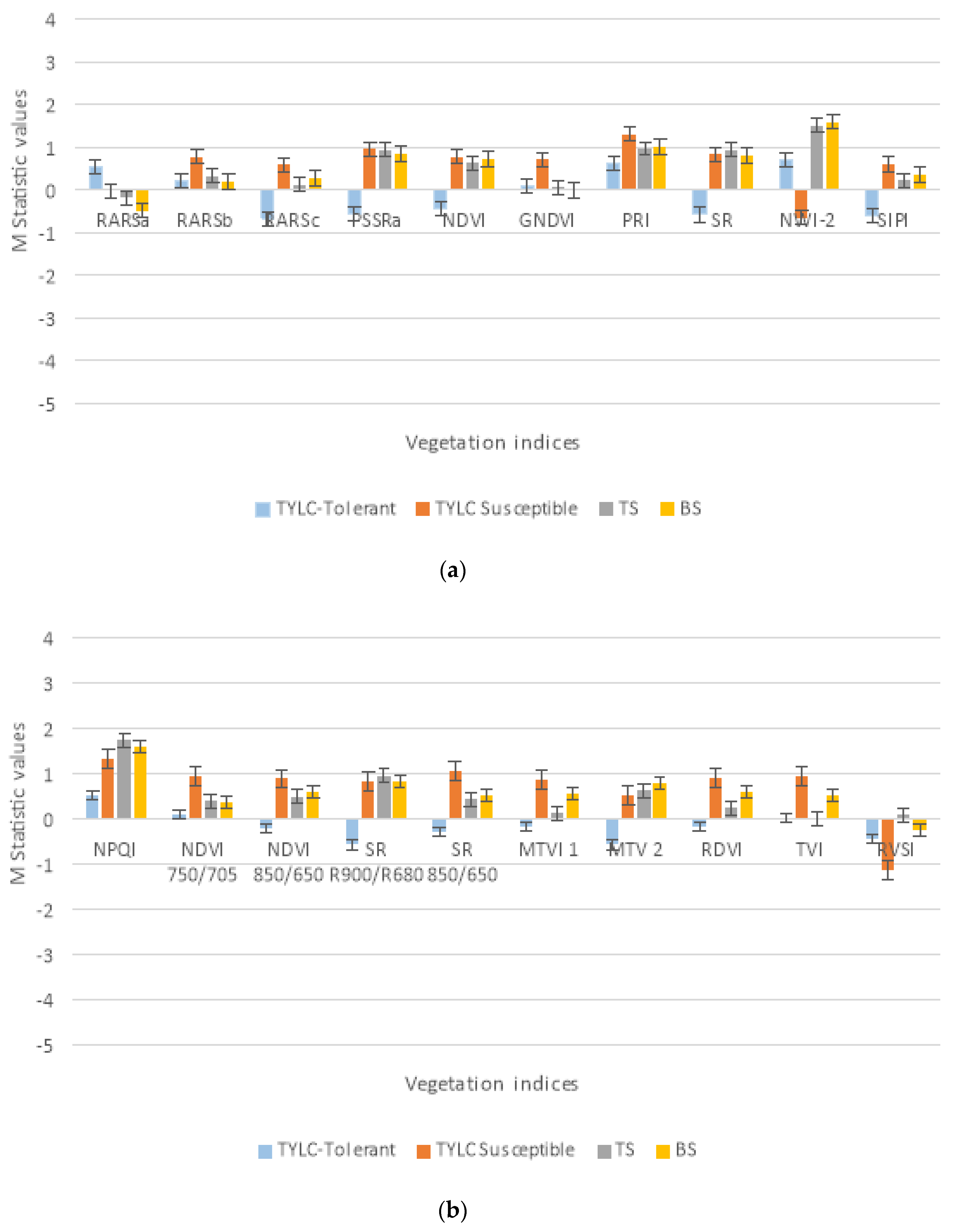
3.3. Significant VIs for Disease Detection: Laboratory-Based Analysis
3.4. Significant VIs for Disease Detection: Field (UAV)-Based Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cruz, A.; Ampatzidis, Y.; Pierro, R.; Materazzi, A.; Panattoni, A.; De Bellis, L.; Luvisi, A. Detection of grapevine yellows symptoms in Vitis vinifera L. with artificial intelligence. Comput. Electron. Agric. 2019, 157, 63–76. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; De Bellis, L.; Luvisi, A. iPathology: Robotic Applications and Management of Plants and Plant Diseases. Sustainability 2017, 9, 1010. [Google Scholar] [CrossRef]
- Luvisi, A.; Ampatzidis, Y.G.; De Bellis, L. Plant Pathology and Information Technology: Opportunity for Management of Disease Outbreak and Applications in Regulation Frameworks. Sustainability 2016, 8, 831. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Kakarla, S.C.; Roberts, P. Detection of target spot and bacterial spot disease in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2019, 21, 955–978. [Google Scholar] [CrossRef]
- Partel, V.; Nunes, L.; Stansley, P.; Ampatzidis, Y. Automated Vision-based System for Monitoring Asian Citrus Psyllid in Orchards utilizing Artificial Intelligence. Comput. Electron. Agric. 2019, 162, 328–336. [Google Scholar] [CrossRef]
- Hariharan, J.; Fuller, J.; Ampatzidis, Y.; Abdulridha, J.; Lerwill, A. Finite difference analysis and bivariate correlation of hyperspectral data for detecting laurel wilt disease and nutritional deficiency in avocado. Remote Sens. 2019, 11, 1748. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection—A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Whetton, R.L.; Hassall, K.L.; Waine, T.W.; Mouazen, A.M. Hyperspectral measurements of yellow rust and fusarium head blight in cereal crops: Part 1: Laboratory study. Biosyst. Eng. 2018, 166, 101–115. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ehsani, R.; Abd-Elrahma, A.; Ampatzidis, Y. A remote sensing technique for detecting laurel wilt disease in avocado in presence of other biotic and abiotic stresses. Comput. Electron. Agric. 2019, 156, 549–557. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V. UAV-Based high throughput phenotyping in citrus utilizing multispectral imaging and artificial intelligence. Remote Sens. 2019, 11, 410. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V.; Meyering, B.; Albrecht, U. Citrus rootstock evaluation utilizing UAV-based remote sensing and artificial intelligence. Comput. Electron. Agric. 2019, 164, 164. [Google Scholar] [CrossRef]
- Albetis, J.; Jacquin, A.; Goulard, M.; Poilve, H.; Rousseau, J.; Clenet, H.; Dedieu, G.; Duthoit, S. On the Potentiality of UAV Multispectral Imagery to Detect Flavescence doree and Grapevine Trunk Diseases. Remote Sens. 2019, 11, 23. [Google Scholar] [CrossRef]
