Application of Deep Learning Architectures for Accurate Detection of Olive Tree Flowering Phenophase
Abstract
1. Introduction
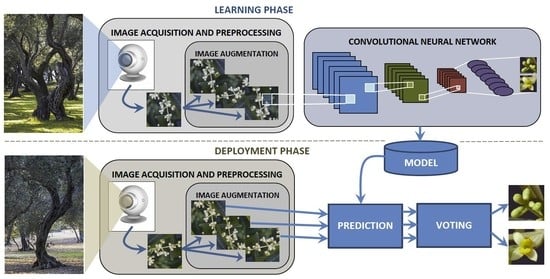
2. Materials and Methods
2.1. Dataset
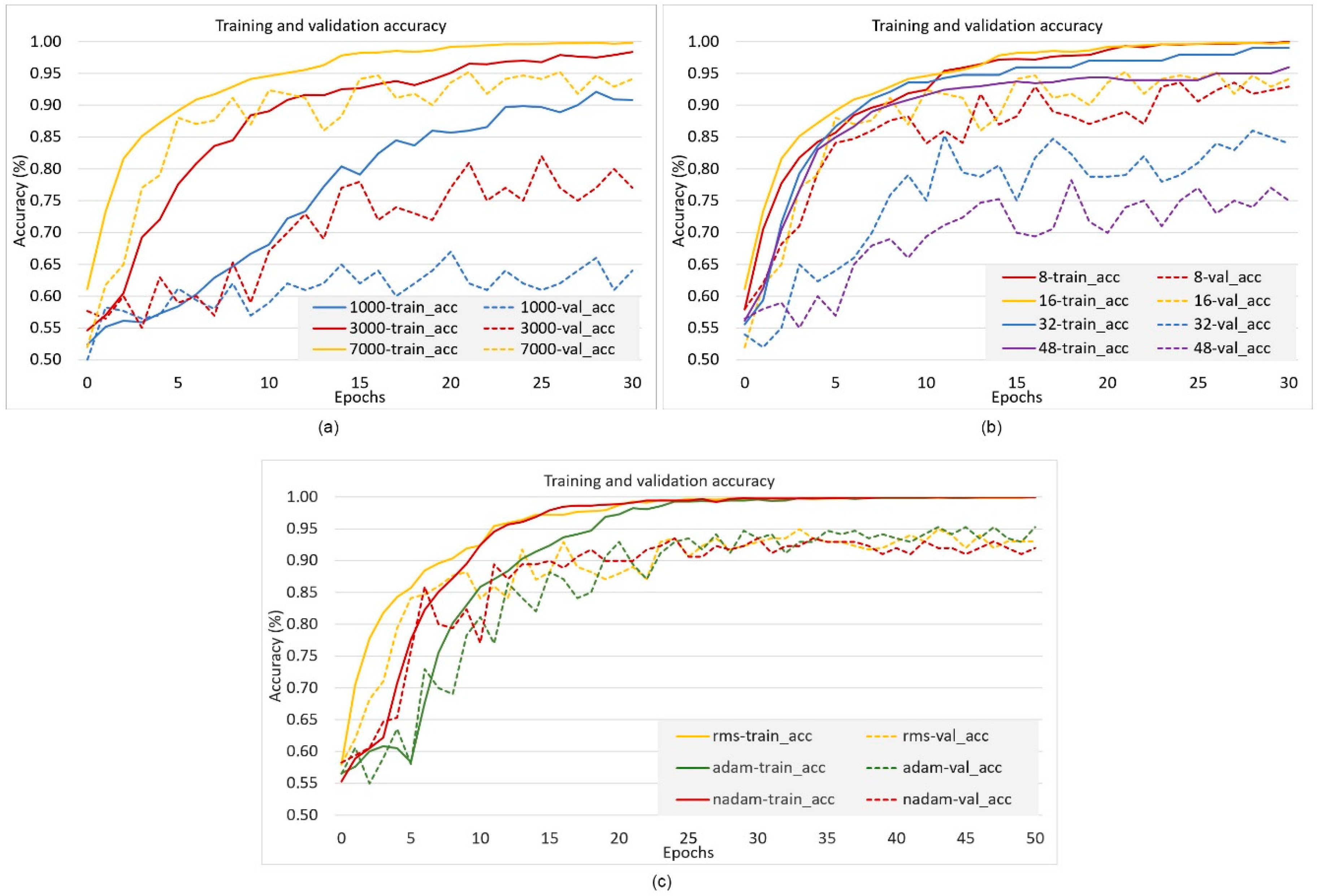
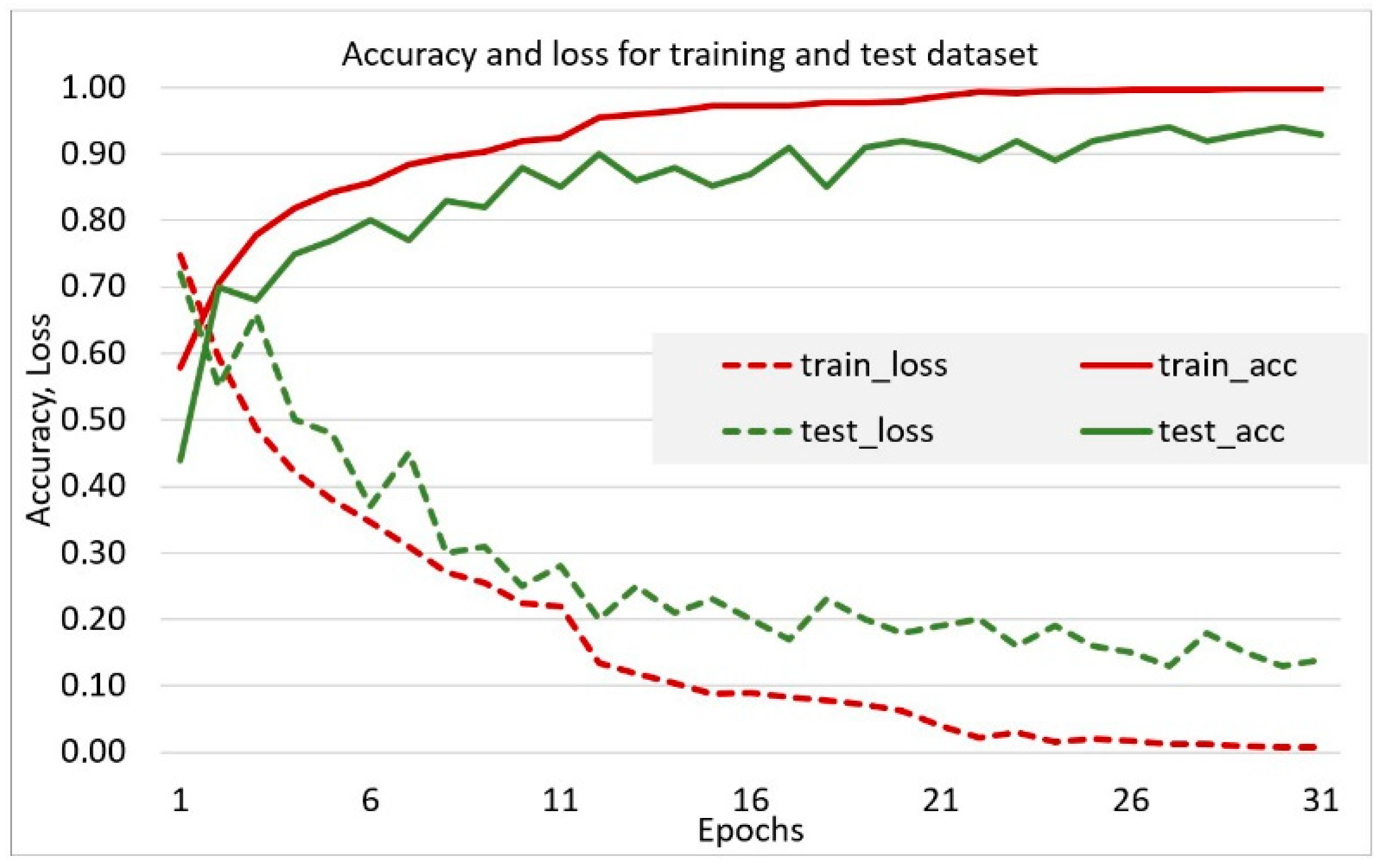
2.2. Experimental Study
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moriondo, M.; Bindi, M. Impact of climate change on the phenology of typical Mediterranean crops. Ital. J. Agrometeorol. 2007, 3, 5–12. [Google Scholar]
- Gutierrez, A.P.; Ponti, L.; Cossu, Q.A. Effects of climate warming on olive and olive fly (Bactrocera oleae (Gmelin)) in California and Italy. Clim. Chang. 2009, 95, 195–217. [Google Scholar] [CrossRef]
- Tang, S.; Tang, G.; Cheke, R.A. Optimum timing for integrated pest management: Modelling rates of pesticide application and natural enemy releases. J. Theor. Biol. 2010, 264, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Chuine, I.; Cour, P.; Rousseau, D.D. Selecting models to predict the timing of flowering of temperate trees: Implications for tree phenology modelling. Plant Cell Environ. 1999, 22, 1–13. [Google Scholar] [CrossRef]
