Mapping at 30 m Resolution of Soil Attributes at Multiple Depths in Midwest Brazil
Abstract
1. Introduction
2. Material and Methods
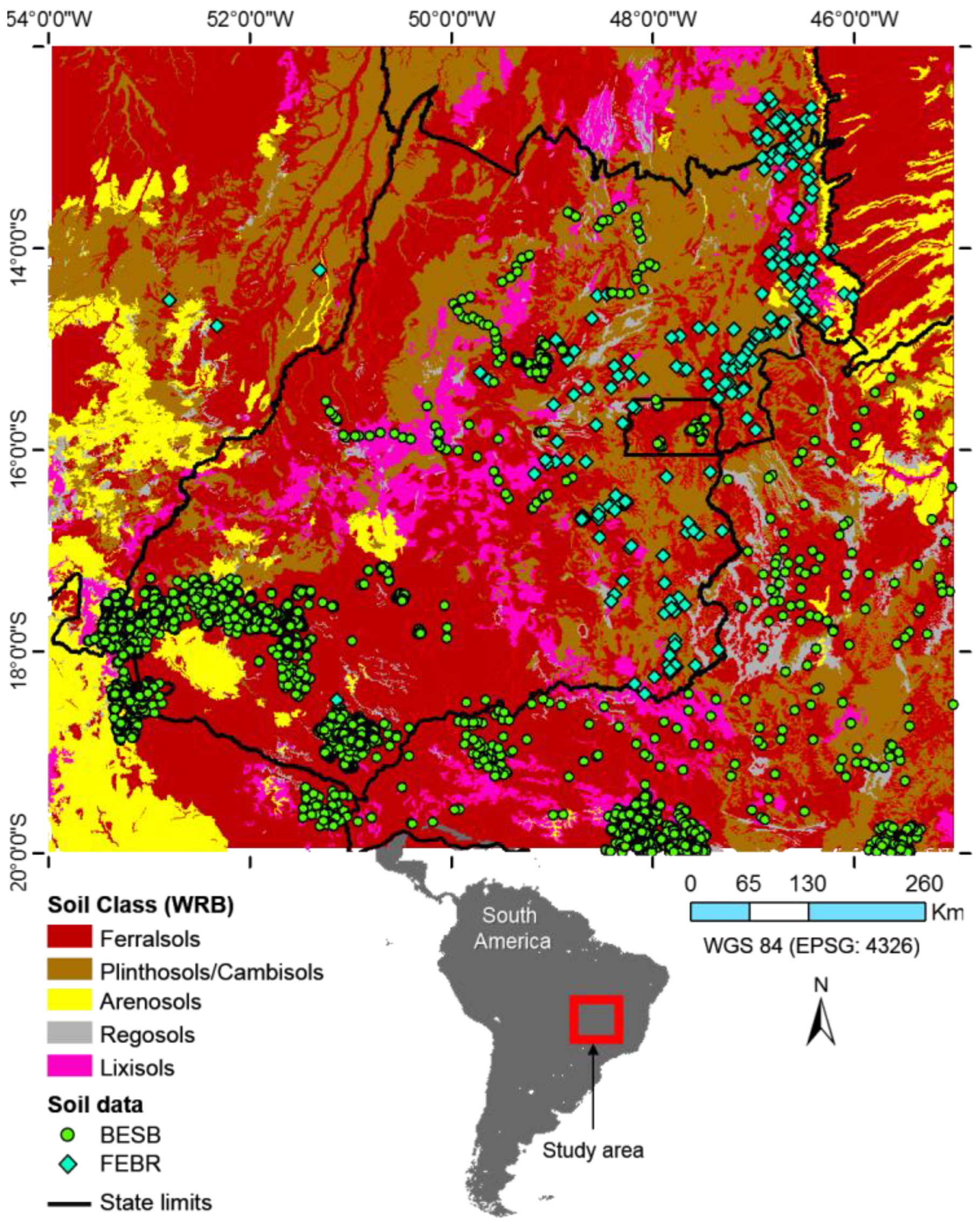
2.1. Study Area and Soil Data
2.2. Preparing Soil Covariates
2.2.1. Climate Data
2.2.2. Relief and Geology Data
2.2.3. Landsat-Derived Data
2.3. Random Forest (RF) Regression
2.3.1. Calibration and Model Tuning
2.3.2. Validation and Variable Importance
2.3.3. Prediction of Continuous Soil Attributes
3. Results
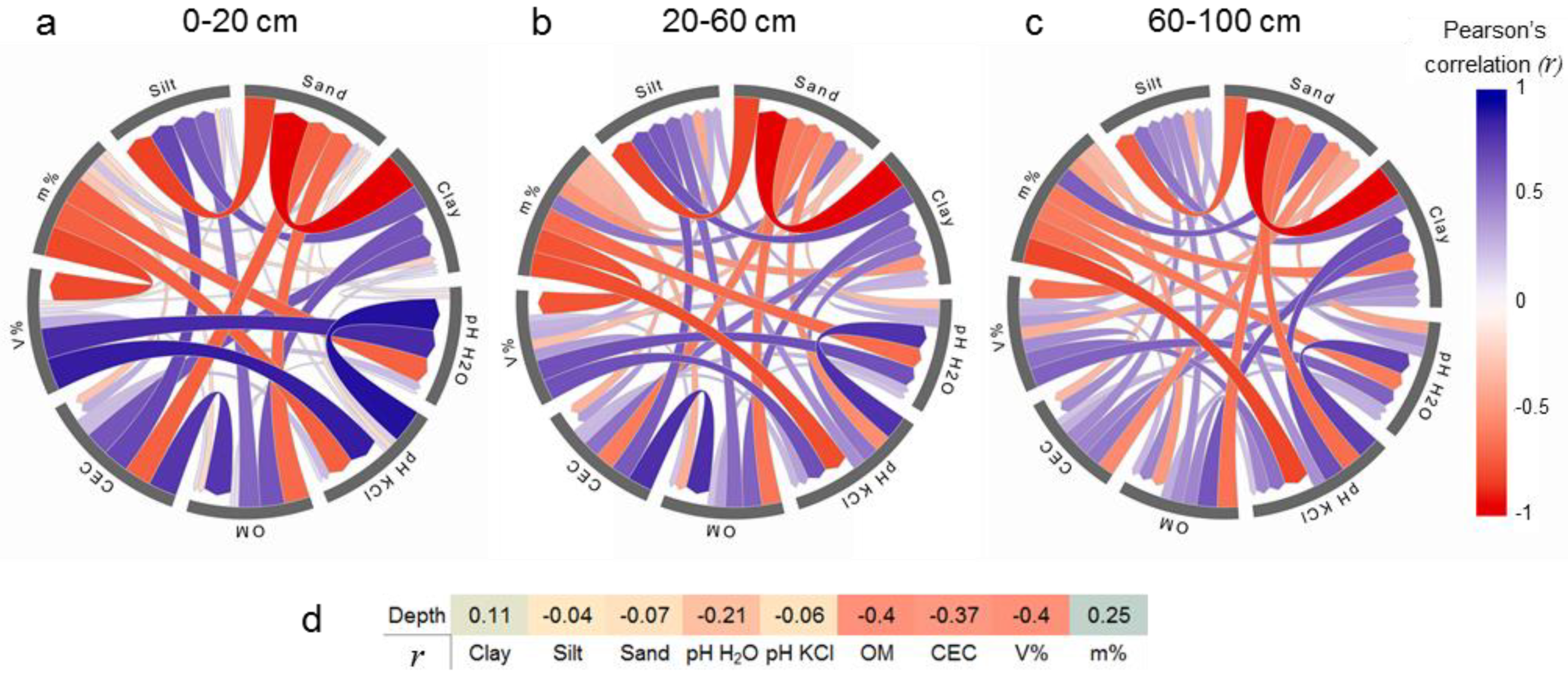
3.1. Summary and Relationships between Soil Attributes
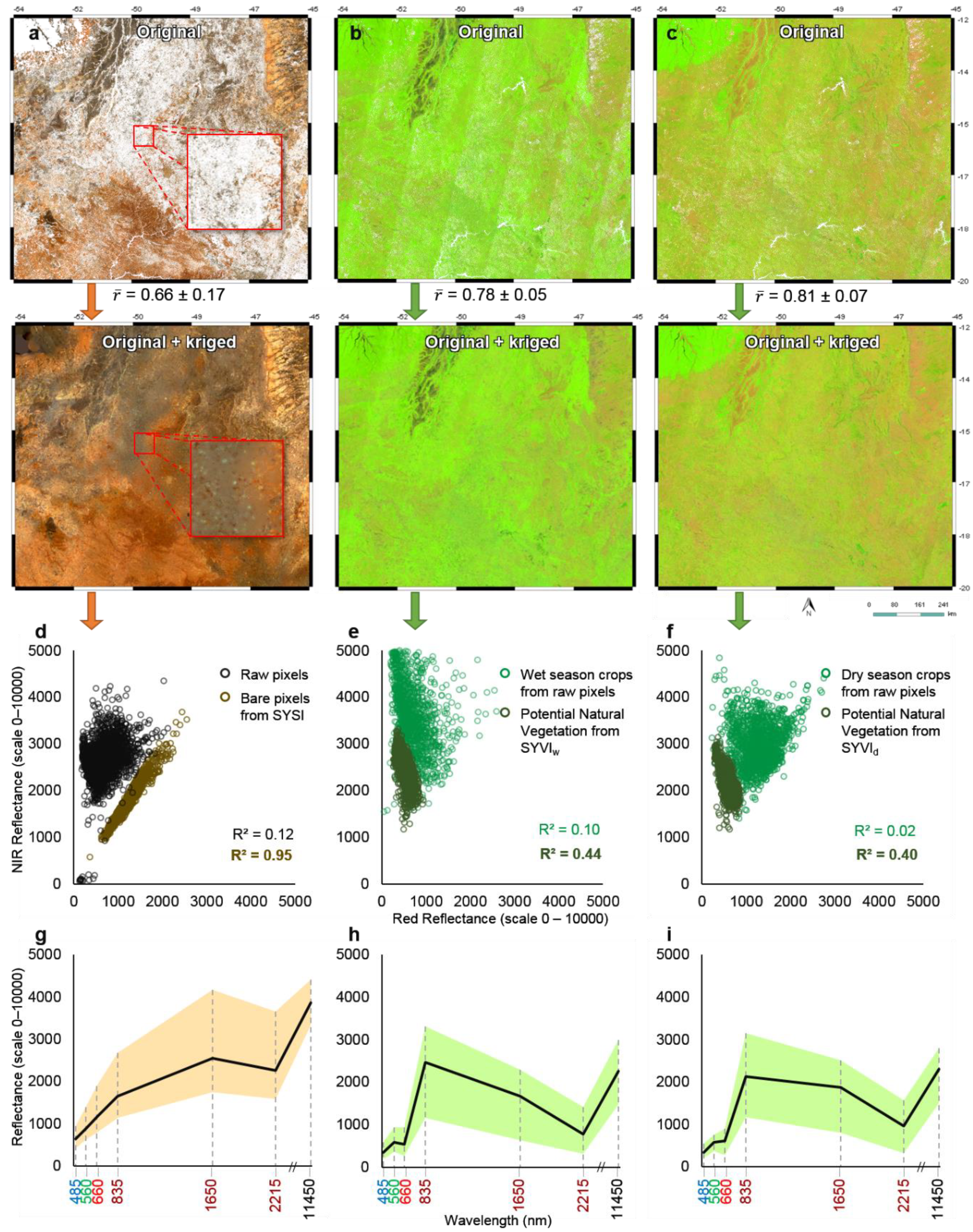
3.2. Synthetic Soil Image (SySI) and Synthetic Vegetation Image (SyVI)
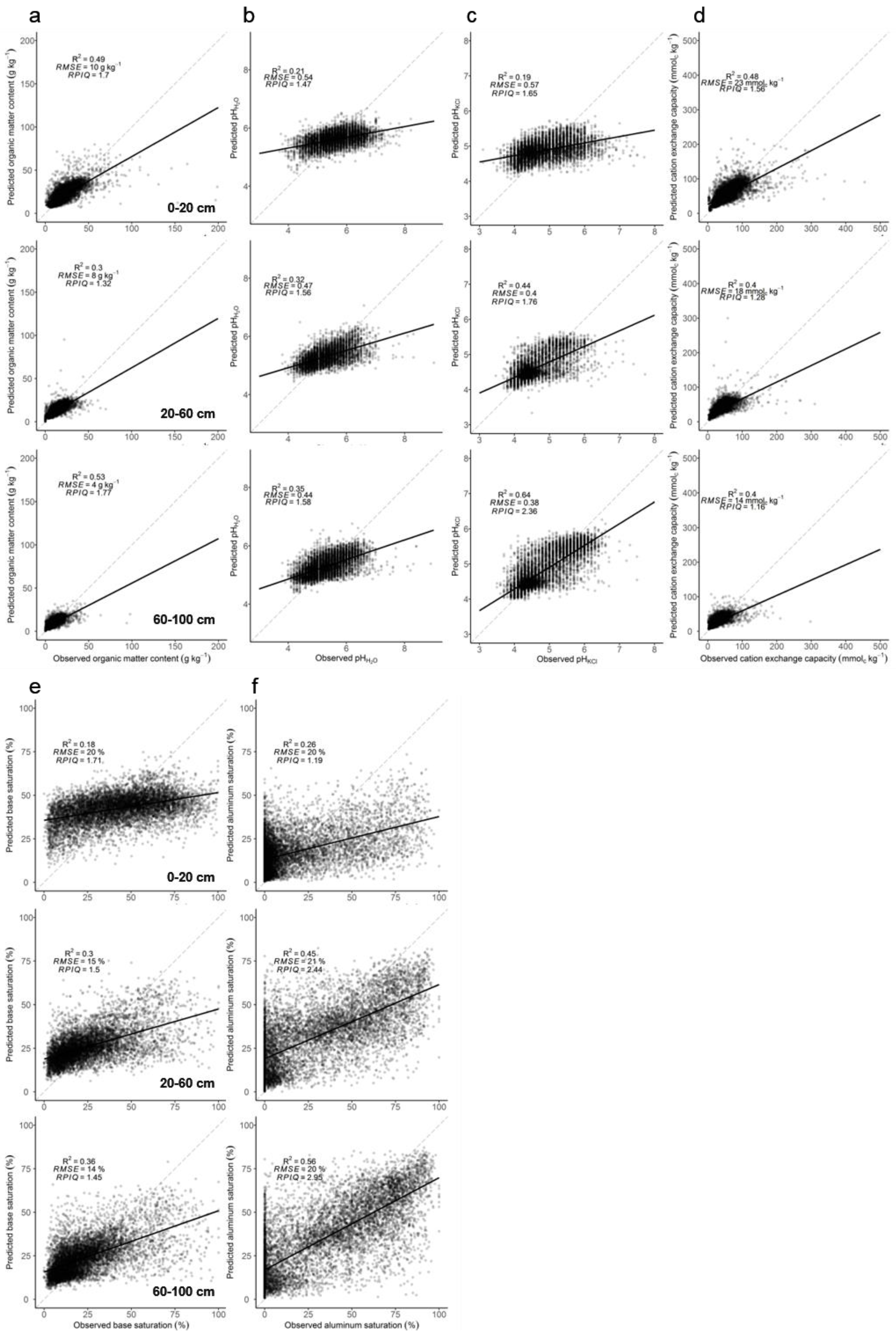
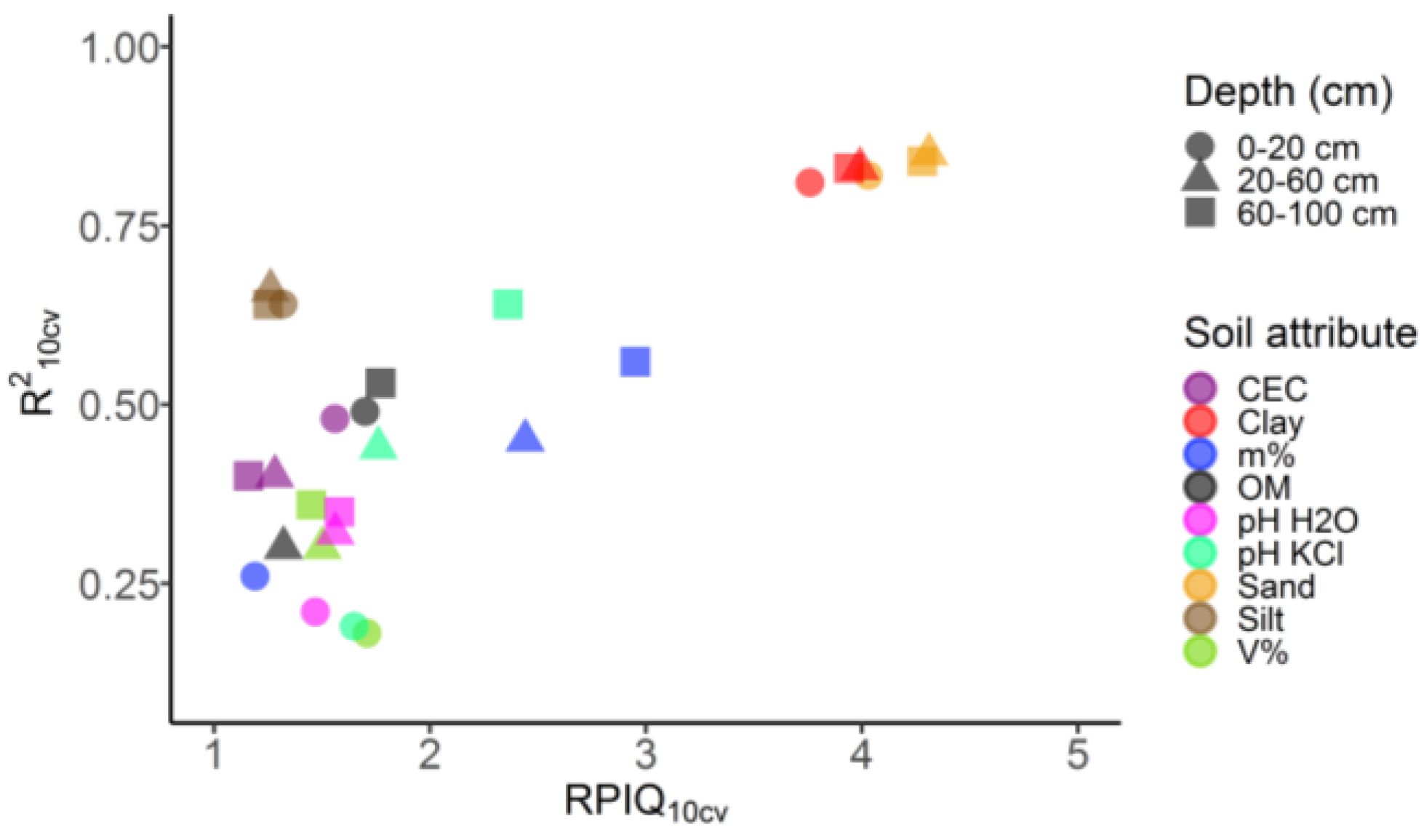
3.3. Model Assessments
3.4. Best Predictors
3.5. Soil Maps at Multiple Depths
4. Discussion
4.1. Soil Data
4.2. Machine Learning
4.3. Interpretation of Covariate Layers
4.4. Reliability and Interpretation of Soil Maps
