Weighted Double-Logistic Function Fitting Method for Reconstructing the High-Quality Sentinel-2 NDVI Time Series Data Set
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Set
2.2. Study Area
2.3. Methods
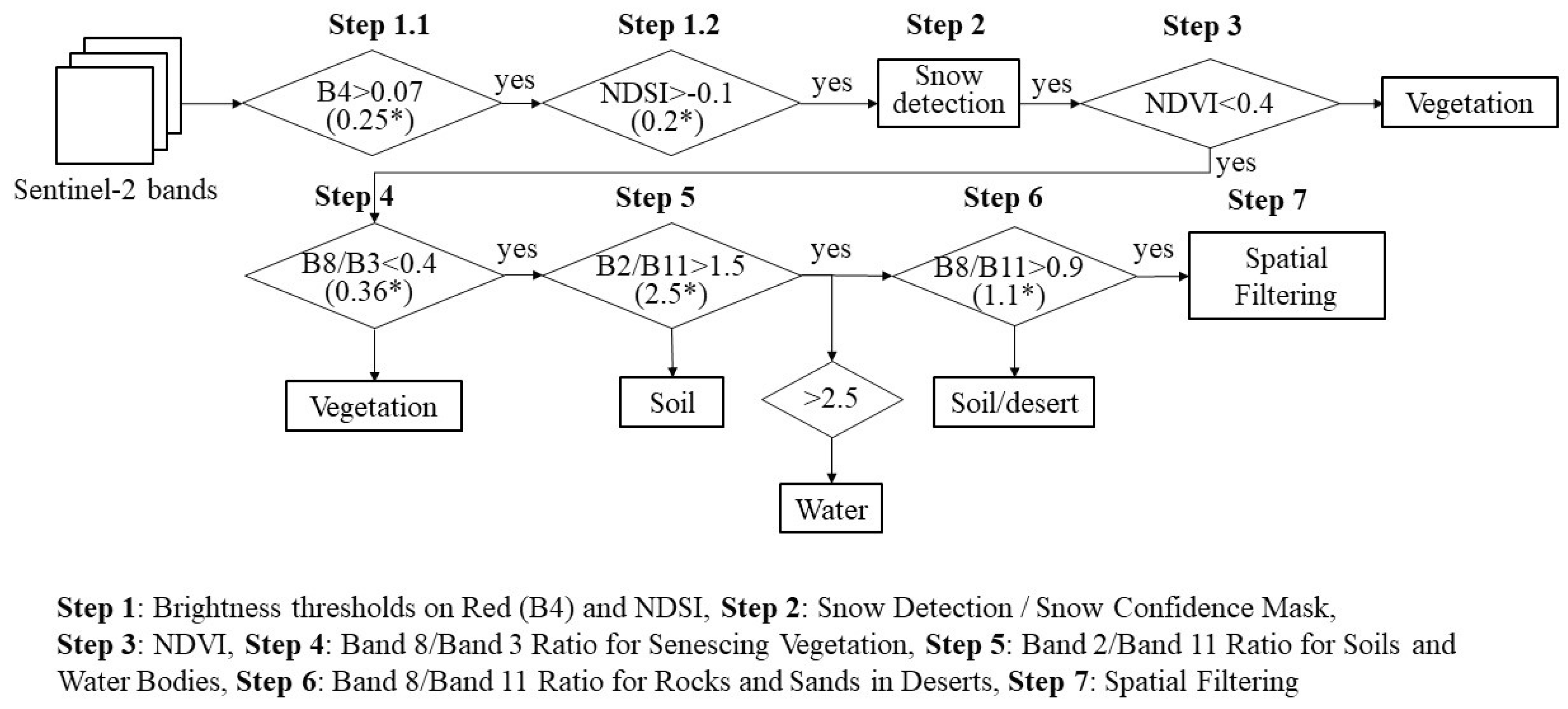
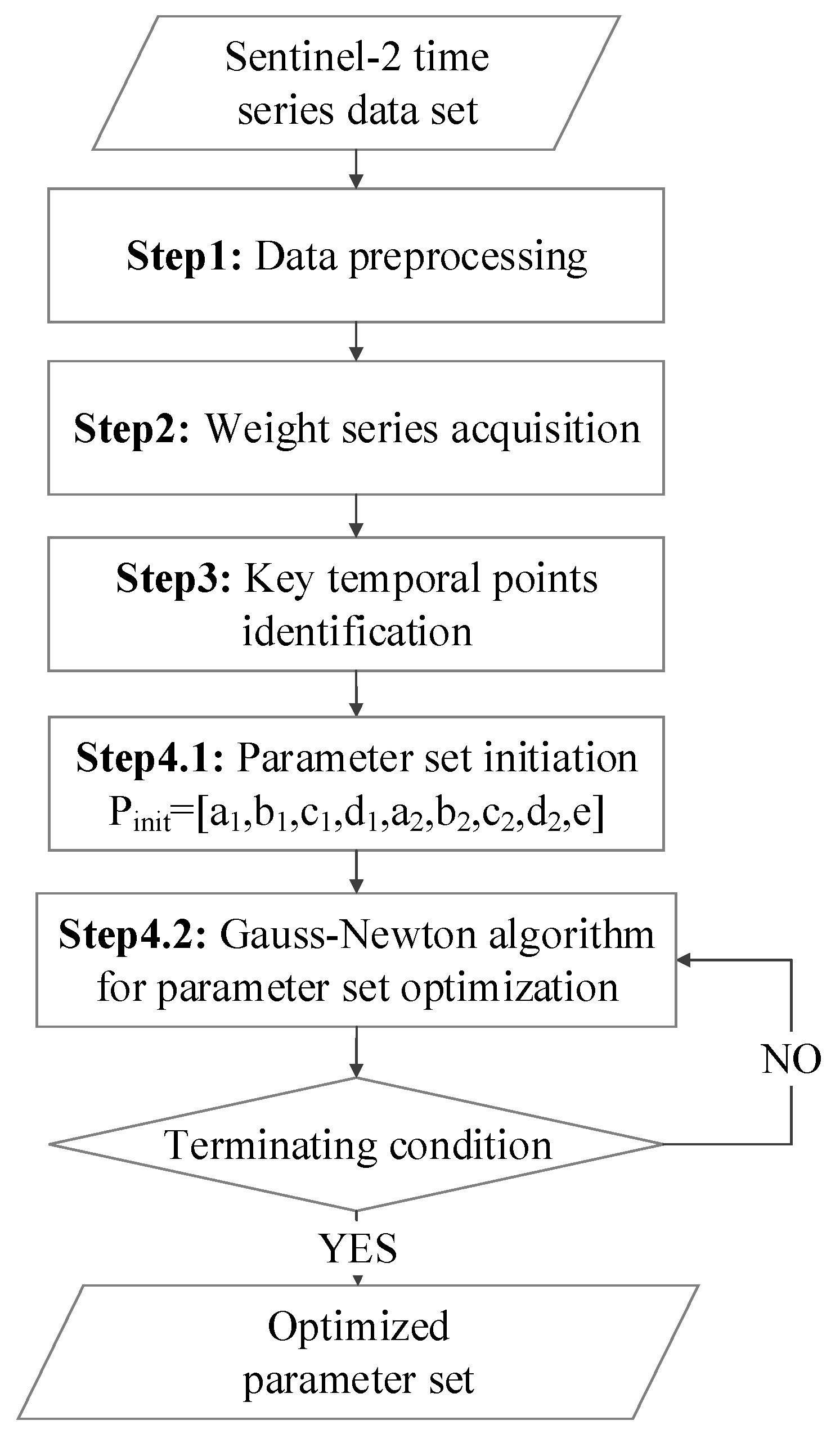
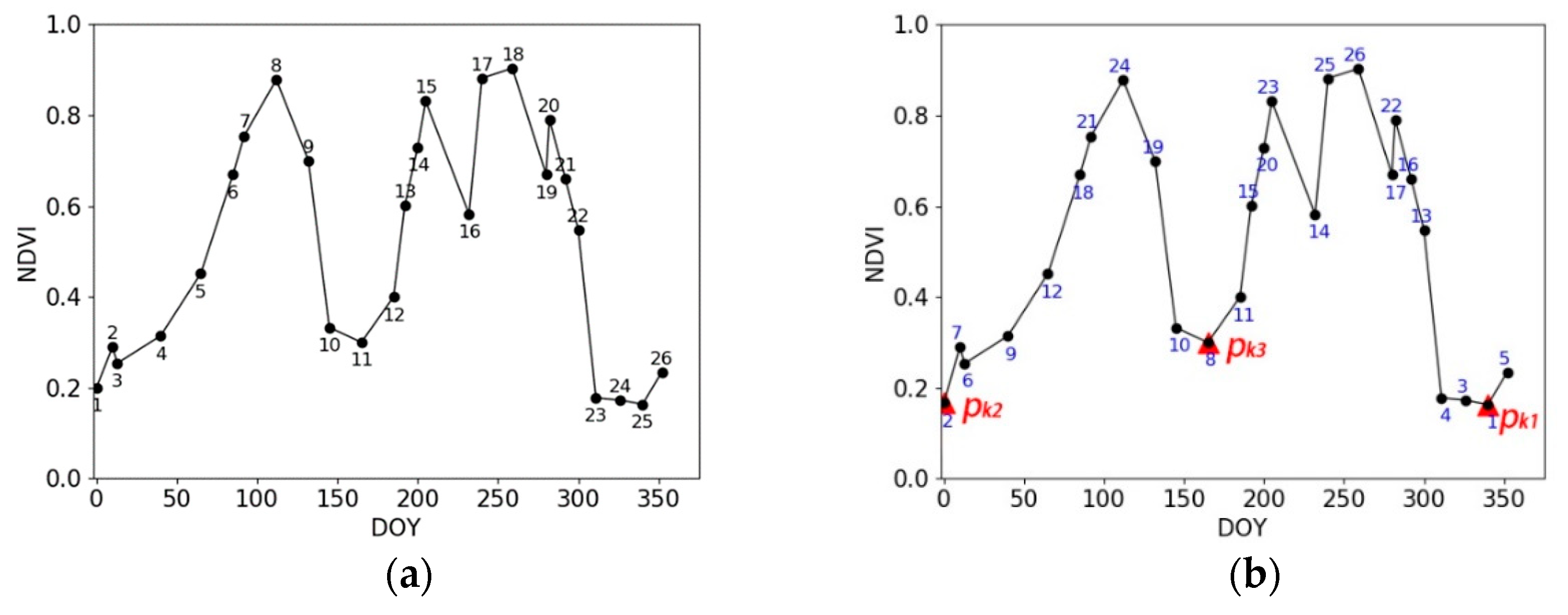
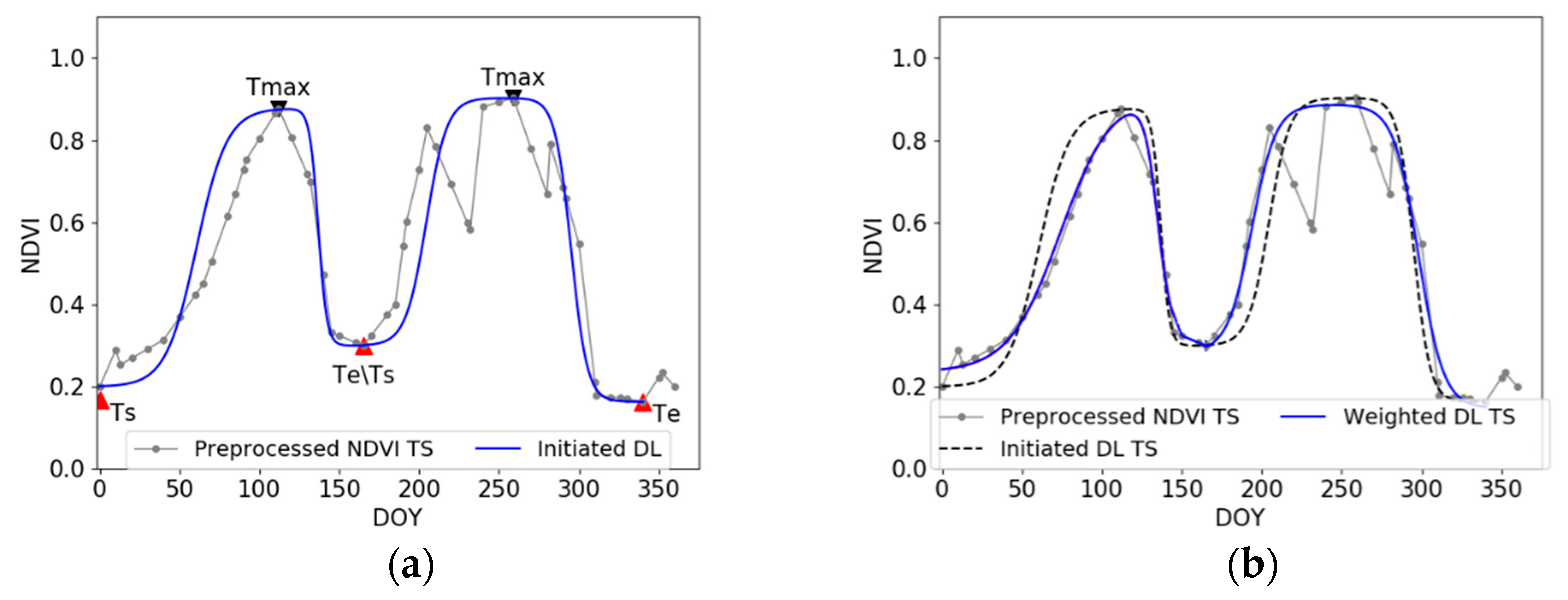
2.3.1. Overview of the proposed method
2.3.2. Description of the Proposed Method
3. Results
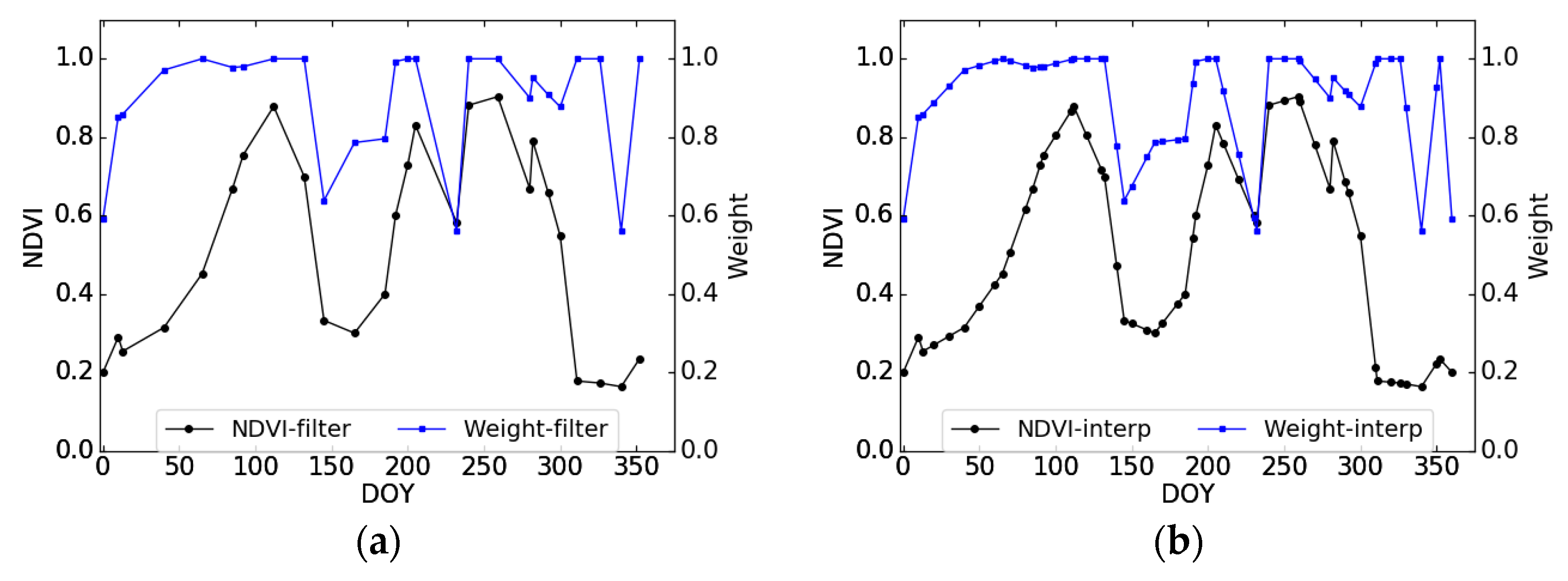
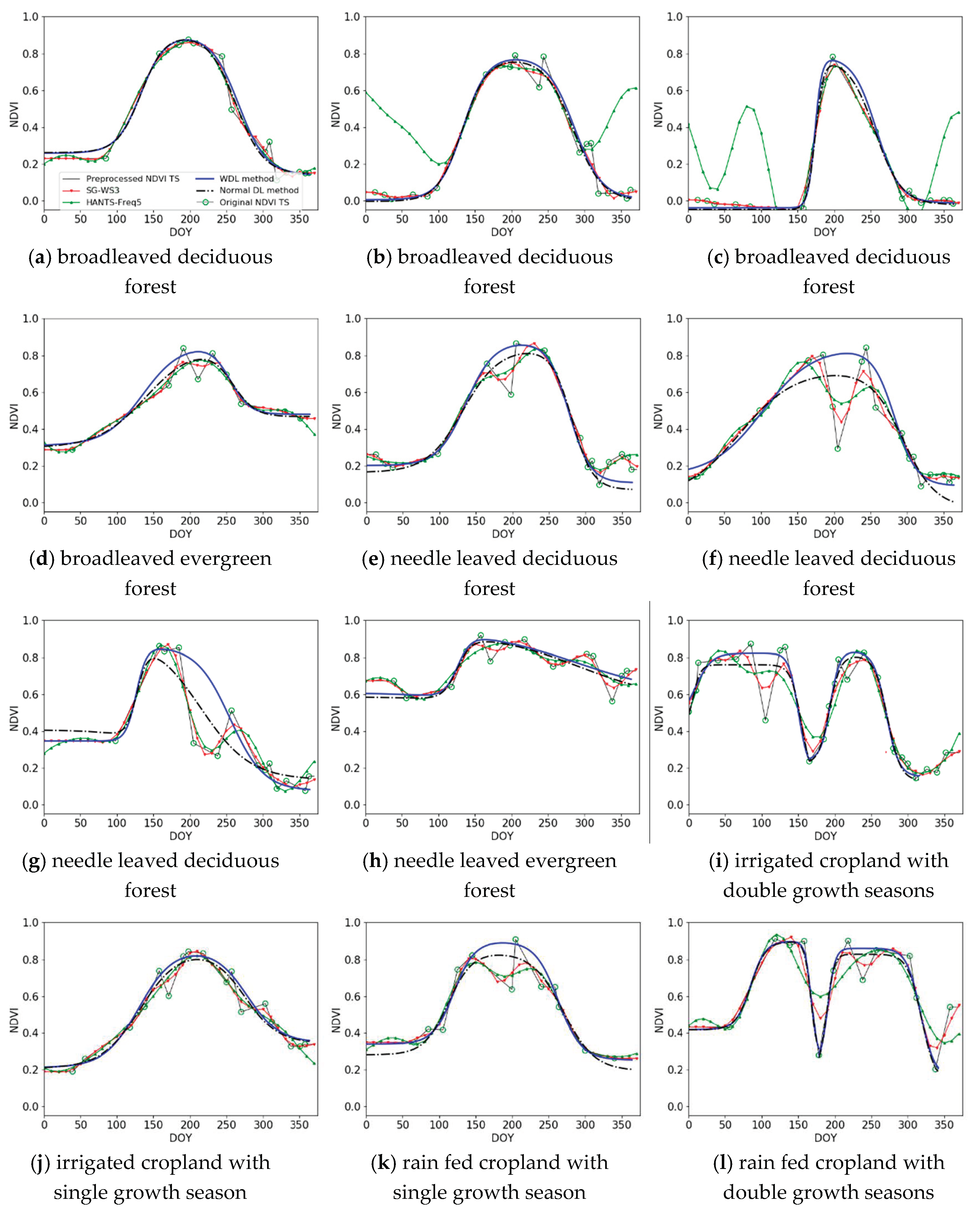
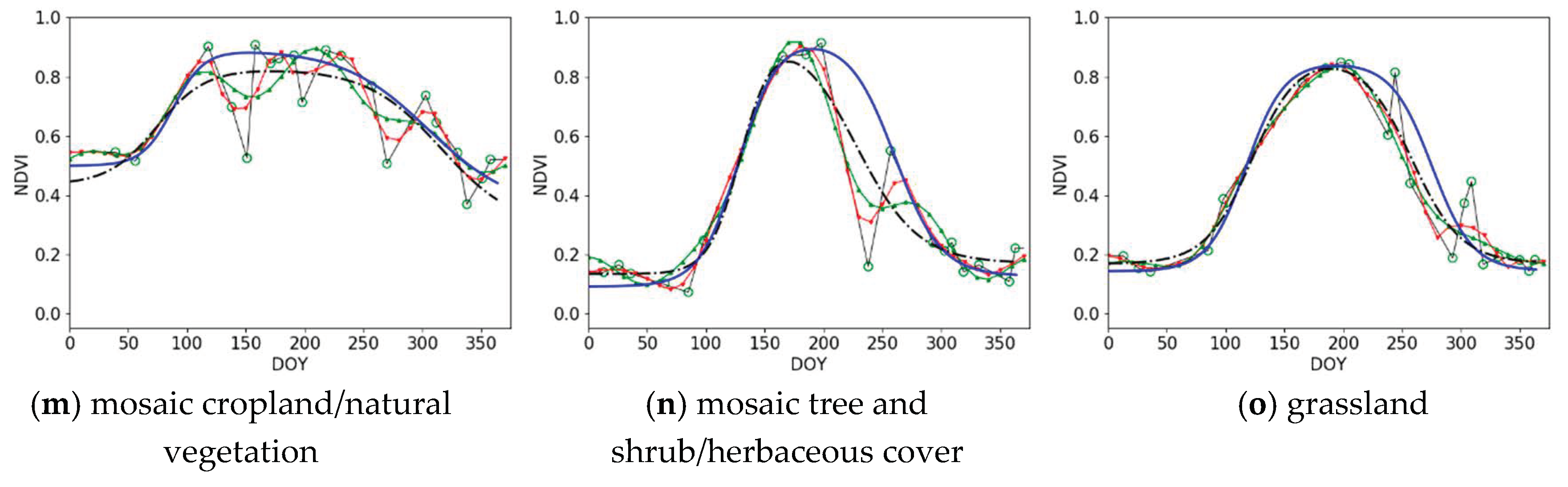
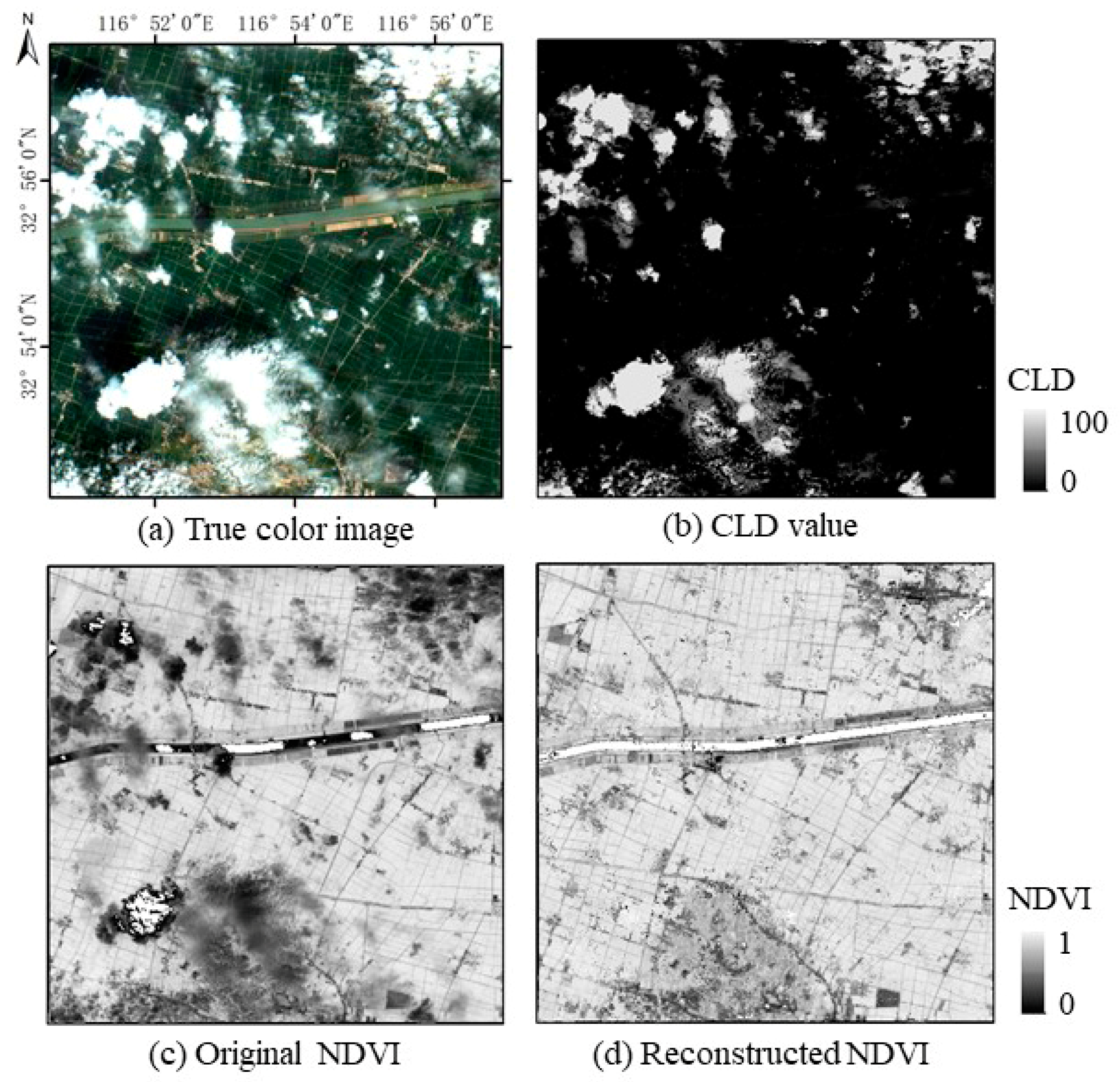
3.1. Qualitative Assessment
3.2. Quantitative Assessment
3.3. Reconstruction of High-Quality NDVI Time Series
4. Discussion
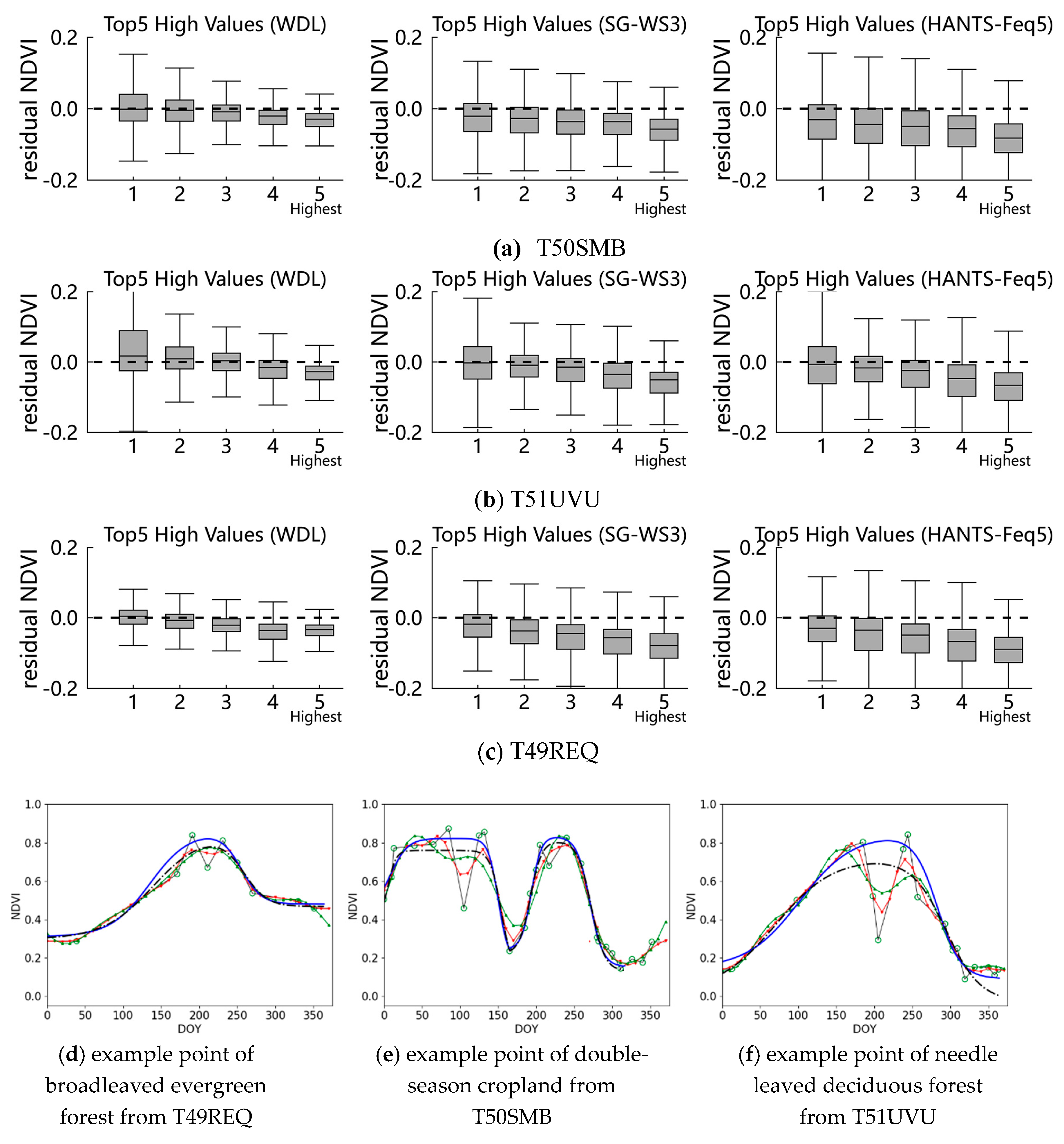
4.1. The Fidelities of High Data Points
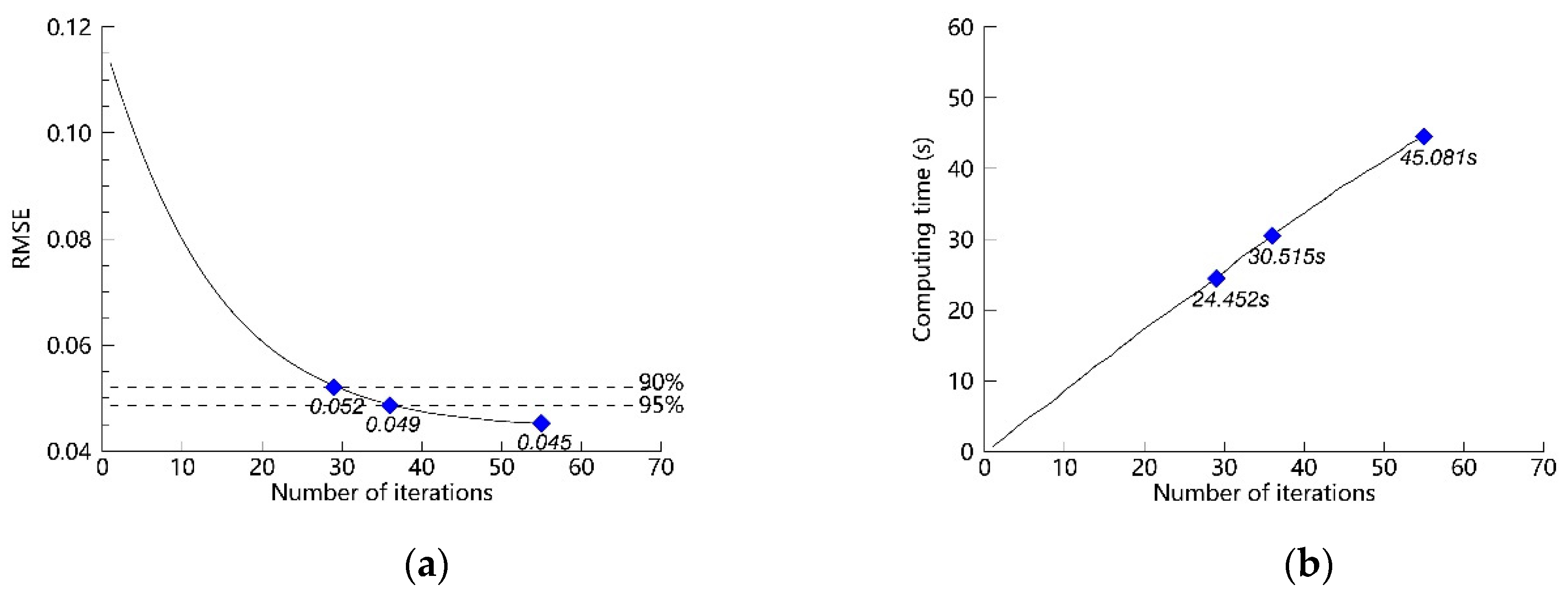
4.2. Discussion of the Number of Iterations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Atzberger, C.; Eilers, P.C. A time series for monitoring vegetation activity and phenology at 10-daily time steps covering large parts of South America. Int. J. Digit. Earth 2011, 4, 365–386. [Google Scholar] [CrossRef]
