1. Introduction
In Europe, approximately 30% to 35% of the agricultural area consists of grasslands [
1]. Mainly permanent grasslands are incredibly variable in species composition, biodiversity, and management practices, as well as in productivity [
2]. Food provisions as forage for ruminants and herbivores and as biomass substrate for energy production are the most comprehensive ecosystem services from grasslands. There exists a multitude of destructive and non-destructive methods to measure or estimate the production and quality of the forage [
3,
4,
5]. Timely and accurate information about forage quality is essential to match available feed on offer with animals’ demand.
Usually, farmers use visual criteria to evaluate forage quality, such as the plants’ phenological stage, leafiness, or colour. Additionally, lab-based chemical analysis and near-infrared reflectance spectroscopy (NIRS) are utilised by agronomists to evaluate forage quality [
6]. Lab-based methods are used to determine chemical forage components that relate to the digestibility of the forage. Acid detergent fibre (ADF), which represents cellulose, lignin, and silica, is an important parameter that is negatively correlated with forage digestibility [
6,
7]. The protein concentration of forages is another parameter essential for the creation of adequate fodder rations. Protein consists mainly of amino acids, which are fundamental elements of all cells and tissues. Protein is an essential component of ruminant nutrition and provides nitrogen for ruminants’ metabolism and production of milk and meat [
8].
Remote sensing (RS) is a non-destructive method for estimating grassland biomass and forage quality [
9,
10,
11]. From satellite RS to field spectroscopy and from optical RS to synthetic aperture radar were employed for forage quality monitoring and mapping [
5,
12]. A literature review on RS based forage quality estimation studies published between 2004 and 2018 shows that more than 60% of studies (21 out of a total of 31 reviewed studies) have utilised field spectroscopy. Often, point-level spectral reflectance data from visible to short-wave infrared regions of the electromagnetic spectrum are collected in field spectroscopy [
13]. Crude protein (CP), nitrogen (N), neutral detergent fibre (NDF), and ADF are the most common forage quality parameters that have been estimated using field spectroscopy data for different grasslands with significant accuracy [
14,
15,
16,
17,
18].
Point-level data observation is laborious and challenging to collect over large areas. The application of imaging spectroscopy (also known as hyperspectral imaging) mounted on higher-altitude platforms (i.e., airborne, space-borne) could overcome these disadvantages. Imaging spectroscopy is the “acquisition of images in many (hundreds) narrow contiguous spectral bands” [
19]. Instead of point observation, imaging spectroscopy provides a two-dimensional image with each pixel containing spectral reflectance data. Airborne imaging spectroscopy has been utilised for forage quality mapping in grassland [
16,
20,
21], but conducting manned airborne mission requires extensive well-ahead planning. On the contrary, an unmanned aerial vehicle (UAV) allows the collection of low altitude images over large areas with less effort and relatively low costs.
UAV as an RS platform for both multi- and hyperspectral imaging has been frequently applied for field-scale vegetation mapping in the last few years owing to miniature sensor availability and the advancement of computer vision techniques. Concerning grassland monitoring and mapping, the UAV-borne RS has been employed mainly for grassland biomass estimation with multispectral imaging [
22], imaging spectroscopy [
23,
24], and consumer-grade cameras [
25,
26]. Only two studies that utilised UAV-borne imaging spectroscopy data for forage quality estimation are known to the authors [
23,
24], and the results showed that UAV-borne imaging spectroscopy data could accurately predict forage quality parameters. However, those studies have only focused on experimental plots, and no study has evaluated the UAV-borne imaging spectroscopy for forage quality estimation under practical field conditions including different grassland types in terms of plant community composition and with a wide range of management practices.
Except for the spectral data collection method, the model development for forage quality estimation employed various techniques. In general, forage quality modelling utilised actual reflectance data or transformed spectral reflectance data, among which normalised spectra, derivative spectra, and continuum removal spectra. The most frequently applied statistical modelling method is the linear regression (simple linear, step-wise linear) with selected highly correlated spectral features, such as wavebands [
20,
27], normalised difference spectral indices (NDSIs) [
18,
24,
28], spectral ratios (SRs) [
15,
29], and other well-known vegetation indices (e.g., NDVI, SAVI, NDRE) [
17,
23]. Predictive modelling (also known as machine learning) algorithms, such as partial least squares regression (PLSR) [
16,
27,
30,
31,
32], random forest regression (RFR) [
24,
33,
34], and artificial neural network [
20,
21,
35], were employed to estimate forage quality parameters using highly correlated spectral reflectance data. Predictive modelling algorithms frequently enhanced the predictive capability compared with the simpler linear regression models [
24,
34]. Moreover, there are several other algorithms in predictive modelling, namely, Gaussian processing regression (GPR), support vector regression (SVR), and cubist regression (CBR), that have not been examined with spectral data to estimate forage quality.
Typically, the models developed to estimate forage quality using spectral data are restricted to single grassland types. Consequently, the transferability of the models to other grassland types is limited due to the low variability of the underlying training data. Therefore, a general model that can estimate forage quality parameters, irrespective of the grassland type, would be a preferable solution. Thus, we hypothesise that the usage of spectral data over different grasslands with predictive modelling algorithms can build models to estimate major forage quality parameters regardless of the grassland type or management practice. The overall objective of this study is to develop comprehensive predictive models to estimate CP and ADF concentrations (from now on referred to as CP and ADF) on the field level, using UAV-borne imaging spectroscopy data from different grasslands. Specifically, to build models that can predict values regardless of the grassland type and to examine the effect of grassland type for the model performances. With this in mind, the specific sub-objectives are as follows:
To understand the relationship between forage quality parameters (CP and ADF) and spectral reflectance;
To develop models for CP and ADF estimation from imaging spectroscopy data and to evaluate models;
To create field-level CP and ADF maps for grasslands and describe the spatial and temporal variation of forage quality.