4.5. Possible Applications of the Maps
5. Conclusions and Final Considerations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALOS | Advanced Land Observing Satellite |
| DSM | Digital Soil Mapping |
| GEE | Google Earth Engine |
| OM | Organic Matter |
| CEC | Cation Exchange Capacity |
| V% | Base Saturation |
| m% | Aluminum Saturation |
| SySI | Synthetic Soil Image |
| SyVIw | Synthetic Vegetation Image of wet season |
| SyVId | Synthetic Vegetation Image of dry season |
| PNV | Potential Natural Vegetation |
| IDW | Inverse Distance Weighted |
| DEM | Digital Elevation Model |
| NIR | Near infrared spectral band |
| SWIR1 | First shortwave infrared spectral band |
| SWIR2 | Second shortwave infrared spectral band |
| LST | Land surface temperature |
| RMSE | Root Mean Square Error |
| RPIQ | Ratio of the Performance to Inter-Quartile distance |
Appendix A
| Band. | L4 TM | L5 TM | L7 ETM+ | L8 OLI/TIRS |
|---|---|---|---|---|
| Blue | 1 (450–520 nm) | 1 (450–520 nm) | 1 (450–520 nm) | 2 (452–512 nm) |
| Green | 2 (520–600 nm) | 2 (520–600 nm) | 2 (520–600 nm) | 3 (533–590 nm) |
| Red | 3 (630–690 nm) | 3 (630–690 nm) | 3 (630–690 nm) | 4 (636–673 nm) |
| NIR | 4 (770–900 nm) | 4 (770–900 nm) | 4 (770–900 nm) | 5 (851–879 nm) |
| SWIR1 | 5 (1550–1750 nm) | 5 (1550–1750 nm) | 5 (1550–1750 nm) | 6 (1566–1651 nm) |
| SWIR2 | 7 (2080–2350 nm) | 7 (2080–2350 nm) | 7 (2080–2350 nm) | 7 (2107–2294 nm) |
| LST | 6 (10,400–12,500 nm) | 6 (10,400–12,500 nm) | 6 (10,400–12,500 nm) | 10 (10,600–11,190 nm) |
| Data | 1982–1993 | 1984–2012 | 1999–present | 2013–present |



References
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- United Nations—Department of Economic and Social Affairs—Population Division. World Population Prospects 2019: Highlights; United Nations: New York, NY, USA, 2019. Available online: https://population.un.org/wpp/Publications/Files/WPP2019_Highlights.pdf (accessed on 20 September 2019).