- Beck, P.S.A.; Atzberger, C.; Høgda, K.A.; Johansen, B.; Skidmore, A.K. Improved monitoring of vegetation dynamics at very high latitudes: A new method using MODIS NDVI. Remote Sens. Environ. 2006, 99, 321–334. [Google Scholar] [CrossRef]
- Tucker, C.J.; Eldin, J.H., Jr.; Iii, M.M.; Fan, C.J. Monitoring corn and soybean crop development with hand-held radiometer spectral data. Remote Sens. Environ. 1979, 8, 237–248. [Google Scholar] [CrossRef]
- Guyon, D.; Guillot, M.; Vitasse, Y.; Cardot, H.; Hagolle, O.; Delzon, S.; Wigneron, J.P. Monitoring elevation variations in leaf phenology of deciduous broadleaf forests from SPOT/VEGETATION time-series. Remote Sens. Environ. 2011, 115, 615–627. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Leeuwen, W.J.D.V.; Hartfield, K.; Miranda, M.; Meza, F.J. Trends and ENSO/AAO Driven Variability in NDVI Derived Productivity and Phenology alongside the Andes Mountains. Remote Sens. 2013, 5, 1177–1203. [Google Scholar] [CrossRef]
- Zeng, H.; Jia, G.; Forbes, B.C. Shifts in Arctic phenology in response to climate and anthropogenic factors as detected from multiple satellite time series. Environ. Res. Lett. 2013, 8, 035036. [Google Scholar] [CrossRef]
- Xiao, X.; Boles, S.; Frolking, S.; Li, C.; Babu, J.Y.; Salas, W.; Iii, B.M. Mapping paddy rice agriculture in South and Southeast Asia using multi-temporal MODIS images. Remote Sens. Environ. 2006, 100, 95–113. [Google Scholar] [CrossRef]
- Chen, C.F.; Huang, S.W.; Son, N.T.; Chang, L.Y. Mapping double-cropped irrigated rice fields in Taiwan using time-series Satellite Pour I’Observation de la Terre data. J. Appl. Remote Sens. 2011, 5, 3528. [Google Scholar] [CrossRef]
- Patakamuri, S.K. Time-Series analysis of MODIS NDVI data along with ancillary data for Land use/Land cover mapping of Uttarakhand. ISPRS-Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, XL-8, 1491–1500. [Google Scholar] [CrossRef]
- Defries, R.S.; Townshend, J.R.G. NDVI-derived land cover classifications at a global scale. Int. J. Remote Sens. 1994, 15, 3567–3586. [Google Scholar] [CrossRef]