- Abdulridha, J.; Battuman, O.; Ampatzidis, Y. UAV-Based Remote Sensing Technique to Detect Citrus Canker Disease Utilizing Hyperspectral Imaging and Machine Learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.X.; Dong, Y.Y.; Shi, Y.; Huang, W.J.; Han, L.H.; Gonzalez-Moreno, P.; Ma, H.Q.; Ye, H.C.; Sobeih, T. A Deep Learning-Based Approach for Automated Yellow Rust Disease Detection from High-Resolution Hyperspectral UAV Images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef]
- Gates, D.M. Citation classic-spectral properties of plants. Curr. Contents-Agric. Biol. Environ. Sci. 1980, 48, 10. [Google Scholar]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Vahtmae, E.; Kotta, J.; Orav-Kotta, H.; Kotta, I.; Parnoja, M.; Kutser, T. Predicting macroalgal pigments (chlorophyll a, chlorophyll b, chlorophyll a plus b, carotenoids) in various environmental conditions using high-resolution hyperspectral spectroradiometers. Int. J. Remote Sens. 2018, 39, 5716–5738. [Google Scholar] [CrossRef]
- Sonobe, R.; Sano, T.; Horie, H. Using spectral reflectance to estimate leaf chlorophyll content of tea with shading treatments. Biosyst. Eng. 2018, 175, 168–182. [Google Scholar] [CrossRef]
- Rustioni, L.; Grossi, D.; Brancadoro, L.; Failla, O. Iron, magnesium, nitrogen and potassium deficiency symptom discrimination by reflectance spectroscopy in grapevine leaves. Sci. Hortic. 2018, 241, 152–159. [Google Scholar] [CrossRef]
- Li, W.G.; Sun, Z.Q.; Lu, S.; Omasa, K. Estimation of the leaf chlorophyll content using multiangular spectral reflectance factor. Plant Cell Environ. 2019, 42, 3152–3165. [Google Scholar] [CrossRef]
- Yang, P.Q.; van der Tol, C.; Verhoef, W.; Damm, A.; Schickling, A.; Kraska, T.; Muller, O.; Rascher, U. Using reflectance to explain vegetation biochemical and structural effects on sun-induced chlorophyll fluorescence. Remote Sens. Environ. 2019, 231, 110996. [Google Scholar] [CrossRef]
- Costa, L.; Nunes, L.; Ampatzidis, Y. A new visible band index (vNDVI) for estimating NDVI values on RGB images utilizing genetic algorithms. Comput. Electron. Agric. 2020, 172, 105334. [Google Scholar] [CrossRef]
- Chen, T.T.; Zhang, J.L.; Chen, Y.; Wan, S.B.; Zhang, L. Detection of peanut leaf spots disease using canopy hyperspectral reflectance. Comput. Electron. Agric. 2019, 156, 677–683. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ehsani, R.; de Castro, A. Detection and differentiation between laurel wilt disease, Phytopthora disease and salinity damage using a hyperspectral sensing technique. Agriculture 2016, 6, 56. [Google Scholar] [CrossRef]
- Shuaibu, M.; Lee, W.S.; Schueller, J.; Gader, P.; Hong, Y.K.; Kim, S. Unsupervised hyperspectral band selection for apple Marssonina blotch detection. Comput. Electron. Agric. 2018, 148, 45–53. [Google Scholar] [CrossRef]
- Lu, J.Z.; Ehsani, R.; Shi, Y.Y.; de Castro, A.I.; Wang, S. Detection of multi-tomato leaf diseases (late blight, target and bacterial spots) in different stages by using a spectral-based sensor. Sci. Rep. 2018, 8, 2739. [Google Scholar] [CrossRef]