- McBratney, A.B.; Mendonça Santos, M.L.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Hengl, T.; Mendes de Jesus, J.; Heuvelink, G.B.M.; Ruiperez Gonzalez, M.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef] [PubMed]
- Hengl, T.; Nussbaum, M.; Wright, M.N.; Heuvelink, G.B.M.; Gräler, B. Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 2018, 6, e5518. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, B.; Viscarra Rossel, R.A.; Mouazen, A.M.; Wetterlind, J. Visible and Near Infrared Spectroscopy in Soil Science. Adv. Agron. 2010, 107, 163–215. [Google Scholar] [CrossRef]
- Diek, S.; Schaepman, M.E.; de Jong, R. Creating multi-temporal composites of airborne imaging spectroscopy data in support of digital soil mapping. Remote Sens. 2016, 8, 906. [Google Scholar] [CrossRef]
- Diek, S.; Fornallaz, F.; Schaepman, M.; de Jong, R. Barest Pixel Composite for Agricultural Areas Using Landsat Time Series. Remote Sens. 2017, 9, 1245. [Google Scholar] [CrossRef]
- Rogge, D.; Bauer, A.; Zeidler, J.; Mueller, A.; Esch, T.; Heiden, U. Building an exposed soil composite processor (SCMaP) for mapping spatial and temporal characteristics of soils with Landsat imagery (1984–2014). Remote Sens. Environ. 2018, 205, 1–17. [Google Scholar] [CrossRef]
- Demattê, J.A.M.; Fongaro, C.T.; Rizzo, R.; Safanelli, J.L. Geospatial Soil Sensing System (GEOS3): A powerful data mining procedure to retrieve soil spectral reflectance from satellite images. Remote Sens. Environ. 2018, 212. [Google Scholar] [CrossRef]
- Fongaro, C.; Demattê, J.; Rizzo, R.; Lucas Safanelli, J.; Mendes, W.; Dotto, A.; Vicente, L.; Franceschini, M.; Ustin, S. Improvement of Clay and Sand Quantification Based on a Novel Approach with a Focus on Multispectral Satellite Images. Remote Sens. 2018, 10, 1555. [Google Scholar] [CrossRef]
- Mendes, W.D.S.; Medeiros Neto, L.G.; Demattê, J.A.M.; Gallo, B.C.; Rizzo, R.; Safanelli, J.L.; Fongaro, C.T. Is it possible to map subsurface soil attributes by satellite spectral transfer models? Geoderma 2019, 343, 269–279. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Machine learning and soil sciences: A review aided by machine learning tools. SOIL Discuss. 2019, 2019, 1–29. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Probst, P.; Wright, M.N.; Boulesteix, A.-L. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1301. [Google Scholar] [CrossRef]
- Loiseau, T.; Chen, S.; Mulder, V.L.; Román Dobarco, M.; Richer-de-Forges, A.C.; Lehmann, S.; Bourennane, H.; Saby, N.P.A.; Martin, M.P.; Vaudour, E.; et al. Satellite data integration for soil clay content modelling at a national scale. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101905. [Google Scholar] [CrossRef]
- Gomes, L.C.; Faria, R.M.; de Souza, E.; Veloso, G.V.; Schaefer, C.E.G.R.; Filho, E.I.F. Modelling and mapping soil organic carbon stocks in Brazil. Geoderma 2019, 340, 337–350. [Google Scholar] [CrossRef]
- Amirian-Chakan, A.; Minasny, B.; Taghizadeh-Mehrjardi, R.; Akbarifazli, R.; Darvishpasand, Z.; Khordehbin, S. Some practical aspects of predicting texture data in digital soil mapping. Soil Tillage Res. 2019, 194, 104289. [Google Scholar] [CrossRef]
- Ma, Y.; Minasny, B.; Wu, C. Mapping key soil properties to support agricultural production in Eastern China. Geoderma Reg. 2017, 10, 144–153. [Google Scholar] [CrossRef]
- Keskin, H.; Grunwald, S.; Harris, W.G. Digital mapping of soil carbon fractions with machine learning. Geoderma 2019, 339, 40–58. [Google Scholar] [CrossRef]
- Nussbaum, M.; Spiess, K.; Baltensweiler, A.; Grob, U.; Keller, A.; Greiner, L.; Schaepman, M.E.; Papritz, A. Evaluation of digital soil mapping approaches with large sets of environmental covariates. SOIL 2018, 4, 1–22. [Google Scholar] [CrossRef]
- Hengl, T.; Heuvelink, G.B.M.; Kempen, B.; Leenaars, J.G.B.; Walsh, M.G.; Shepherd, K.D.; Sila, A.; MacMillan, R.A.; Mendes de Jesus, J.; Tamene, L.; et al. Mapping Soil Properties of Africa at 250 m Resolution: Random Forests Significantly Improve Current Predictions. PLoS ONE 2015, 10, e0125814. [Google Scholar] [CrossRef] [PubMed]
- Parente, L.; Mesquita, V.; Miziara, F.; Baumann, L.; Ferreira, L. Assessing the pasturelands and livestock dynamics in Brazil, from 1985 to 2017: A novel approach based on high spatial resolution imagery and Google Earth Engine cloud computing. Remote Sens. Environ. 2019, 232, 111301. [Google Scholar] [CrossRef]
- IBGE—Instituto Brasileiro de Geografia e Estatística Produção Agrícola Municipal [Municipal Agricultural Production]. Available online: https://sidra.ibge.gov.br/pesquisa/pam/tabelas (accessed on 29 September 2019).
- IBGE—Instituto Brasileiro de Geografia e Estatística Pedologia [Pedological maps of Brazil]. Available online: https://www.ibge.gov.br/geociencias/informacoes-ambientais/pedologia/10871-pedologia.html?=&t=downloads (accessed on 30 September 2019).
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; Food and Agriculture Organization: Rome, Italy, 2015; Available online: http://www.fao.org/3/i3794en/I3794EN.pdf (accessed on 01 September 2019).
- CPRM—Companhia de Pesquisa de Recursos Minerais. Carta Geológica do Brasil ao Milionésimo: Sistema de Informações Geográficas-SIG [Geological Map of Brazil 1:1.000.000 Scale: Geographic Information System-GIS]; CPRM: Brasília, Brazil, 2004. Available online: http://www.cprm.gov.br/publique/Geologia/Geologia-Basica/Carta-Geologica-do-Brasil-ao-Milionesimo-298.html (accessed on 02 September 2019).
- Demattê, J.A.M.; Dotto, A.C.; Paiva, A.F.S.; Sato, M.V.; Dalmolin, R.S.D.; Araújo, M.S.B.; Silva, E.B.; Nanni, M.R.; ten Caten, A.; Noronha, N.C.; et al. The Brazilian Soil Spectral Library (BSSL): A general view, application and challenges. Geoderma 2019, 354, 113793. [Google Scholar] [CrossRef]
- Samuel-Rosa, A.; Dalmolin, R.S.D.; Moura-Bueno, J.M.; Teixeira, W.G.; Alba, J.M.F. Open legacy soil survey data in Brazil: Geospatial data quality and how to improve it. Sci. Agric. 2017, 77, e20170430. [Google Scholar] [CrossRef]
- Canadell, J.; Jackson, R.B.; Ehleringer, J.B.; Mooney, H.A.; Sala, O.E.; Schulze, E.-D. Maximum rooting depth of vegetation types at the global scale. Oecologia 1996, 108, 583–595. [Google Scholar] [CrossRef]
- Embrapa—Brazilian Agricultural Research Corporation—National Soils Research Center. Manual of Soil Analysis Methods, 3rd ed.; Teixeira, P.C., Donagemma, G.K., Fontana, A., Teixeira, W.G., Eds.; Embrapa Solos: Brasilia, DF, USA, 2017; Available online: http://ainfo.cnptia.embrapa.br/digital/bitstream/item/171907/1/Manual-de-Metodos-de-Analise-de-Solo-2017.pdf (accessed on 15 September 2019).
- Gu, Z. Circlize: Circular Visualization. 2019. Available online: https://cran.r-project.org/web/packages/circlize/index.html (accessed on 15 September 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 15 September 2019).