4. Discussion
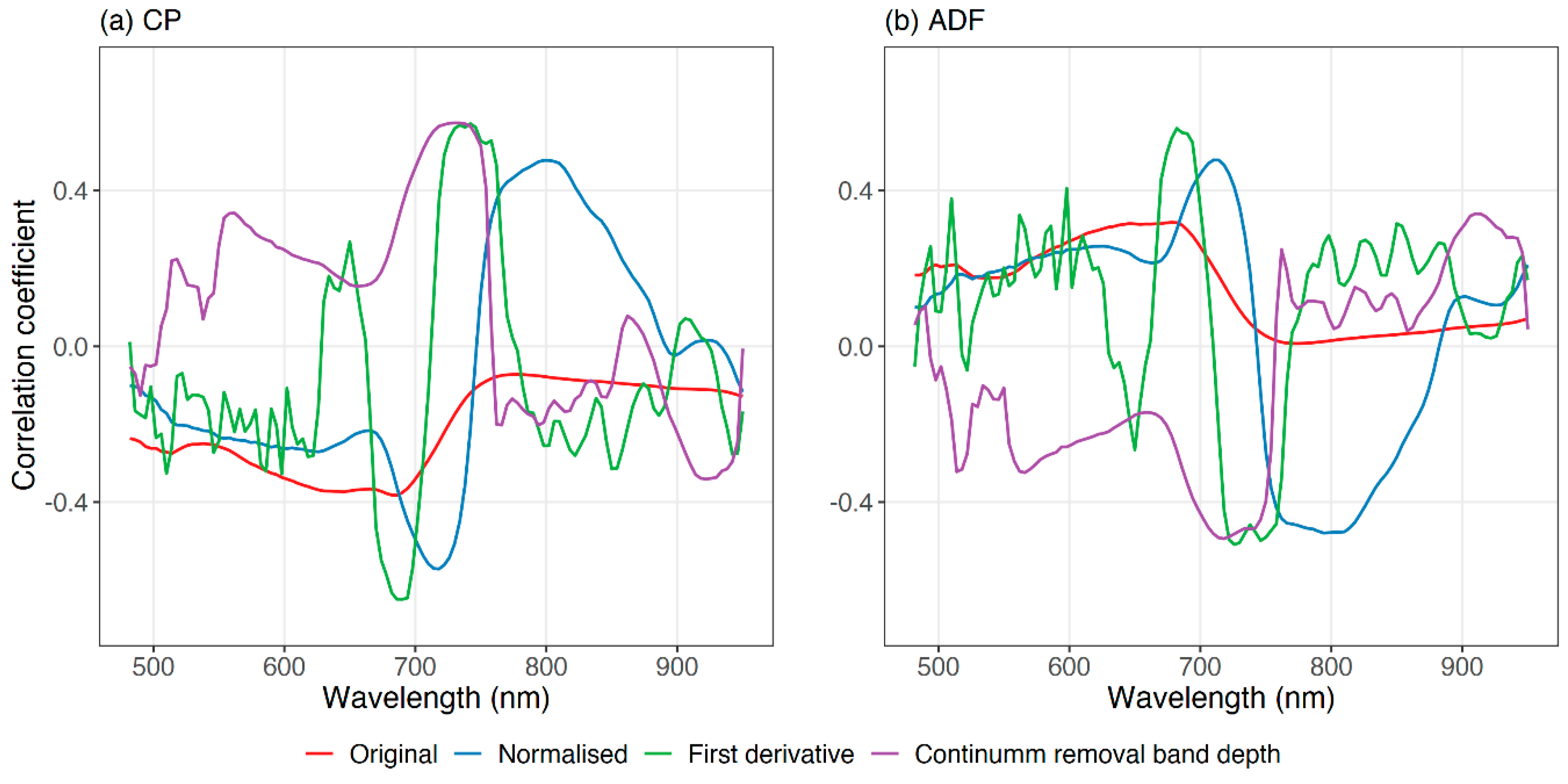
Correlograms were used to understand the relationship between grassland CP and ADF and spectral reflectance (
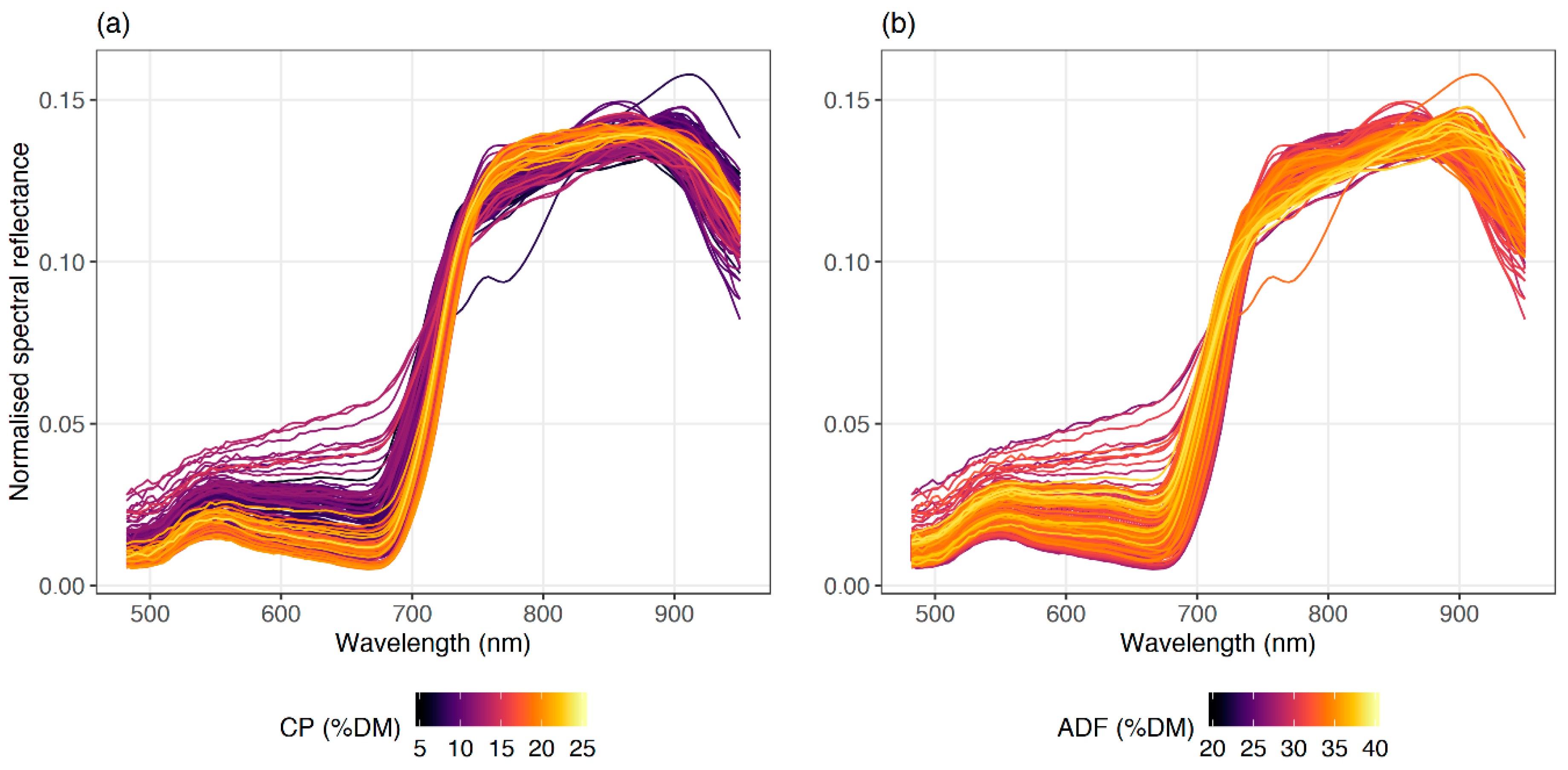
Figure 7). For both quality parameters, normalised spectra indicated a stronger relationship with forage quality parameters than the original spectra or other spectra transformations (derivatives and continuum removal band depth). The normalisation process reduced the temporal noise of the data, which strengthened the relationship between spectral features and forage quality. Further, two correlograms from CP and ADF showed an inverse pattern, confirmed by the negative correlation between them. Nevertheless, both forage quality parameters had a stronger relationship in the red-edge and the near-infrared regions of the spectrum.