- Thompson, B. Stepwise regression and stepwise discriminant-analysis need not apply here—A guidelines editorial. Educ. Psychol. Meas. 1995, 55, 525–534. [Google Scholar] [CrossRef]
- Huberty, C.J. Issues in the use and interpretation of discriminant-analysis. Psychol. Bull. 1984, 95, 156–171. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Kim, M.S.; McMurtrey, J.E. Ration analysis of reflectance spectra (RARS)-An algorithm for the remote estimation concentration of chlorophyll-a, chlorophyll-b, and carotenoid soybean leaves. Remote Sens. Environ. 1992, 39, 239–247. [Google Scholar] [CrossRef]
- Blackburn, G.A. Spectral indices for estimating photosynthetic pigment concentrations: A test using senescent tree leaves. Int. J. Remote Sens. 1998, 19, 657–675. [Google Scholar] [CrossRef]
- Raun, W.R.; Solie, J.B.; Johnson, G.V.; Stone, M.L.; Lukina, E.V.; Thomason, W.E.; Schepers, J.S. In-season prediction of potential grain yield in winter wheat using canopy reflectance. Agron. J. 2001, 93, 131–138. [Google Scholar] [CrossRef]
- Peñuela, J.; Baret, F.; Filella, I. Semiempirical indexes to assess carotenoids chlorophyll-a ratio from leaf spectral reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Barnes, J.D.; Balaguer, L.; Manrique, E.; Elvira, S.; Davison, A.W. A Reappraisal of the Use of Dmso for the Extraction and Determination of Chlorophylls-A and Chlorophylls-B in Lichens and Higher-Plants. Environ. Exp. Bot. 1992, 32, 85–100. [Google Scholar] [CrossRef]
- Merton, R. Monitoring Community Hysteresis Using Spectral Shift Analysis and the Red-Edge Vegetation Stress Index. In Proceedings of the Seventh Annual JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 12–16 January 1998. [Google Scholar]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating Par Absorbed by Vegetation from Bidirectional Reflectance Measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Babar, M.A.; Reynolds, M.P.; Van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Sci. 2006, 46, 1046–1057. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Signature analysis of leaf reflectance spectra: Algorithm development for remote sensing of chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Gamon, J.A.; Penuelas, J.; Field, C.B. A Narrow-Waveband Spectral Index that Tracks Diurnal Changes in Photosynthetic Efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Remer, L.A. Detection of Forests Using Mid-IR Reflectance—An Application for Aerosol Studies. IEEE Trans. Geosci. Remote Sens. 1994, 32, 672–683. [Google Scholar] [CrossRef]
- Rascher, U.; Damm, A.; van der Linden, S.; Okujeni, A.; Pieruschka, R.; Schickling, A.; Gerhards, R.; Menz, G.; Sikora, R.A. Sensing of photosynthetic activity of crop. In Precision Crop Protection—The Challenge and Use of Heterogeneity; Springer: Dordrecht, The Netherlands, 2010; pp. 87–100. [Google Scholar]
- Penuelas, J.; Filella, I. Visible and near-infrared reflectance techniques for diagnosing plant physiological status. Trends Plant Sci. 1998, 3, 151–156. [Google Scholar] [CrossRef]