- Viscarra Rossel, R.A.; Chen, C.; Grundy, M.J.; Searle, R.; Clifford, D.; Campbell, P.H. The Australian three-dimensional soil grid: Australia’s contribution to the GlobalSoilMap project. Soil Res. 2015, 53, 845–864. [Google Scholar] [CrossRef]
- Hengl, T.; de Jesus, J.M.; MacMillan, R.A.; Batjes, N.H.; Heuvelink, G.B.M.; Ribeiro, E.; Samuel-Rosa, A.; Kempen, B.; Leenaars, J.G.B.; Walsh, M.G.; et al. SoilGrids1km—Global Soil Information Based on Automated Mapping. PLoS ONE 2014, 9, e105992. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, S.; Yang, Y.; Zhou, Y.; Shi, Z. High-resolution three-dimensional mapping of soil organic carbon in China: Effects of SoilGrids products on national modeling. Sci. Total Environ. 2019, 685, 480–489. [Google Scholar] [CrossRef]
- Ballabio, C.; Lugato, E.; Fernández-Ugalde, O.; Orgiazzi, A.; Jones, A.; Borrelli, P.; Montanarella, L.; Panagos, P. Mapping LUCAS topsoil chemical properties at European scale using Gaussian process regression. Geoderma 2019, 355, 113912. [Google Scholar] [CrossRef]
- Akinyemi, F.; Adejuwon, J. A GIS-Based Procedure for Downscaling Climate Data for West Africa. Trans. GIS 2008, 12, 613–631. [Google Scholar] [CrossRef]
- Bailey, R.G. Suggested hierarchy of criteria for multi-scale ecosystem mapping. Landsc. Urban Plan. 1987, 14, 313–319. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Tadono, T.; Ishida, H.; Oda, F.; Naito, S.; Minakawa, K.; Iwamoto, H. Precise global DEM generation by ALOS PRISM. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, II-4, 71–76. [Google Scholar] [CrossRef]
- Florinsky, I.V. Digital Terrain Analysis in Soil Science and Geology; Academic press: Sydney, Australia, 2016. [Google Scholar] [CrossRef]
- USGS—United States Geological Survey. Landsat 4–7 Surface Reflectance Code LEDAPS Product Guide; Department of the Interior, USGS: Lawrence, KS, USA, 2019. Available online: https://www.usgs.gov/media/files/landsat-4-7-surface-reflectance-code-ledaps-product-guide (accessed on 17 September 2019).
- USGS—United States Geological Survey. Landsat 8 Surface Reflectance Code LaSRC Product Guide; Department of the Interior, USGS: Lawrence, KS, USA, 2019. Available online: https://www.usgs.gov/media/files/land-surface-reflectance-code-lasrc-product-guide (accessed on 17 September 2019).
- Vandegriend, A.; Owe, M.; Vugts, H.; Ramothwa, G. Botswana Water and Surface Energy Balance Research Program. Part 1: Integrated Approach and Field Campaign Results; NASA Goddard Space Flight Center: Greenbelt, MD, USA, 1992. Available online: https://ntrs.nasa.gov/search.jsp?R=19930011702 (accessed on 18 September 2019).
- Al-Gaadi, K.A.; Hassaballa, A.A.; Tola, E.; Kayad, A.G.; Madugundu, R.; Assiri, F.; Alblewi, B. Characterization of the spatial variability of surface topography and moisture content and its influence on potato crop yield. Int. J. Remote Sens. 2018, 39, 8572–8590. [Google Scholar] [CrossRef]
- Gallo, B.; Demattê, J.; Rizzo, R.; Safanelli, J.; Mendes, W.; Lepsch, I.; Sato, M.; Romero, D.; Lacerda, M. Multi-Temporal Satellite Images on Topsoil Attribute Quantification and the Relationship with Soil Classes and Geology. Remote Sens. 2018, 10, 1571. [Google Scholar] [CrossRef]
- McBratney, A.B.; Webster, R. Choosing functions for semi-variograms of soil properties and fitting them to sampling estimates. J. Soil Sci. 1986, 37, 617–639. [Google Scholar] [CrossRef]
- Baret, F.; Jacquemoud, S.; Hanocq, J.F. About the soil line concept in remote sensing. Adv. Space Res. 1993, 13, 281–284. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- FAO. Soil Organic Carbon Mapping Cookbook, 2nd ed.; FAO: Rome, Italy, 2018; Available online: http://www.fao.org/documents/card/en/c/I8895EN (accessed on 20 September 2019).
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. arXiv 2017, arXiv:1508.04409. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and Regression Training. 2019. Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 21 September 2019).
- Bellon-Maurel, V.; Fernandez-Ahumada, E.; Palagos, B.; Roger, J.-M.; McBratney, A. Critical review of chemometric indicators commonly used for assessing the quality of the prediction of soil attributes by NIR spectroscopy. TrAC Trends Anal. Chem. 2010, 29, 1073–1081. [Google Scholar] [CrossRef]
- Poppiel, R.R.; Lacerda, M.P.C.; Safanelli, J.L.; Rizzo, R.; Oliveira, M.P., Jr.; Novais, J.J.; Demattê, J.A.M. 250 m-gridded soil texture at multiple depths of Midwest Brazil. Data Mendeley 2019. [Google Scholar] [CrossRef]
- Ma, Y.; Minasny, B.; Malone, B.P.; Mcbratney, A.B. Pedology and digital soil mapping (DSM). Eur. J. Soil Sci. 2019, 70, 216–235. [Google Scholar] [CrossRef]
- Goebes, P.; Schmidt, K.; Seitz, S.; Both, S.; Bruelheide, H.; Erfmeier, A.; Scholten, T.; Kühn, P. The strength of soil-plant interactions under forest is related to a Critical Soil Depth. Sci. Rep. 2019, 9, 8635. [Google Scholar] [CrossRef] [PubMed]
- Vaudour, E.; Gomez, C.; Fouad, Y.; Lagacherie, P. Sentinel-2 image capacities to predict common topsoil properties of temperate and Mediterranean agroecosystems. Remote Sens. Environ. 2019, 223, 21–33. [Google Scholar] [CrossRef]
- Bui, E.N.; Henderson, B.L.; Viergever, K. Knowledge discovery from models of soil properties developed through data mining. Ecol. Model. 2006, 191, 431–446. [Google Scholar] [CrossRef]
- Bui, E.; Henderson, B.; Viergever, K. Using knowledge discovery with data mining from the Australian Soil Resource Information System database to inform soil carbon mapping in Australia. Global Biogeochem. Cycles 2009, 23. [Google Scholar] [CrossRef]
- Miller, B.A.; Koszinski, S.; Wehrhan, M.; Sommer, M. Impact of multi-scale predictor selection for modeling soil properties. Geoderma 2015, 239, 97–106. [Google Scholar] [CrossRef]
- Poppiel, R.R.; Lacerda, M.P.C.; Demattê, J.A.M.; Oliveira, M.P., Jr.; Gallo, B.C.; Safanelli, J.L. Pedology and soil class mapping from proximal and remote sensed data. Geoderma 2019, 348, 189–206. [Google Scholar] [CrossRef]
- Savin, I.Y.; Zhogolev, A.V.; Prudnikova, E.Y. Modern Trends and Problems of Soil Mapping. Eurasian Soil Sci. 2019, 52, 471–480. [Google Scholar] [CrossRef]
- Maynard, J.J.; Levi, M.R. Hyper-temporal remote sensing for digital soil mapping: Characterizing soil-vegetation response to climatic variability. Geoderma 2017, 285, 94–109. [Google Scholar] [CrossRef]
- Serteser, A.; Kargιoğlu, M.; Içağa, Y.; Konuk, M. Vegetation as an Indicator of Soil Properties and Water Quality in the Akarçay Stream (Turkey). Environ. Manag. 2008, 42, 764. [Google Scholar] [CrossRef] [PubMed]
- Hengl, T.; Walsh, M.G.; Sanderman, J.; Wheeler, I.; Harrison, S.P.; Prentice, I.C. Global mapping of potential natural vegetation: An assessment of machine learning algorithms for estimating land potential. PeerJ 2018, 6, e5457. [Google Scholar] [CrossRef] [PubMed]
- Das, S. Comparison among influencing factor, frequency ratio, and analytical hierarchy process techniques for groundwater potential zonation in Vaitarna basin, Maharashtra, India. Groundw. Sustain. Dev. 2019, 8, 617–629. [Google Scholar] [CrossRef]
- Soil Survey Staff. Keys to Soil Taxonomy; United States Department of Agriculture: Washington, DC, USA, 2014; Volume 12. Available online: http://www.nrcs.usda.gov/Internet/FSE_DOCUMENTS/nrcs142p2_051546.pdf (accessed on 22 September 2019).
