1. Introduction
The impact of climate change, specifically global warming, has intensified the challenges faced by precision farming. Major crops like rice, wheat, and maize, which collectively account for 50% of global caloric intake and 20% of protein, are highly inclined to environmental stresses such as heat, drought, and salinity [
1]. In recent decades, changes in climatic patterns have led to a significant decrease in crop yield, raising issues about food security. Studies indicate that climate change could reduce global food output by 3–12% by the mid-21st century, with more severe growth and reduction to 25% projected by the end of the century if current warming trends carry on [
2,
3]. The crop sensitivity to intense environmental conditions highlights the need for advanced research to develop more stable plants capable of overcoming stress [
4]. In sustainable agriculture, prompt and precise identification of plant stress mitigates pesticide exploitation, decreases fertilizing requirements, and minimizes yield loss. Intelligent stress monitoring systems, such as HARPS, facilitate protective measures that immediately reduce environmental footprints and enhance long-term agricultural sustainability.
Proteins containing SAPs, particularly those with A20 and AN1 domains, are important for improving plant resistance to different abiotic stresses, including cold, drought, heat, and salinity. The AN1 domain, originally identified in
Anopheles gambiae, functions similarly to ubiquitin, supporting the regulation of protein degradation and managing damaged proteins accumulated under stress conditions [
5]. The A20 domain, initially found in the human TNFα-induced protein, limits cell death and regulates inflammatory responses in animals, and in plants, it modulates stress signaling to mitigate extensive cellular damage.
Heat stress significantly affects plant growth, physiology, and yield, especially under the increasing global temperatures driven by climate change. Traditional methods of assessing plant stress rely on manual observation of physiological markers, which are often time-consuming, subjective, and require expert knowledge. In recent years, ML has emerged as a powerful tool in agricultural research, enabling the automatic identification and evaluation of stress responses through analysis of high-dimensional data such as spectral images, gene expression profiles, and physiological measurements [
6,
7].
Several studies have demonstrated the effectiveness of ML algorithms in detecting heat stress symptoms in crops by analyzing morphological, thermal, and chlorophyll fluorescence imaging data, for example, developing a deep learning model using convolutional neural networks (CNNs) to classify tomato plants under heat stress based on RGB and thermal images, achieving high accuracy and robustness. Such approaches enable rapid and non-invasive screening of plant stress levels, providing valuable support for crop breeding and stress management strategies. Integrating ML into plant phenotyping pipelines holds significant promise for advancing precision agriculture and climate-resilient crop development [
8,
9].
SAPs comprised of these domains have been extensively studied in plant species like
Oryza sativa (rice),
Arabidopsis thaliana,
Triticum aestivum (wheat), and
Zea mays (maize), where they significantly increase tolerance to adverse environmental conditions [
10,
11]. By understanding and harnessing the molecular properties of SAPs containing AN1 and A20 domains, researchers aim to develop resilient crops capable of growing among climate change, therefore supporting agricultural sustainability and enhancing global food security [
1].
Considering these challenges, ML techniques, specifically deep learning (DL) and hybrid models, have become reliable methods for analyzing and predicting plant stress responses. ML has significantly transformed our awareness of biological processes by offering novel solutions to challenges in the fields of protein science and stress biology [
12]. These computational techniques are progressively being utilized to study, predict, and design proteins, as well as to examine cellular responses to a wide range of stressors [
13,
14]. ML models are especially good at identifying slight trends in vast biological datasets, which has greatly contributed to progress in areas such as gene identification, regulatory network mapping, and pathway analysis under stress conditions [
15]. These advancements are crucial in biotechnology, agricultural trade, and medicine, enhancing our understanding of biological responses to external stimuli and aiding in new drug discovery. These approaches have demonstrated notable accuracy in interpreting biological data and forecasting plant reactions to various stress conditions, including water scarcity and extreme temperatures. Recent studies have effectively employed ML algorithms, for example, support vector machines (SVMs), to identify and classify stress-tolerant plants, particularly in essential crops like corn and wheat [
16,
17].
In this study, we propose a novel ML-based methodology for identifying and evaluating heat stress in plants. Our hybrid approach integrates key parameters derived from Random Forest (RF), SVM, Decision Tree (DT), and Gradient Boosting (GB) algorithms. The objective is to develop a predictive model leveraging data from multiple plant species possessing known SAP domains (AN1 and A20), enabling accurate forecasting of plant responses to abiotic stress, especially heat stress. By refining key molecular features, such as molecular weight, isoelectric point, and instability index, we aim to classify plant proteins based on their stability under heat-stress conditions. This research holds the potential to provide valuable insights into plant stress biology and contribute to the development of crops with enhanced resilience to climate change. Future directions could involve further optimizing DL models by exploring different weight initialization strategies, activation functions, and learning optimizers to improve classification accuracy.