- Daub, M.E.; Ehrenshaft, M. The photoactivated Cercospora toxin cercosporin: Contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol. 2000, 38, 461–490. [Google Scholar] [CrossRef] [PubMed]
- Bruning, B.; Liu, H.J.; Brien, C.; Berger, B.; Lewis, M.; Garnett, T. The Development of Hyperspectral Distribution Maps to Predict the Content and Distribution of Nitrogen and Water in Wheat (Triticum aestivum). Front. Plant Sci. 2019, 10, 1380. [Google Scholar] [CrossRef]
- Kovar, M.; Brestic, M.; Sytar, O.; Barek, V.; Hauptvogel, P.; Zivcak, M. Evaluation of Hyperspectral Reflectance Parameters to Assess the Leaf Water Content in Soybean. Water 2019, 11, 443. [Google Scholar] [CrossRef]
- Pagano, M.; Baldacci, L.; Ottomaniello, A.; de Dato, G.; Chianucci, F.; Masini, L.; Carelli, G.; Toncelli, A.; Storchi, P.; Tredicucci, A.; et al. THz Water Transmittance and Leaf Surface Area: An Effective Nondestructive Method for Determining Leaf Water Content. Sensors 2019, 19, 4838. [Google Scholar] [CrossRef]
- Barhom, H.; Machnev, A.A.; Noskov, R.E.; Goncharenko, A.; Gurvitz, E.A.; Timin, A.S.; Shkoldin, V.A.; Koniakhin, S.V.; Koval, O.Y.; Zyuzin, M.V.; et al. Biological Kerker Effect Boosts Light Collection Efficiency in Plants. Nano Lett. 2019, 19, 7062–7071. [Google Scholar] [CrossRef]
- Heim, R.H.J.; Wright, I.J.; Chang, H.C.; Carnegie, A.J.; Pegg, G.S.; Lancaster, E.K.; Falster, D.S.; Oldeland, J. Detecting myrtle rust (Austropuccinia psidii) on lemon myrtle trees using spectral signatures and machine learning. Plant Pathol. 2018, 67, 1114–1121. [Google Scholar] [CrossRef]
- Romer, C.; Burling, K.; Hunsche, M.; Rumpf, T.; Noga, G.; Plumer, L. Robust fitting of fluorescence spectra for pre-symptomatic wheat leaf rust detection with Support Vector Machines. Comput. Electron. Agric. 2011, 79, 180–188. [Google Scholar] [CrossRef]
- Moshou, D.; Bravo, C.; Wahlen, S.; West, J.; McCartney, A.; Baerdemaeker, J.D.; Ramon, H.; de Baerdemaeker, J. Simultaneous identification of plant stresses and diseases in arable crops based on a proximal sensing system and Self-Organising Neural Networks. In Proceedings of the the 4th European Conference on Precision Agriculture, Berlin, Germany, 15–19 June 2003; pp. 425–431. [Google Scholar]
- West, J.S.; Bravo, C.; Oberti, R.; Lemaire, D.; Moshou, D.; McCartney, H.A. The potential of optical canopy measurement for targeted control of field crop diseases. Annu. Rev. Phytopathol. 2003, 41, 593–614. [Google Scholar] [CrossRef]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Abdulridha, J.; Ampatzidis, Y.; Ehsani, R.; de Castro, A.I. Evaluating the performance of spectral features and multivariate analysis tools to detect laurel wilt disease and nutritional deficiency in avocado. Comput. Electron. Agric. 2018, 155, 203–211. [Google Scholar] [CrossRef]


| Vegetation Indices | Equations | References |
|---|---|---|
| Ratio Analysis of reflectance Spectral Chlorophyll-a (RARSa) | [29] | |
| Ratio Analysis of reflectance Spectral Chlorophyll b (RARSb) | [29] | |