- Vieira, B.C.; Salgado, A.A.R.; Santos, L.J.C. Landscapes and Landforms of Brazil; Springer: Berlin/Heidelberg, Germany, 2015; Available online: https://link.springer.com/book/10.1007%2F978-94-017-8023-0#editorsandaffiliations (accessed on 22 September 2019).
- Moraes, J.M. Geodiversidade do Estado de Goiás e do Distrito Federal [Geodiversity of Goiás State and the Federal District, Brazil]; CPRM: Goiânia, GO, USA, 2014. Available online: http://rigeo.cprm.gov.br/jspui/handle/doc/16732 (accessed on 21 September 2019).

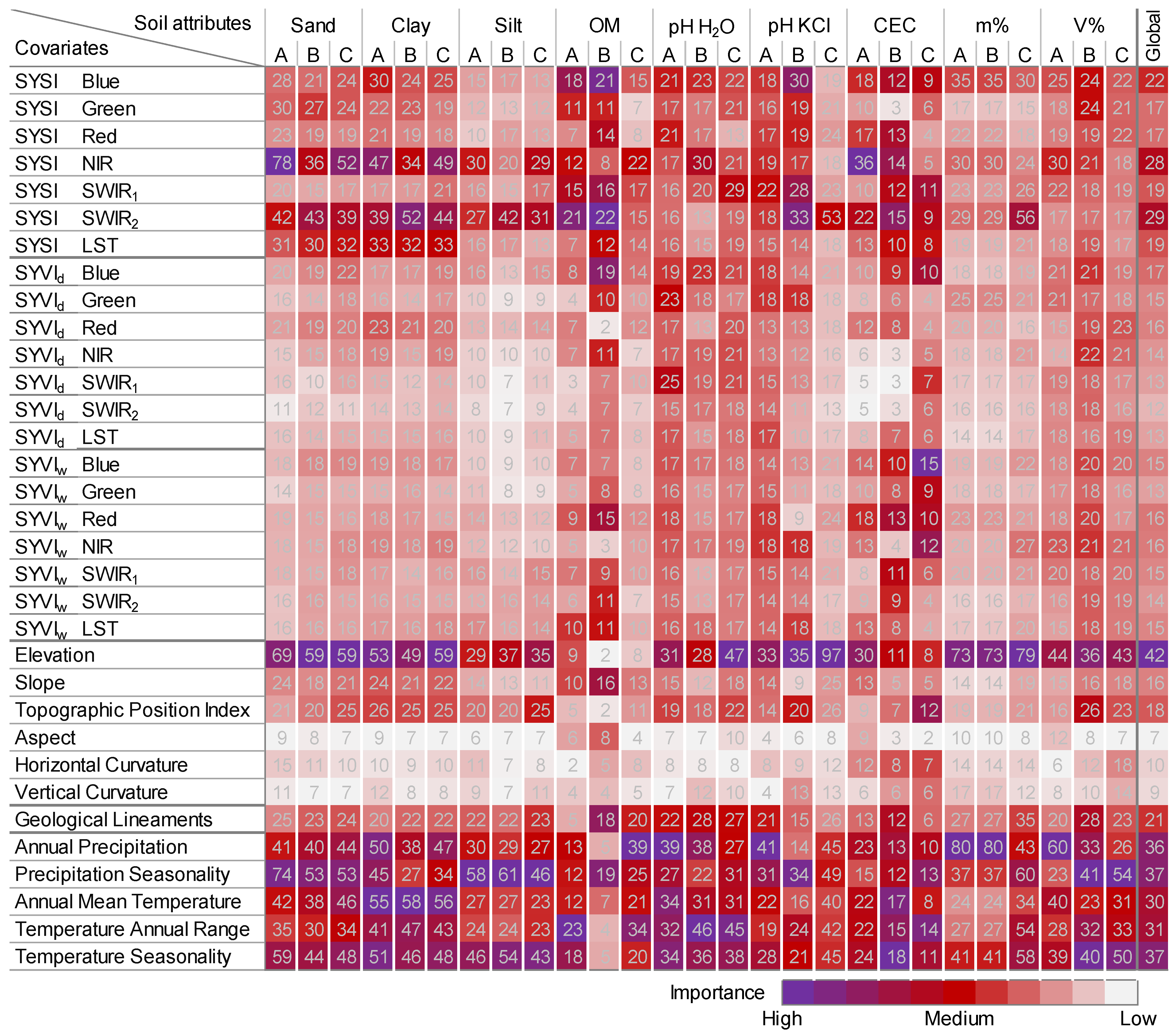
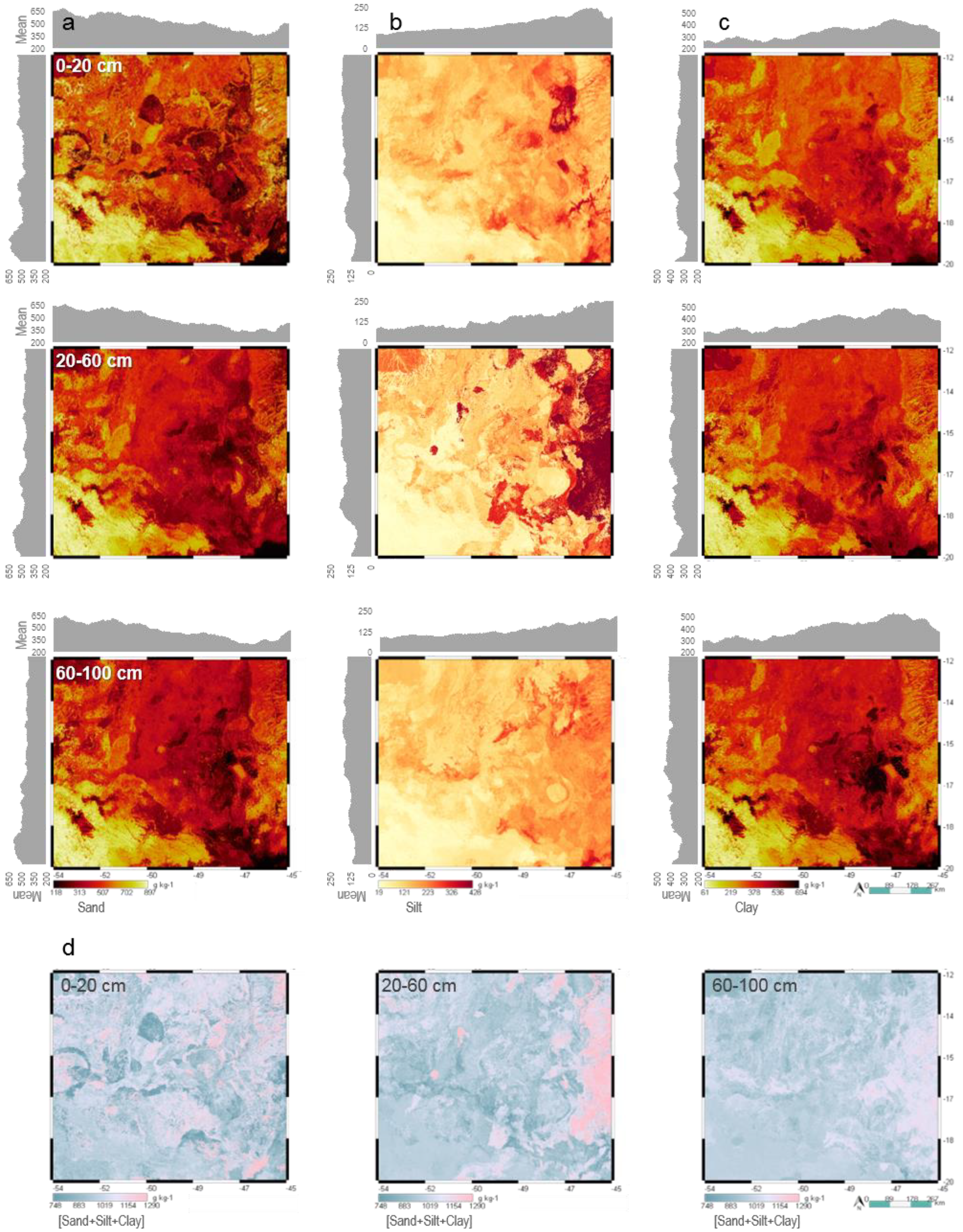
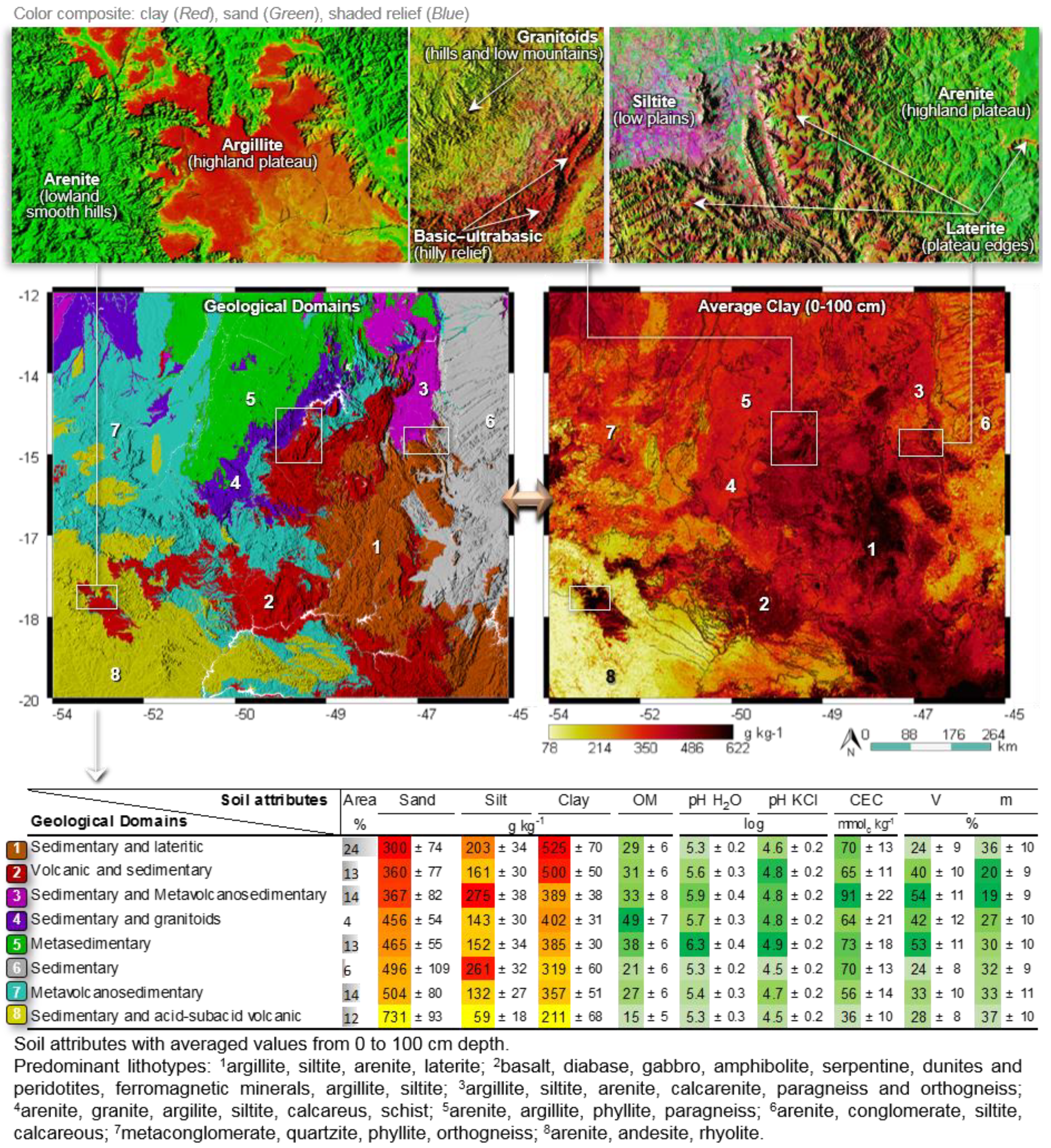
| Covariate | Description | Native Scale or Resolution | Source |
|---|---|---|---|
| Soil, Parent Material and Age | |||
| Synthetic Soil Image (SySI) | Bare soil reflectance covering VNIR-SWIR-TIR range (7 bands) | 30 m | Landsat 4, 5, 7 and 8 |
| Geological Lineaments Density | Meters of structural features per km2 | 1:1,000,000 a | CPRM |
| Organisms | |||
| Synthetic Vegetation Image of dry season (SyVId) | Potential natural vegetation reflectance from November to March covering VNIR-SWIR-TIR range (7 bands) | 30 m | Landsat 4 and 5 |
| Synthetic Vegetation Image of wet season (SyVIw) | Potential natural vegetation reflectance from May to September covering VNIR-SWIR-TIR range (7 bands) | 30 m | Landsat 4 and 5 |
| Climate | |||
| Annual Precipitation (mm) | Bioclimatic variables obtained from the monthly temperature and rainfall in order to generate more biologically meaningful values. | 1 km b | WorldClim |
| Precipitation Seasonality (CV) | 1 km b | WorldClim | |
| Annual Mean Temperature (°C) | 1 km b | WorldClim | |
| Temperature Annual Range (°C) | 1 km b | WorldClim | |
| Temperature Seasonality (°C) | 1 km b | WorldClim | |
| Relief and Age | |||
| Elevation (m) | Height of terrain above sea level | 30 m | ALOS |
| Slope (degree) | Slope gradient | 30 m | ALOS |
| Aspect (degree) | Compass direction | 30 m | ALOS |
| Topographic Position Index (m) | Distinguishes ridge from valley forms | 30 m | ALOS |
| Horizontal Curvature (m) | Curvature tangent to the contour line | 30 m | ALOS |
| Vertical Curvature (m) | Curvature tangent to the slope line | 30 m | ALOS |
| Soil Attribute | Depth (cm) | n | Min. | Q1 | Mean | Median | Q3 | Max. | Sd | IQR | Skew. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clay (g kg−1) | 0–20 | 7930 | 10 | 87 | 271 | 176 | 450 | 920 | 221 | 363 | 0.8 |