Linear regression models with spectral features (single waveband, NDSI, SR) obtained a maximum adj.R
2 of 0.42 for CP and 0.34 for ADF. The best correlated NDSIs comprised wavebands in the chlorophyll absorption features in the visible domain of the spectrum (470–518 nm and 550–750 nm), which relates to N and other biochemicals in green canopies [
14]. These results paralleled with the results from previous studies that come from the same geographical region [
15,
18]. The study from Safari et al. [
32] obtained linear regression models with NDSI with R
2 of 0.58 and 0.49 for CP and ADF, respectively. Moreover, the results from Biewer et al. [
15] reported R
2 values of 0.33 and 0.13 R
2 for CP and ADF models with SR, respectively. Nonetheless, the results from the studies mentioned above were obtained with field spectroscopy data, which had a more comprehensive spectral range (305–1700 nm) than the camera utilised in the present study.
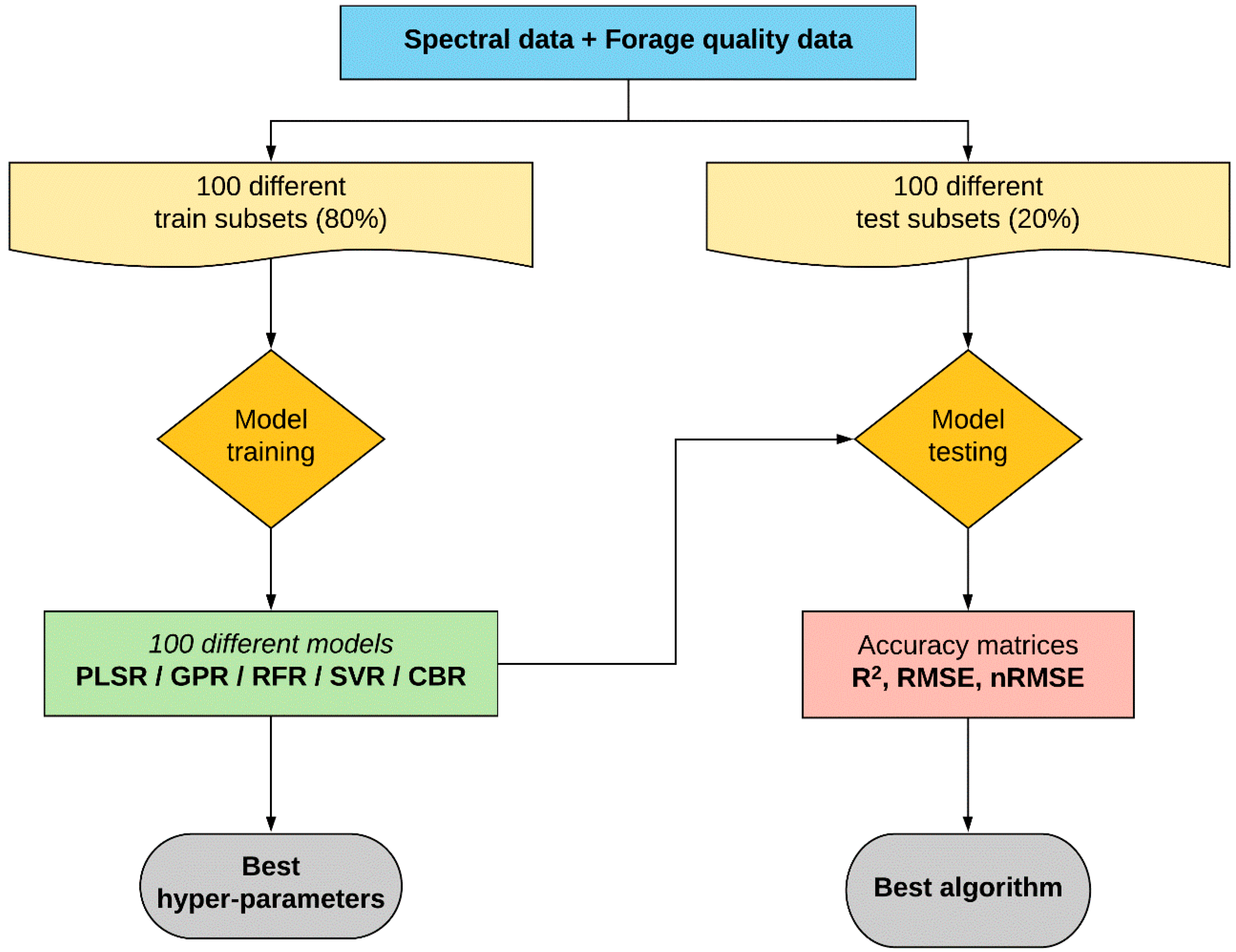
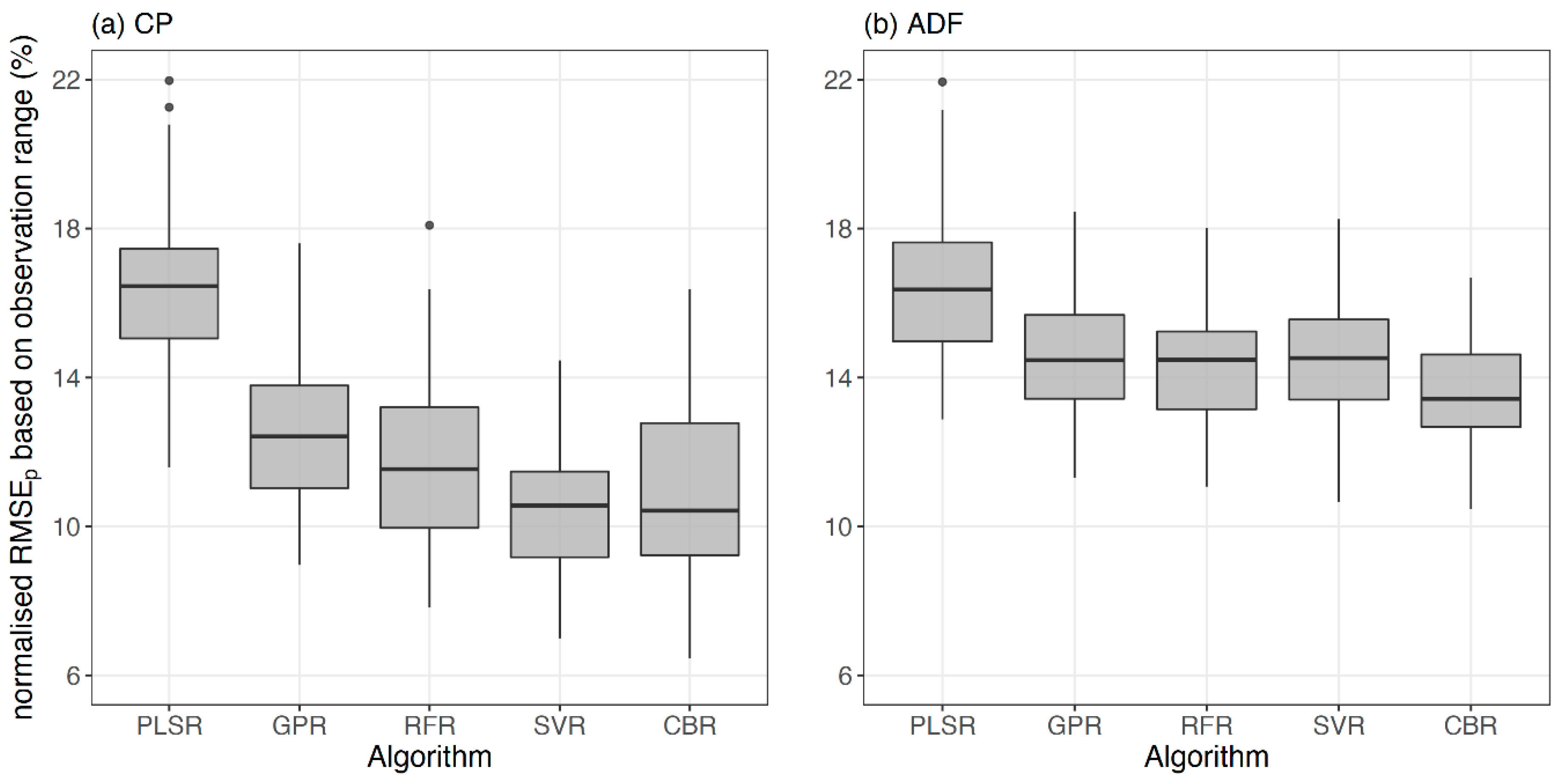
Several predictive modelling algorithms were tested to identify the best algorithms to estimate CP and ADF from the full spectral data, as no single consistent algorithm was shown to surpass all the given circumstances every time in a study by the authors of [