- De Melo-Abreu, J.P.; Barranco, D.; Cordeiro, A.M.; Tous, J.; Rogado, B.M.; Villalobos, F.J. Modelling olive flowering date using chilling for dormancy release and thermal time. Agric. For. Meteorol. 2004, 125, 117–127. [Google Scholar] [CrossRef]
- Osborne, C.P.; Chuine, I.; Viner, D.; Woodward, F.I. Olive phenology as a sensitive indicator of future climatic warming in the Mediterranean. Plant Cell Environ. 2000, 23, 701–710. [Google Scholar] [CrossRef]
- Aguilera, F.; Fornaciari, M.; Ruiz-Valenzuela, L.; Galán, C.; Msallem, M.; Dhiab, A.B.; Díaz-de La Guardia, C.; del Mar Trigo, M.; Bonofiglio, T.; Orlandi, F. Phenological models to predict the main flowering phases of olive (Olea europaea L.) along a latitudinal and longitudinal gradient across the Mediterranean region. Int. J. Biometeorol. 2015, 59, 629–641. [Google Scholar] [CrossRef]
- Orlandi, F.; Vazquez, L.M.; Ruga, L.; Bonofiglio, T.; Fornaciari, M.; Garcia-Mozo, H.; Domínguez, E.; Romano, B.; Galan, C. Bioclimatic requirements for olive flowering in two Mediterranean regions located at the same latitude (Andalucia, Spain, and Sicily, Italy). Ann. Agric. Environ. Med. 2005, 12, 47. [Google Scholar]
- Garcia-Mozo, H.; Orlandi, F.; Galan, C.; Fornaciari, M.; Romano, B.; Ruiz, L.; de la Guardia, C.D.; Trigo, M.M.; Chuine, I. Olive flowering phenology variation between different cultivars in Spain and Italy: Modeling analysis. Theor. Appl. Climatol. 2009, 95, 385. [Google Scholar] [CrossRef]