- Arvor, D.; Jonathan, M.; Dubreuil, V.; Durieux, L. Classification of MODIS EVI time series for crop mapping in the state of Mato Grosso, Brazil. Int. J. Remote Sens. 2011, 32, 7847–7871. [Google Scholar] [CrossRef]
- Rodrigues, A.; Marcal, A.R.S.; Furlan, D.; Ballester, M.V.; Cunha, M. Land cover map production for Brazilian Amazon using NDVI SPOT VEGETATION time series. Can. J. Remote Sens. 2013, 39, 277–289. [Google Scholar] [CrossRef]
- Sellers, P.J.; Tucker, C.J.; Collatz, G.J.; Los, S.O.; Justice, C.O.; Dazlich, D.A.; Randall, D.A. A global 1° by 1° NDVI data set for climate studies. Part 2: The generation of global fields of terrestrial biophysical parameters from the NDVI. Int. J. Remote Sens. 1994, 15, 3519–3545. [Google Scholar] [CrossRef]
- Möller, M.; Gerstmann, H.; Dahms, T. Phenological NDVI time series for the dynamic derivation of soil coverage information. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Fort Worth, TX, USA, 23–28 July 2017; pp. 4278–4281. [Google Scholar]
- Hird, J.N.; Mcdermid, G.J. Noise reduction of NDVI time series: An empirical comparison of selected techniques. Remote Sens. Environ. 2009, 113, 248–258. [Google Scholar] [CrossRef]