61]. Moreover, model consistency was evaluated by training and testing 100 models using 100 different random train and test data sets, which allowed to disclose the model performance irrespective of the calibration data set. Except for the PLSR, other tested predictive algorithms (GPR, RFR, SVR, and CBR) proved promising for the underlying data. The SVR and CBR models for CP and ADF estimation showed the maximum precision (highest R
2p) and prediction accuracy (lowest nRMSE
p) followed by the RFR, GPR, and PLSR models, respectively. SVR and CBR predictive modelling algorithms were previously utilised to estimate water quality parameters based on spectral data [
62,
63], however, to our knowledge, such algorithms have not been employed so far to estimate forage quality parameters.
According to the literature, the PLSR and RFR were the most prominent predictive modelling algorithms in forage quality parameter estimation using spectral data. For example, Safari et al. (2016) report on PLSR models, which obtained nRMSE values of 8.5% and 7.3% for CP and ADF, respectively. However, PLSR models resulted in the lowest accuracies both for CP and ADF in this study. Moreover, Pullanagari et al. [
34] achieved an nRMSE of 11.2% for CP with the RFR model, and Singh et al. [
33] obtained an nRMSE of 21.7% for ADF with the same modelling algorithm. It is noteworthy that the studies mentioned above only tested one predictive modelling algorithm; thus, no conclusions are possible considering the comparison with other algorithms.