- Oteros, J.; García-Mozo, H.; Vázquez, L.; Mestre, A.; Domínguez-Vilches, E.; Galán, C. Modelling olive phenological response to weather and topography. Agric. Ecosyst. Environ. 2013, 179, 62–68. [Google Scholar] [CrossRef]
- Herz, A.; Hassan, S.A.; Hegazi, E.; Nasr, F.N.; Youssef, A.A.; Khafagi, W.E.; Agamy, E.; Ksantini, M.; Jardak, T.; Mazomenos, B.E.; et al. Towards sustainable control of Lepidopterous pests in olive cultivation. Gesunde Pflanz. 2005, 57, 117–128. [Google Scholar] [CrossRef]
- Haniotakis, G.E. Olive pest control: Present status and prospects. IOBC/WPRS Bull. 2005, 28, 1. [Google Scholar]
- Hilal, A.; Ouguas, Y. Integrated control of olive pests in Morocco. IOBC/WPRS Bull. 2005, 28, 101. [Google Scholar]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Raschka, S.; Mirjalili, V. Python Machine Learning: Machine Learning and Deep Learning with Python, Scikit-Learn, and TensorFlow 2; Packt Publishing Ltd.: Birmingham, UK, 2019. [Google Scholar]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv preprint 2017, arXiv:1712.04621. Available online: https://arxiv.org/abs/1712.04621 (accessed on 11 October 2019).