- Chen, J.; Jönsson, P.; Tamura, M. A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky–Golay filter. Remote Sens. Environ. 2004, 91, 332–344. [Google Scholar] [CrossRef]
- Julien, Y.; Sobrino, J.A. Global land surface phenology trends from GIMMS database. Int. J. Remote Sens. 2009, 30, 3495–3513. [Google Scholar] [CrossRef]
- Zhou, J.; Jia, L.; Menenti, M.; Gorte, B. On the performance of remote sensing time series reconstruction methods—A spatial comparison. Remote Sens. Environ. 2016, 187, 367–384. [Google Scholar] [CrossRef]
- Holben, B. Characteristics of maximum-value composite images from temporal AVHRR data. Int. J. Remote Sens. 1986, 7, 1417–1434. [Google Scholar] [CrossRef]
- Yang, G.; Shen, H.; Zhang, L.; He, Z.; Li, X. A Moving Weighted Harmonic Analysis Method for Reconstructing High-Quality SPOT VEGETATION NDVI Time-Series Data. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6008–6021. [Google Scholar] [CrossRef]
- Viovy, N.; Arino, O.; Belward, A.S. The Best Index Slope Extraction ( BISE): A method for reducing noise in NDVI time-series. Int. J. Remote Sens. 1992, 13, 1585–1590. [Google Scholar] [CrossRef]