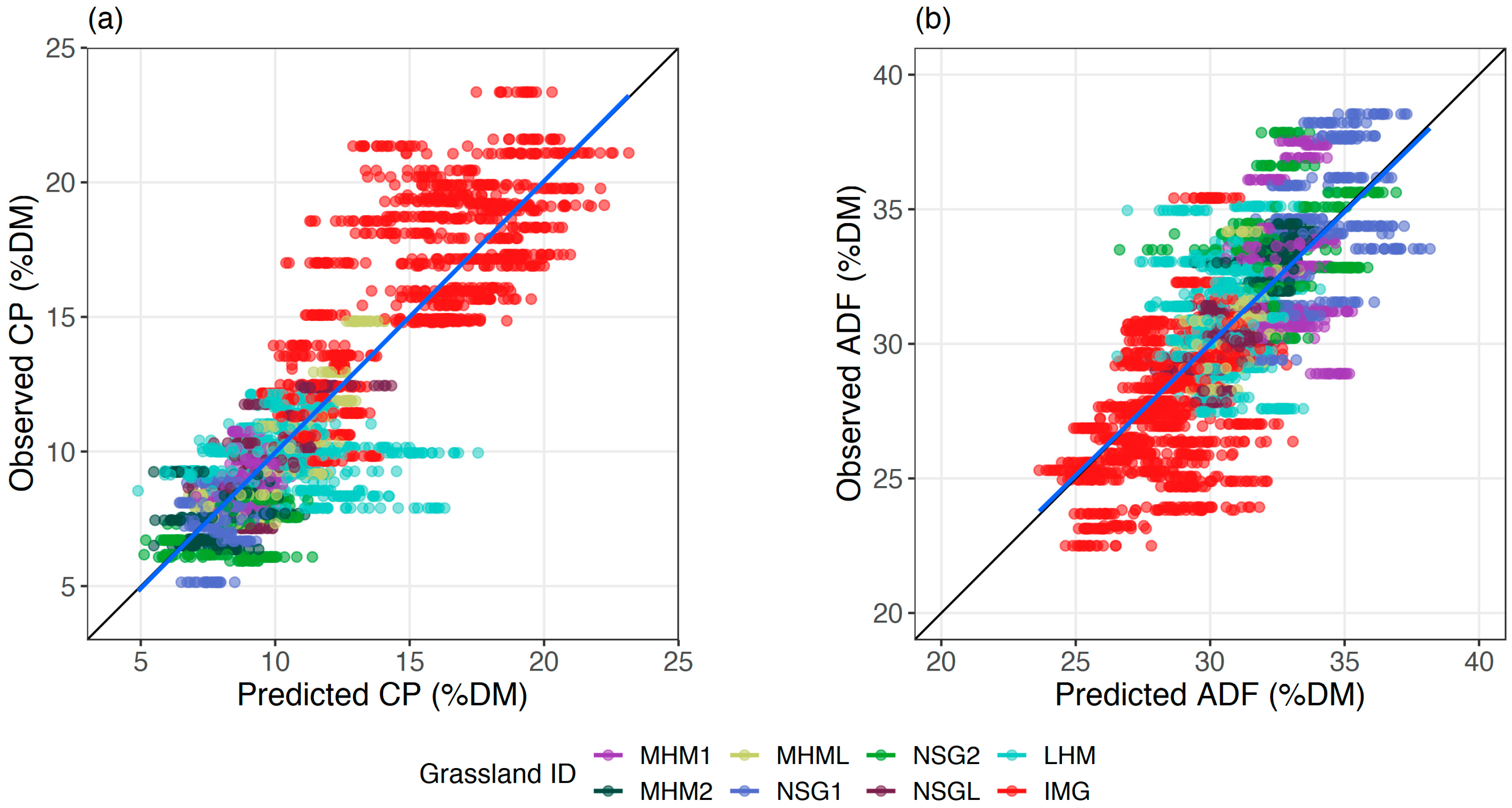
Both CP and ADF estimation models resulted in less than 15% relative prediction error. However, CP estimation had a slightly lower relative error (median nRMSE
p = 10.6%) than ADF estimation (median nRMSE
p = 13.4%). A similar relative error pattern for CP and ADF estimation models was obtained in previous studies that utilised field spectroscopy data [
16,
18,
64]. Moreover, the data points from the IMG were clustered out and acted as the driver of the CP model according to the plot of fit (
Figure 9a). Although, ADF data points in the observed against predicted plot did not highlight a similar pattern (
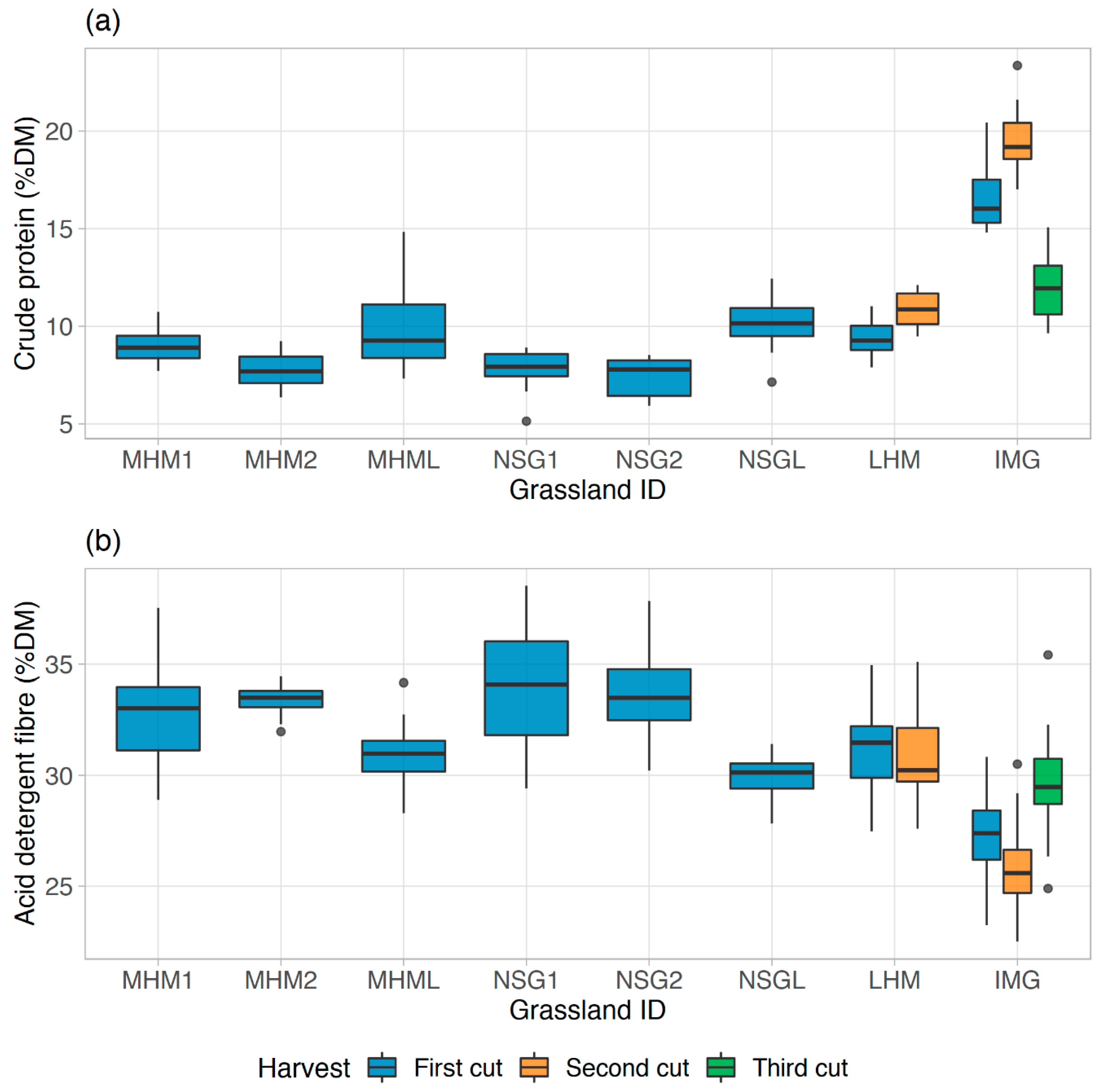
Figure 9b). The high variation of CP between the cuts in IMG due to intensive management practice might be the reason for the mentioned pattern. However, the data points from other grasslands were mostly grouped in both plots because they almost experienced similar management practice.