| 20–60 | 6908 | 10 | 100 | 287 | 176 | 482 | 930 | 233 | 382 | 0.8 | |
| 60–100 | 7520 | 12 | 125 | 314 | 225 | 500 | 950 | 231 | 375 | 0.8 | |
| Silt (g kg−1) | 0–20 | 7930 | 1 | 24 | 77 | 38 | 94 | 816 | 89 | 70 | 2.3 |
| 20–60 | 6907 | 1 | 24 | 71 | 37 | 82 | 760 | 79 | 58 | 2.3 | |
| 60–100 | 7520 | 1 | 24 | 67 | 37 | 80 | 794 | 75 | 56 | 2.6 | |
| Sand (g kg−1) | 0–20 | 7930 | 1 | 409 | 652 | 783 | 883 | 975 | 280 | 474 | -0.8 |
| 20–60 | 6907 | 1 | 393 | 643 | 783 | 873 | 973 | 284 | 480 | -0.8 | |
| 60–100 | 7520 | 1 | 377 | 619 | 741 | 848 | 967 | 276 | 471 | -0.7 | |
| Organic Matter (g kg−1) | 0–20 | 7242 | 0 | 11 | 21 | 17 | 28 | 393 | 14 | 17 | 4.8 |
| 20–60 | 6021 | 0 | 7 | 13 | 11 | 17 | 412 | 9 | 10 | 15.2 | |
| 60–100 | 6808 | 0 | 4 | 9 | 8 | 12 | 98 | 6 | 7 | 2.3 | |
| pH H2O (log) | 0–20 | 6200 | 3.7 | 5.2 | 5.6 | 5.6 | 6.0 | 8.2 | 0.6 | 0.8 | 0.1 |
| 20–60 | 5149 | 3.8 | 4.9 | 5.3 | 5.2 | 5.6 | 9.0 | 0.6 | 0.7 | 0.7 | |
| 60–100 | 7511 | 3.8 | 4.9 | 5.3 | 5.2 | 5.6 | 9.1 | 0.5 | 0.7 | 0.7 | |
| pH KCl (log) | 0–20 | 5596 | 3.1 | 4.6 | 4.9 | 4.8 | 5.3 | 7.7 | 0.6 | 1.0 | 0.6 |
| 20–60 | 4707 | 0.4 | 4.2 | 4.6 | 4.4 | 4.9 | 7.7 | 0.5 | 0.7 | 1.1 | |
| 60–100 | 7384 | 3.5 | 4.3 | 4.8 | 4.5 | 5.2 | 7.5 | 0.6 | 0.9 | 0.9 | |
| CEC (mmolc kg−1) | 0–20 | 8010 | 2 | 32 | 53 | 45 | 68 | 641 | 33 | 37 | 3.0 |
| 20–60 | 6852 | 2 | 22 | 36 | 32 | 45 | 696 | 23 | 23 | 5.7 | |
| 60–100 | 7655 | 1 | 16 | 26 | 22 | 32 | 582 | 18 | 16 | 6.2 | |
| Base Saturation (V%) | 0–20 | 8018 | 0 | 24 | 42 | 42 | 58 | 100 | 22 | 34 | 0.2 |
| 20–60 | 6860 | 0 | 12 | 25 | 21 | 34 | 100 | 18 | 23 | 1.2 | |
| 60–100 | 7655 | 0 | 10 | 23 | 18 | 31 | 100 | 17 | 20 | 1.5 | |
| Aluminum Saturation (m%) | 0–20 | 7964 | 0 | 0 | 16 | 4 | 24 | 100 | 23 | 24 | 1.6 |
| 20–60 | 6841 | 0 | 5 | 33 | 28 | 57 | 100 | 29 | 52 | 0.4 | |
| 60–100 | 7635 | 0 | 3 | 36 | 34 | 62 | 100 | 30 | 59 | 0.3 |
| Soil Attribute | Depth (cm) | mTry | minNS | RMSEcal | RPIQcal | R2cal | RMSE10cv | RPIQ10cv | R210cv |
|---|---|---|---|---|---|---|---|---|---|
| Clay (g kg−1) | 0–20 | 24 | 5 | 39 | 9.4 | 0.97 | 96 | 3.8 | 0.81 |
| 20–60 | 24 | 5 | 38 | 10.0 | 0.97 | 96 | 4.0 | 0.83 | |
| 60–100 | 24 | 5 | 38 | 9.9 | 0.97 | 95 | 4.0 | 0.83 | |
| Silt (g kg−1) | 0–20 | 24 | 5 | 21 | 3.3 | 0.94 | 53 | 1.3 | 0.64 |
| 20–60 | 33 | 5 | 18 | 3.2 | 0.95 | 46 | 1.3 | 0.66 | |
| 60–100 | 24 | 5 | 18 | 3.1 | 0.94 | 45 | 1.3 | 0.64 | |
| Sand (g kg−1) | 0–20 | 33 | 5 | 47 | 10.1 | 0.97 | 118 | 4.0 | 0.82 |