- Lovell, J.L.; Graetz, R.D. Filtering Pathfinder AVHRR Land NDVI data for Australia. Int. J. Remote Sens. 2001, 22, 2649–2654. [Google Scholar] [CrossRef]
- Ma, M.; Veroustraete, F. Reconstructing pathfinder AVHRR land NDVI time-series data for the Northwest of China. Adv. Space Res. 2006, 37, 835–840. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Jonsson, P.; Eklundh, L. Seasonality extraction by function fitting to time-series of satellite sensor data. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- Fisher, J.I.; Mustard, J.F.; Vadeboncoeur, M.A. Green leaf phenology at Landsat resolution: Scaling from the field to the satellite. Remote Sens. Environ. 2006, 100, 265–279. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Hermance, J.F.; Jacob, R.W.; Bradley, B.A.; Mustard, J.F. Extracting Phenological Signals From Multiyear AVHRR NDVI Time Series: Framework for Applying High-Order Annual Splines With Roughness Damping. IEEE Trans. Geosci. Remote Sens. 2007, 45, 3264–3276. [Google Scholar] [CrossRef]
- Geerken, R.; Zaitchik, B.; Evans, J.P. Classifying rangeland vegetation type and coverage from NDVI time series using Fourier Filtered Cycle Similarity. Int. J. Remote Sens. 2005, 26, 5535–5554. [Google Scholar] [CrossRef]
- Verhoef, W. Application of harmonic analysis of NDVI time series (HANTS). Fourier Anal. Temporal NDVI South. Afr. Am. Cont. 1966, 108, 19–24. [Google Scholar]
- Evans, J.P.; Geerken, R. Classifying rangeland vegetation type and coverage using a Fourier component based similarity measure. Remote Sens. Environ. 2006, 105, 1–8. [Google Scholar] [CrossRef]
- Crist, E.P.; Malila, W.A. A temporal-spectral analysis technique for vegetation applications of Landsat. In Proceedings of the International Symposium on Remote Sensing of Environment, San Jose, CA, USA, 23–30 April 1980. [Google Scholar]
- Fischer, A.; Kergoat, L.; Dedieu, G. Coupling satellite data with vegetation functional models: Review of different approaches and perspectives suggested by the assimilation strategy. Remote Sens. Rev. 1997, 15, 283–303. [Google Scholar] [CrossRef]
- Available online: https://earth.esa.int/web/sentinel/technical-guides/sentinel-2-msi/level-2a/algorithm (accessed on 6 October 2019).