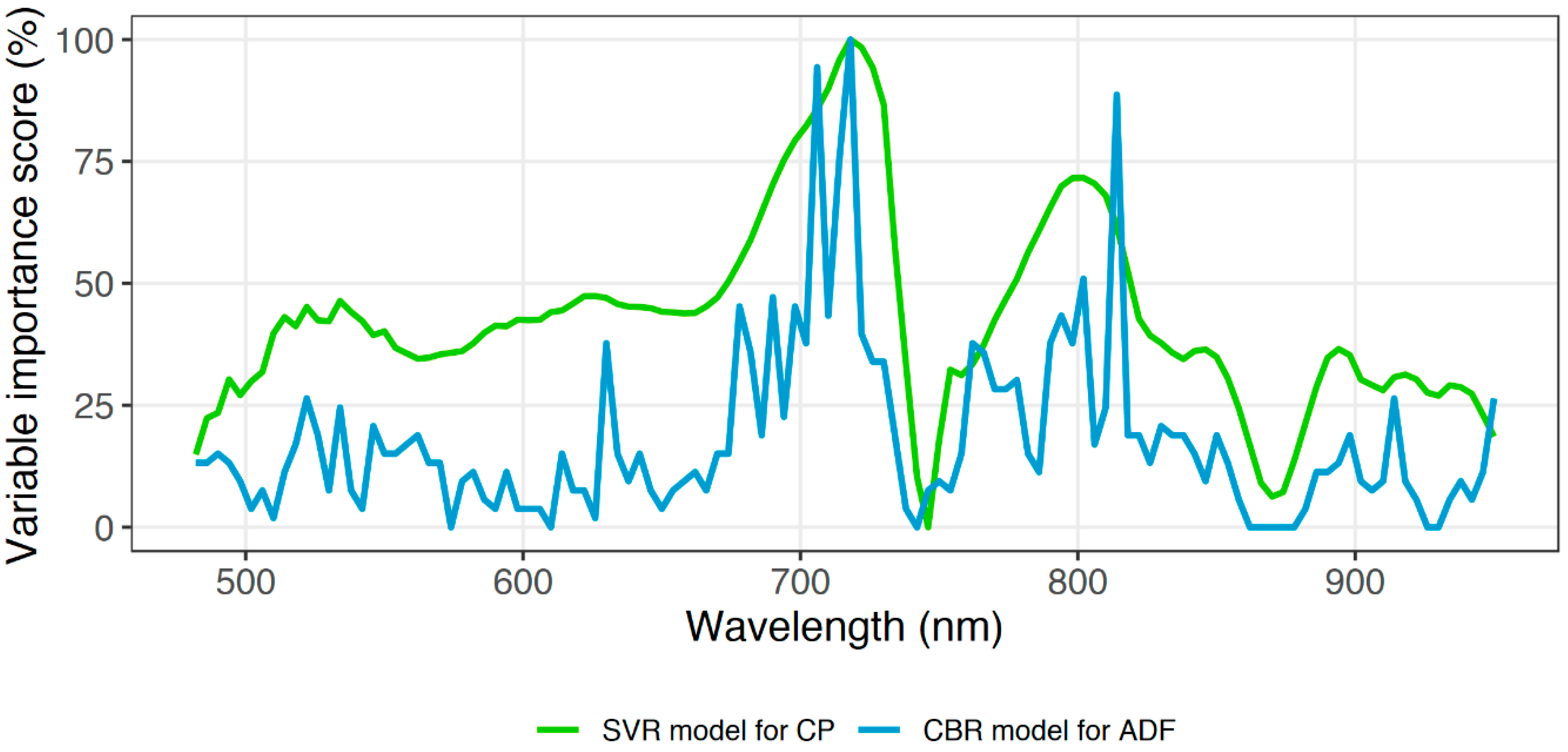
Remarkably, the wavelength (718 nm) that obtained the highest linear correlation with CP turned out to be the most fundamental wavelength for the final CP and ADF models (
Figure A1). However, the identified first most important wavelengths were not related to any foliar chemical absorption feature, such as chlorophyll, protein, or cellulose [
65]. According to Curran [
65], the absorption features that relate to CP and ADF can be found at wavelengths in the shortwave infrared region of the spectrum (1400–3000 nm). Singh et al. [
33] also confirmed that important wavelengths for ADF were in the shortwave infrared region of the spectrum. The camera used in the present study did not have the sensitivity to the shortwave infrared regions, even though the study showed a superior accuracy for grassland CP and ADF.