- Meier, U.; Bleiholder, H.; Buhr, L.; Feller, C.; Hack, H.; Heß, M.; Lancashire, P.D.; Schnock, U.; Stauß, R.; Van Den Boom, T.; et al. The BBCH system to coding the phenological growth stages of plants–history and publications. J. Kult. 2009, 61, 41–52. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint 2014, arXiv:1409.1556. Available online: https://arxiv.org/abs/1409.1556 (accessed on 13 October 2019).
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-first AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; pp. 4278–4284. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning Mit Python und Keras: Das Praxis-Handbuch vom Entwickler der Keras-Bibliothek; MITP-Verlags GmbH & Co. KG.: Bonn, Germany, 2018. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint 2016, arXiv:1603.04467. Available online: https://arxiv.org/abs/1603.04467 (accessed on 20 October 2019).
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. In Proceedings of the ICML, Atlanta, GA, USA, 16–31 June 2013; Volume 30, p. 3. [Google Scholar]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. COURSERA Neural Netw. Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint 2015, arXiv:1502.03167. Available online: https://arxiv.org/abs/1502.03167 (accessed on 10 October 2019).
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Asghari, M.H.; Jalali, B. Edge detection in digital images using dispersive phase stretch transform. Int. J. Biomed. Imaging 2015. [Google Scholar] [CrossRef]
- Jalali, B.; Suthar, M.; Asghari, M.; Mahjoubfar, A. Time Stretch Inspired Computational Imaging. arXiv preprint 2017, arXiv:1706.07841. Available online: https://arxiv.org/abs/1706.07841 (accessed on 20 September 2019).
- Ang, R.B.Q.; Nisar, H.; Khan, M.B.; Tsai, C.Y. Image segmentation of activated sludge phase contrast images using phase stretch transform. Microscopy 2019, 68, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Masters, D.; Luschi, C. Revisiting small batch training for deep neural networks. arXiv preprint 2018, arXiv:1804.07612. Available online: https://arxiv.org/abs/1804.07612 (accessed on 20 October 2019).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv preprint 2014, arXiv:1412.6980. Available online: https://arxiv.org/abs/1412.6980 (accessed on 10 October 2019).
- Dozat, T. Incorporating Nesterov Momentum into Adam. In Proceedings of the 4th International Conference on Learning Representations, Workshop Track, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Rojo, J.; Pérez-Badia, R. Models for forecasting the flowering of Cornicabra olive groves. Int. J. Biometeorol. 2015, 59, 1547–1556. [Google Scholar] [CrossRef]
- Mancuso, S.; Pasquali, G.; Fiorino, P. Phenology modelling and forecasting in olive (Olea europaea L.) using artificial neural networks. Adv. Hortic. Sci. 2002, 16, 155–164. [Google Scholar]
- Barzman, M.; Bàrberi, P.; Birch, A.N.E.; Boonekamp, P.; Dachbrodt-Saaydeh, S.; Graf, B.; Hommel, B.; Jensen, J.E.; Kiss, J.; Kudsk, P.; et al. Eight principles of integrated pest management. Agron. Sustain. Dev. 2015, 35, 1199–1215. [Google Scholar] [CrossRef]








| CNN | Depth | Dataset | Parameters | Accuracy (%) |