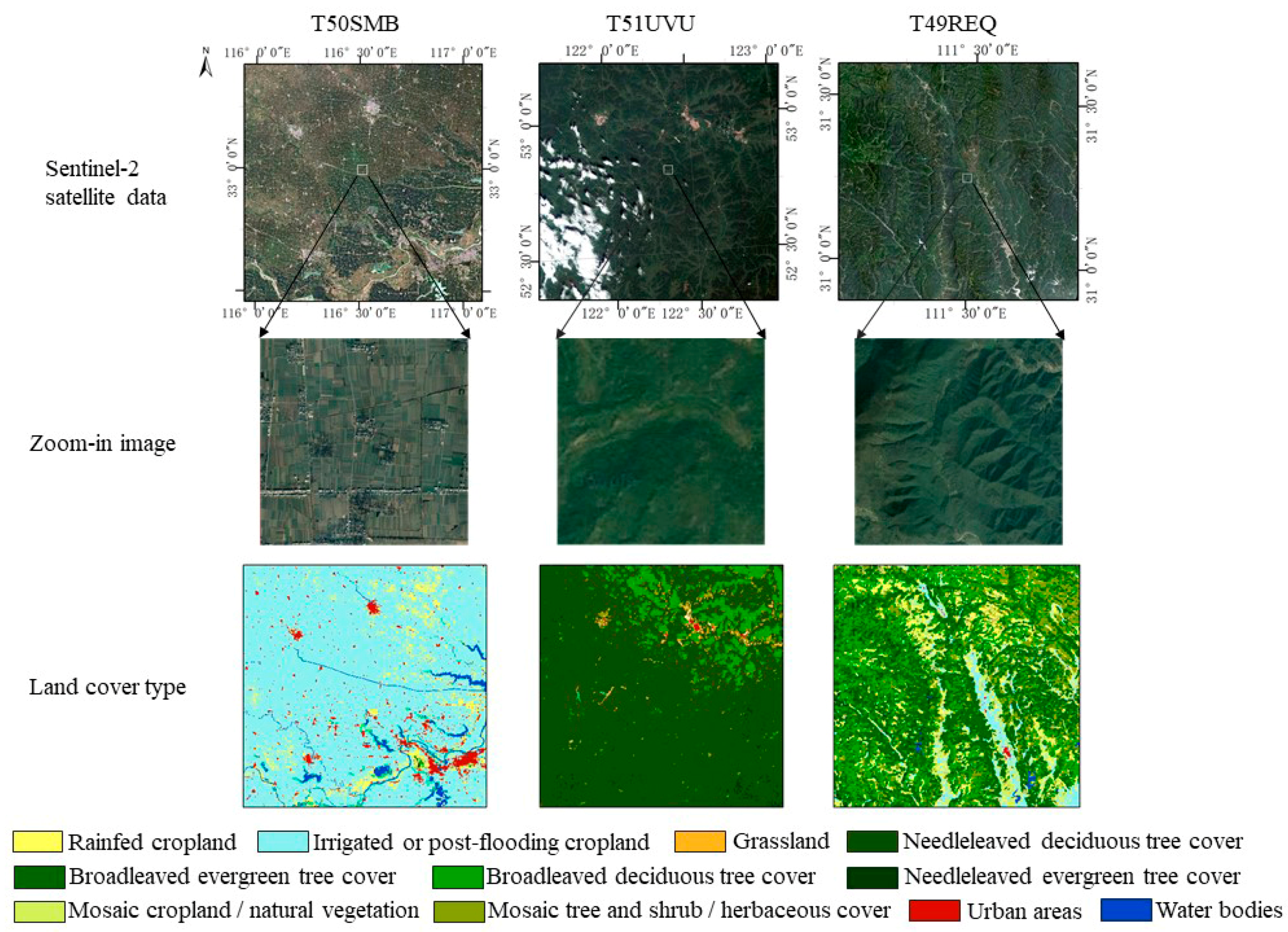

| Land Cover Type | T50SMB | T51UVU | T49REQ |
|---|---|---|---|
| Rain fed cropland | 223 | 1 | 294 |
| Irrigated cropland | 1676 | 415 | |
| Mosaic cropland/natural vegetation | 192 | ||
| Broadleaved evergreen forest | 651 | ||
| Broadleaved deciduous forest | 228 | 214 | |
| Needle leaved evergreen forest | 39 | 4 | |
| Needle leaved deciduous forest | 1701 | ||
| Mosaic tree and shrub/herbaceous cover | 18 | 145 | |
| Grassland | 8 | 3 |
| Study Area | Method | Levels of Noise | ||
|---|---|---|---|---|
| Low | Medium | High | ||
| T50SMB | WDL | 0.046 | 0.069 | 0.089 |
| S-G | 0.051 | 0.088 | 0.115 | |
| HANTS | 0.058 | 0.088 | 0.114 | |
| T51UVU | WDL | 0.039 | 0.061 | 0.083 |
| S-G | 0.043 | 0.081 | 0.112 | |
| HANTS | 0.055 | 0.084 | 0.119 | |
| T49REQ | WDL | 0.049 | 0.074 | 0.098 |
| S-G | 0.053 | 0.095 | 0.129 | |
| HANTS | 0.053 | 0.093 | 0.126 | |
| Study Area | Method | RMSE for Top5 High Points | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 a | ||
| T50SMB | WDL | 0.053 | 0.044 | 0.033 | 0.037 | 0.040 |
| S-G | 0.059 | 0.060 | 0.059 | 0.059 | 0.073 | |
| HANTS | 0.077 | 0.084 | 0.086 | 0.088 | 0.101 | |
| T51UVU | WDL | 0.093 | 0.046 | 0.034 | 0.041 | 0.041 |
| S-G | 0.073 | 0.049 | 0.050 | 0.064 | 0.073 | |
| HANTS | 0.081 | 0.059 | 0.067 | 0.082 | 0.089 | |
| T49REQ | WDL | 0.028 | 0.030 | 0.033 | 0.047 | 0.042 |
| S-G | 0.052 | 0.061 | 0.074 | 0.082 | 0.094 | |
| HANTS | 0.062 | 0.078 | 0.087 | 0.099 | 0.107 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Luo, J.; Huang, Q.; Wu, W.; Sun, Y. Weighted Double-Logistic Function Fitting Method for Reconstructing the High-Quality Sentinel-2 NDVI Time Series Data Set. Remote Sens. 2019, 11, 2342. https://doi.org/10.3390/rs11202342
Yang Y, Luo J, Huang Q, Wu W, Sun Y. Weighted Double-Logistic Function Fitting Method for Reconstructing the High-Quality Sentinel-2 NDVI Time Series Data Set. Remote Sensing. 2019; 11(20):2342. https://doi.org/10.3390/rs11202342
Chicago/Turabian StyleYang, Yingpin, Jiancheng Luo, Qiting Huang, Wei Wu, and Yingwei Sun. 2019. "Weighted Double-Logistic Function Fitting Method for Reconstructing the High-Quality Sentinel-2 NDVI Time Series Data Set" Remote Sensing 11, no. 20: 2342. https://doi.org/10.3390/rs11202342
APA StyleYang, Y., Luo, J., Huang, Q., Wu, W., & Sun, Y. (2019). Weighted Double-Logistic Function Fitting Method for Reconstructing the High-Quality Sentinel-2 NDVI Time Series Data Set. Remote Sensing, 11(20), 2342. https://doi.org/10.3390/rs11202342