| Ratio analysis of reflectance spectra (RARSc) | [29] | |
| Pigment specific simple ratio (PSSRa) | [30] | |
| Normalized difference vegetation index 780 (NDVI 780) | [31] | |
| Structure Insensitive Pigment Index (SIPI) | [32] | |
| Normalized phaeophytinization index (NPQI) | [33] | |
| Red-Edge Vegetation Stress Index 1 (RVS1) | [34] | |
| Triangle Vegetation Index (TVI) | [35] | |
| Renormalized Difference Vegetation Index (RDVI) | [36] | |
| Normalized difference vegetation index 850 (NDVI850) | [31] | |
| Simple Ratio Index (SR 761) | [37] | |
| Simple Ratio Index (SR 850) | This study | |
| Simple Ratio Index (SR 900) | This study | |
| Water Stress and Canopy Temperature (NWI 2) | [38] | |
| Green NDVI (GNDVI) | [39] | |
| Photochemical Reflectance Index (PRI) | [40] | |
| Modified Chlorophyll Absorption in Reflectance Index (mCARI 1) | [41] | |
| Modified Triangular Vegetation Index1 (MTVI 1) | [41] | |
| Modified Triangular Vegetation Index2 (MTVI 2) | [41] |
| Parameter | STDA | RBF (%) | |||
|---|---|---|---|---|---|
| Overall Percent (%) | Cross Validation (%) | Wilks Lambda | Chi-Square | ||
| Laboratory based | |||||
| H vs. TYLC Tolerant-Asy | 100 | 100 | 0.014 | 388.0 | 89 |
| H vs. TYLC Tolerant-Sym | 100 | 100 | 0.028 | 374.1 | 100 |
| H vs. TYLC Susceptible-Asy | 100 | 100 | 0.023 | 320.2 | 83 |
| H vs. TYLC Susceptible -Sym | 100 | 100 | 0.044 | 321.4 | 100 |
| Tolerant-Asy vs. Susceptible-Asy | 100 | 100 | 0.086 | 91.8 | 100 |
| Tolerant-Sym vs. Susceptible-Sym | 100 | 100 | 0.096 | 111.1 | 66.7 |
| H vs. TS-Asy | 95 | 95 | 0.045 | 523.6 | 82 |
| H vs. TS-Sym | 95 | 95 | 0.005 | 422.4 | 90 |
| H vs. BS-Asy | 94 | 94 | 0.005 | 924.1 | 95 |
| H vs. BS-Sym | 95 | 94 | 0.005 | 523.6 | 89 |
| TS-Asy vs. BS-Asy | 88 | 87 | 0.306 | 145.0 | 83 |
| Ts-Sym vs. BS-Sym | 82 | 82 | 0.456 | 120.3 | 46 |
| Field (UAV) based | |||||
| H vs. TYLC Tolerant | 100 | 100 | 0.006 | 539.9 | 100 |
| H vs. TYLC Susceptible | 100 | 100 | 0.018 | 378.1 | 97 |
| TYLC Tolerant vs. Susceptible | 100 | 100 | 0.104 | 146.2 | 76 |
| H vs. TS | 98 | 96 | 0.026 | 597.8 | 98 |
| H vs. BS | 96 | 96 | 0.013 | 541.5 | 93 |
| Ts vs. BS | 82 | 80 | 0.457 | 141.9 | 64 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdulridha, J.; Ampatzidis, Y.; Qureshi, J.; Roberts, P. Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning. Remote Sens. 2020, 12, 2732. https://doi.org/10.3390/rs12172732
Abdulridha J, Ampatzidis Y, Qureshi J, Roberts P. Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning. Remote Sensing. 2020; 12(17):2732. https://doi.org/10.3390/rs12172732
Chicago/Turabian StyleAbdulridha, Jaafar, Yiannis Ampatzidis, Jawwad Qureshi, and Pamela Roberts. 2020. "Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning" Remote Sensing 12, no. 17: 2732. https://doi.org/10.3390/rs12172732
APA StyleAbdulridha, J., Ampatzidis, Y., Qureshi, J., & Roberts, P. (2020). Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning. Remote Sensing, 12(17), 2732. https://doi.org/10.3390/rs12172732