| 20–60 | 24 | 5 | 45 | 10.7 | 0.98 | 111 | 4.3 | 0.85 | |
| 60–100 | 24 | 5 | 44 | 10.6 | 0.97 | 110 | 4.3 | 0.84 | |
| Organic Matter (g kg−1) | 0–20 | 33 | 5 | 4 | 4.1 | 0.91 | 10 | 1.7 | 0.49 |
| 20–60 | 33 | 5 | 3 | 3.4 | 0.90 | 8 | 1.3 | 0.30 | |
| 60–100 | 24 | 5 | 2 | 4.3 | 0.92 | 4 | 1.8 | 0.53 | |
| pH H2O (log) | 0–20 | 33 | 5 | 0.21 | 3.7 | 0.88 | 0.54 | 1.5 | 0.21 |
| 20–60 | 33 | 5 | 0.19 | 3.9 | 0.89 | 0.47 | 1.6 | 0.32 | |
| 60–100 | 33 | 5 | 0.18 | 3.9 | 0.90 | 0.44 | 1.6 | 0.35 | |
| pH KCl (log) | 0–20 | 33 | 5 | 0.23 | 4.2 | 0.87 | 0.57 | 1.7 | 0.19 |
| 20–60 | 33 | 5 | 0.16 | 4.3 | 0.91 | 0.40 | 1.8 | 0.44 | |
| 60–100 | 24 | 5 | 0.15 | 5.9 | 0.94 | 0.38 | 2.4 | 0.64 | |
| CEC (mmolc kg−1) | 0–20 | 33 | 5 | 10 | 3.7 | 0.91 | 23 | 1.6 | 0.48 |
| 20–60 | 24 | 5 | 8 | 3.0 | 0.89 | 18 | 1.3 | 0.40 | |
| 60–100 | 24 | 5 | 6 | 2.7 | 0.89 | 14 | 1.2 | 0.40 | |
| Base Saturation (V%) | 0–20 | 33 | 5 | 8 | 4.4 | 0.87 | 20 | 1.7 | 0.18 |
| 20–60 | 33 | 5 | 6 | 3.7 | 0.89 | 15 | 1.5 | 0.30 | |
| 60–100 | 33 | 5 | 6 | 3.6 | 0.89 | 14 | 1.5 | 0.36 | |
| Aluminum Saturation (m%) | 0–20 | 33 | 5 | 8 | 2.9 | 0.88 | 20 | 1.2 | 0.26 |
| 20–60 | 33 | 5 | 9 | 6.1 | 0.91 | 21 | 2.4 | 0.45 | |
| 60–100 | 24 | 5 | 8 | 7.4 | 0.93 | 20 | 3.0 | 0.56 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poppiel, R.R.; Lacerda, M.P.C.; Safanelli, J.L.; Rizzo, R.; Oliveira, M.P., Jr.; Novais, J.J.; Demattê, J.A.M. Mapping at 30 m Resolution of Soil Attributes at Multiple Depths in Midwest Brazil. Remote Sens. 2019, 11, 2905. https://doi.org/10.3390/rs11242905
Poppiel RR, Lacerda MPC, Safanelli JL, Rizzo R, Oliveira MP Jr., Novais JJ, Demattê JAM. Mapping at 30 m Resolution of Soil Attributes at Multiple Depths in Midwest Brazil. Remote Sensing. 2019; 11(24):2905. https://doi.org/10.3390/rs11242905
Chicago/Turabian StylePoppiel, Raúl R., Marilusa P. C. Lacerda, José L. Safanelli, Rodnei Rizzo, Manuel P. Oliveira, Jr., Jean J. Novais, and José A. M. Demattê. 2019. "Mapping at 30 m Resolution of Soil Attributes at Multiple Depths in Midwest Brazil" Remote Sensing 11, no. 24: 2905. https://doi.org/10.3390/rs11242905
APA StylePoppiel, R. R., Lacerda, M. P. C., Safanelli, J. L., Rizzo, R., Oliveira, M. P., Jr., Novais, J. J., & Demattê, J. A. M. (2019). Mapping at 30 m Resolution of Soil Attributes at Multiple Depths in Midwest Brazil. Remote Sensing, 11(24), 2905. https://doi.org/10.3390/rs11242905