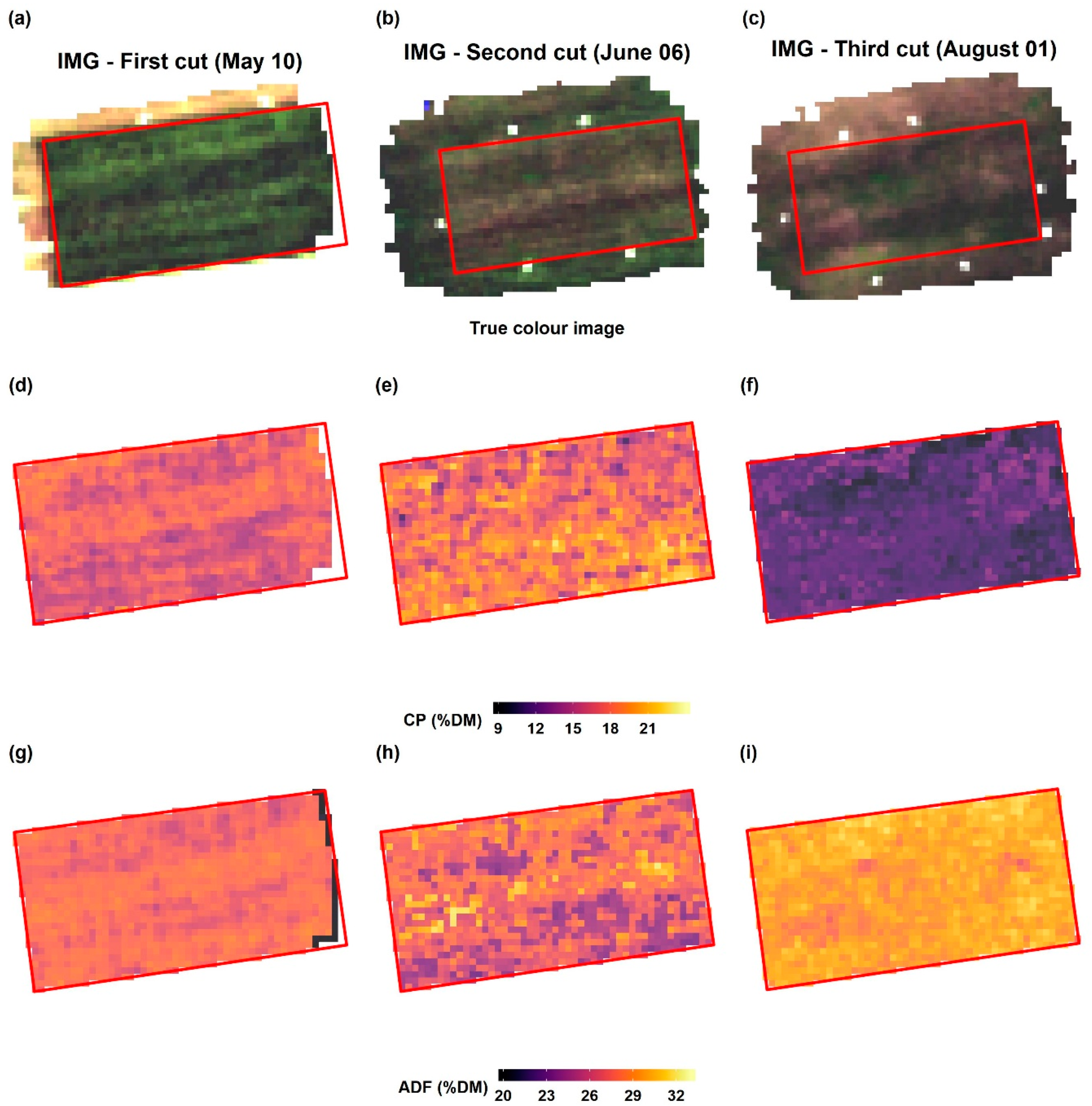
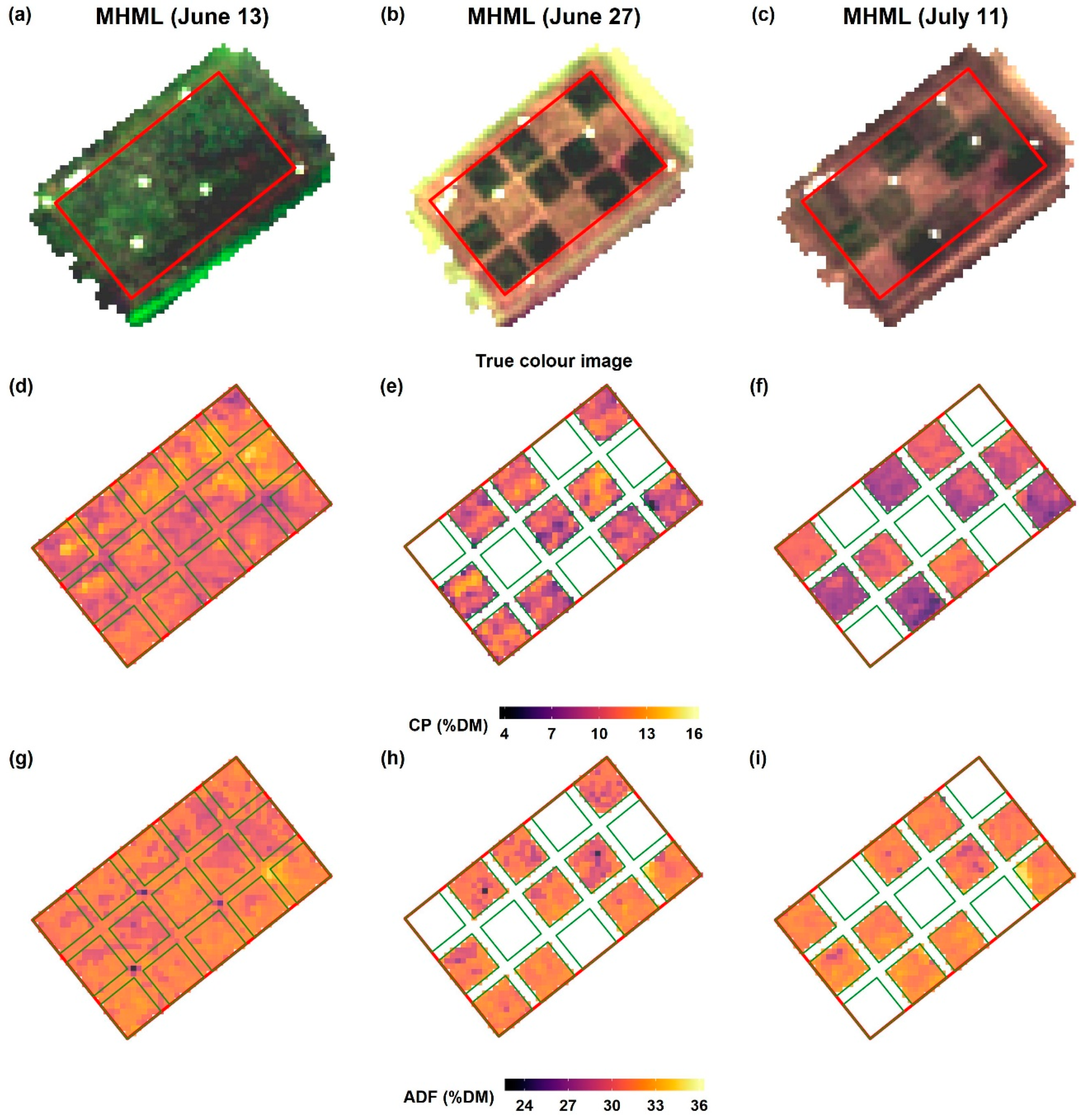
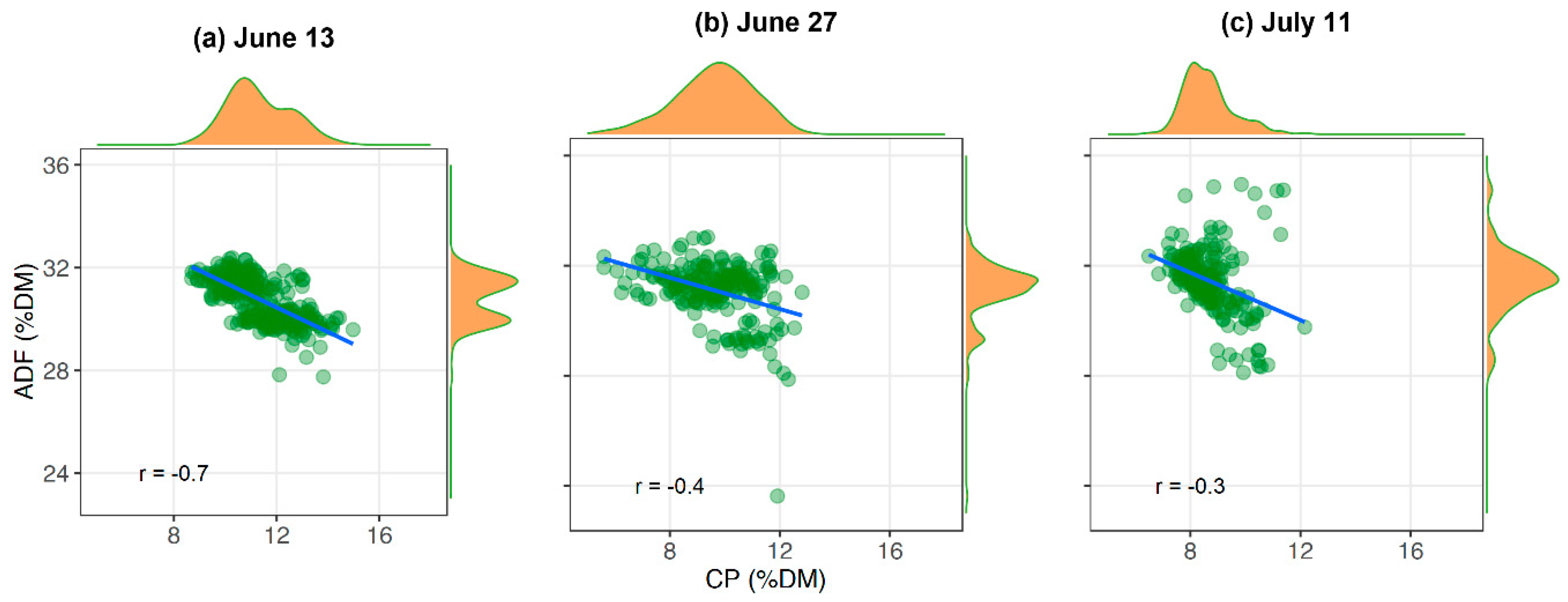
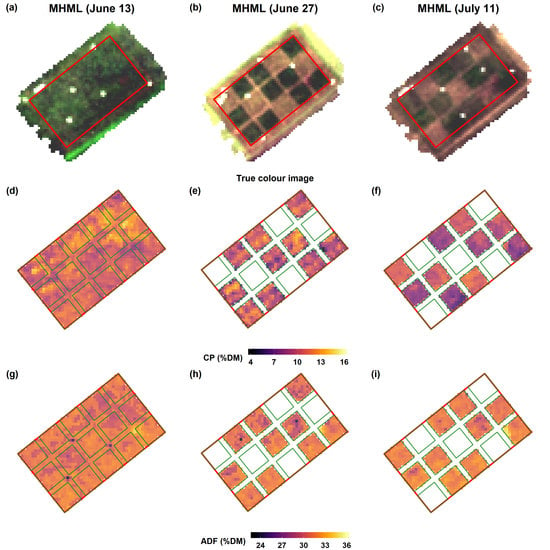
Imaging spectroscopy provides the advantage to generate maps by direct application of the model to recorded spectral images without the need for spatial interpolation. The prediction maps in our study displayed a substantial spatial variation in forage quality on the grasslands. While there is an inverse relationship between CP and ADF at the early cutting date of the MHML grassland, it vanishes with the increasing maturity of plants (
Figure 12). Thus, biomass from such grasslands (MHML, NSGL) may display a much more considerable variation in CP than in ADF content, which probably depends very much on the degree of
Lupinus polyphyllus invasion. Such information may help farmers to identify areas with contrasting forage quality, which is a prerequisite for targeted site-specific management of larger fields, for example, to determine a sound scheduling of mowing or grazing activities. Further, biomass from grassland areas with contrasting quality may be collected and ensiled separately to match the requirements of animals, which usually differ sharply among different animal species and performance categories.
To summarise, predictive modelling algorithms allow an adequate forage quality estimation, regardless of the grassland type and cutting regimes applied. Ultimately, fine-tuning of the calibrated models with data from further diverse grassland types could increase the robustness of the models to generate forage quality maps for grasslands with different vegetation types and management practices.