|---|---|---|---|---|
| VGG19 | 19 | Augm. | 24,219,106 | 69.50 |
| VGG19 (pretrained) | 19 | Augm. | 24,219,106 | *54.00 |
| InceptionResNetV2 | 164 | Augm. | 55,912,674 | 65.50 |
| InceptionResNetV2 (pretrained) | 164 | Augm. | 55,912,674 | 66.50 |
| Xception (pretrained) | 71 | Augm. | 22,910,480 | 67.00 |
| ResNet50 | 50 | Augm. | 25,687,938 | 64.00 |
| ResNet50 (pretrained) | 50 | Augm. | 25,687,938 | *54.00 |
| Custom CNN | 12 | Augm. | 3,614,768 | 91.50 |
| Custom CNN | 13 | Augm. | 3,746,402 | 93.50 |
| Custom CNN | 14 | Orig. | 6,106,242 | 88.50 |
| Custom CNN | 14 | Augm. | 6,106,242 | 94.50 |
| Custom CNN | 15 | Augm. | 6,892,130 | 93.50 |
| Custom CNN | 16 | Augm. | 10,039,458 | 91.50 |
| Layer Type | Output Shape | #Param |
|---|---|---|
| Input | 256 × 256×3 | - |
| Conv2D(64) | 256 × 256 × 64 | 640 |
| ACT(LReLu)-BN | 256 × 256 × 64 | 1,024 |
| Conv2D(64) | 256 × 256 × 64 | 36,928 |
| ACT(LReLu)-BN | 256 × 256 × 64 | 1,024 |
| MP(2,2)+DR(0.1) | 128 × 128 × 64 | - |
| Conv2D(64) | 128 × 128 × 64 | 36,928 |
| ACT(LReLu)-BN | 128 × 128 × 64 | 512 |
| Conv2D(64) | 128 × 128 × 64 | 36,928 |
| ACT(LReLu)-BN | 128 × 128 × 64 | 512 |
| MP(2,2)+DR(0.2) | 64 × 64 × 64 | - |
| Conv2D(128) | 64 × 64 × 128 | 73,856 |
| ACT(LReLu)-BN | 64 × 64 × 128 | 256 |
| Conv2D(128) | 64 × 64 × 128 | 147,584 |
| ACT(LReLu)-BN | 64 × 64 × 128 | 256 |
| MP(2,2)+DR(0.1) | 32 × 32 × 128 | - |
| Conv2D(128) | 32 × 32 × 128 | 147,584 |
| ACT(LReLu)-BN | 32 × 32 × 128 | 128 |
| Conv2D(128) | 32 × 32 × 128 | 147,584 |
| ACT(LReLu)-BN | 32 × 32 × 128 | 128 |
| MP(2,2)+DR(0.2) | 16 × 16 × 128 | - |
| Conv2D(256) | 16 × 16 × 256 | 295,168 |
| ACT(LReLu)-BN | 16 × 16 × 256 | 64 |
| Conv2D(256) | 16 × 16 × 256 | 590,080 |
| ACT(LReLu)-BN | 16 × 16 × 256 | 64 |
| MP(2,2)+DR(0.2) | 8 × 8 × 256 | - |
| Conv2D(256) | 8 × 8 × 512 | 1,180,160 |
| ACT(LReLu)-BN | 8 × 8 × 512 | 32 |
| Conv2D(256) | 8 × 8 × 512 | 2,359,808 |
| ACT(LReLu)-BN | 8 × 8 × 512 | 32 |
| MP(2,2)+DR(0.2) | 4 × 4 × 512 | - |
| Flatten | 8192 | - |
| Dense(128) | 128 | 1,048,704 |
| ACT(LReLu)-DR(0.3) | 128 | - |
| Dense(2) | 2 | 258 |
| ACT(softmax) | 2 | - |
| TOTAL | 6,106,242 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milicevic, M.; Zubrinic, K.; Grbavac, I.; Obradovic, I. Application of Deep Learning Architectures for Accurate Detection of Olive Tree Flowering Phenophase. Remote Sens. 2020, 12, 2120. https://doi.org/10.3390/rs12132120
Milicevic M, Zubrinic K, Grbavac I, Obradovic I. Application of Deep Learning Architectures for Accurate Detection of Olive Tree Flowering Phenophase. Remote Sensing. 2020; 12(13):2120. https://doi.org/10.3390/rs12132120
Chicago/Turabian StyleMilicevic, Mario, Krunoslav Zubrinic, Ivan Grbavac, and Ines Obradovic. 2020. "Application of Deep Learning Architectures for Accurate Detection of Olive Tree Flowering Phenophase" Remote Sensing 12, no. 13: 2120. https://doi.org/10.3390/rs12132120
APA StyleMilicevic, M., Zubrinic, K., Grbavac, I., & Obradovic, I. (2020). Application of Deep Learning Architectures for Accurate Detection of Olive Tree Flowering Phenophase. Remote Sensing, 12(13), 2120. https://doi.org/10.3390/rs12132120