The main contributions of this manuscript are as follows:
Evaluate the SAP AN1 and A20 domains across various plant species using ML algorithms.
Using the UniProt and SMART databases and a Python-based pipeline, extract different heat stress-related metrics from particular species and genera associated with the zinc finger domain.
Propose a novel optimized ML approach (HARPS) to enhance the performance under heat stress in terms of accuracy and time complexity.
Conduct rigorous simulations to evaluate Precision, Recall and F1-Measure for all existing and proposed algorithms. Further, we also performed ablation analysis to extract valuable insights from the dataset.
3. Proposed HARPS Framework
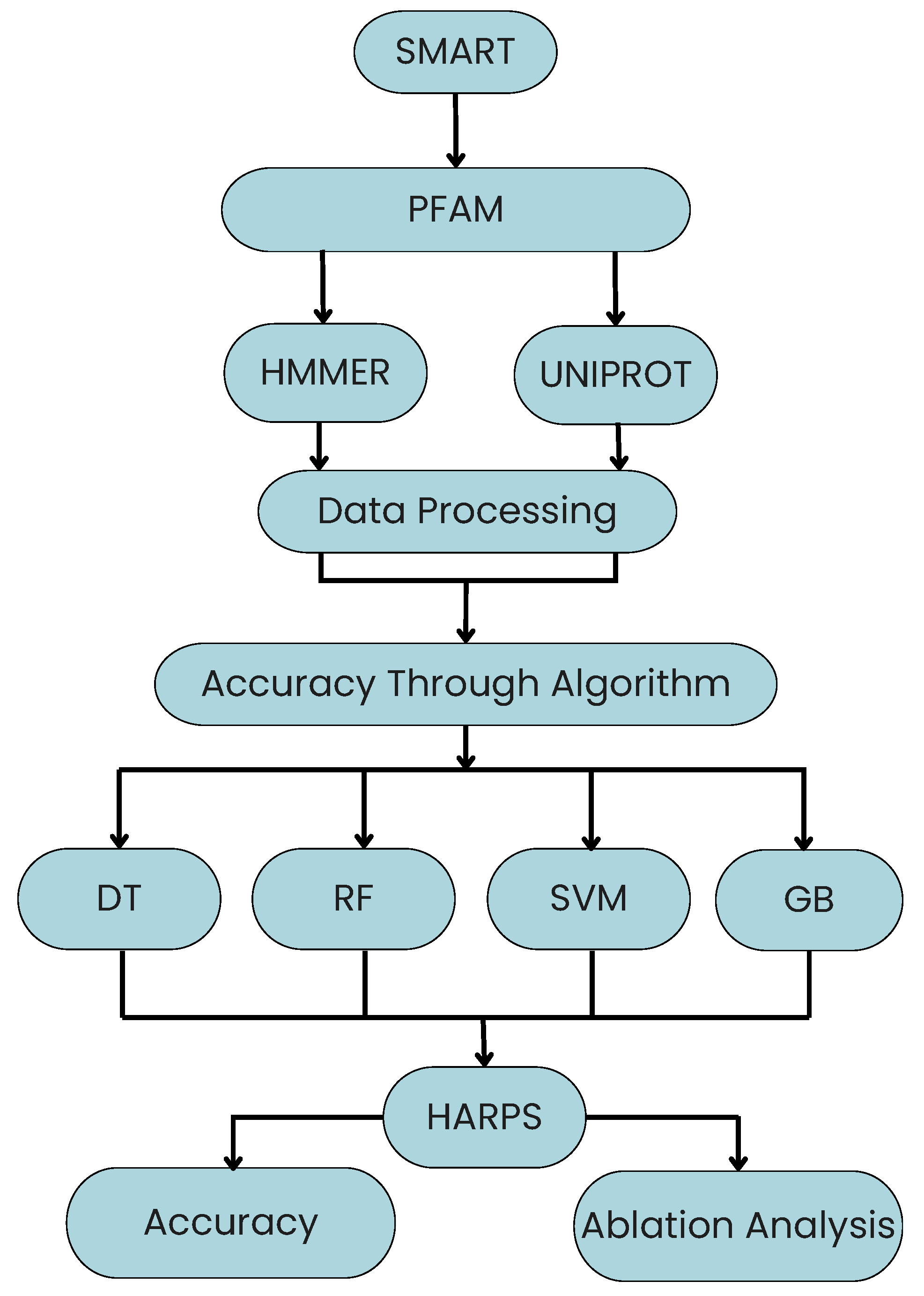
A Hybrid Algorithm for Robust Plant Stress (HARPS) is proposed in this work to enhance the performance in terms of classification accuracy. HARPS is created by extracting the promising features from existing ML algorithms such as DT, RF, SVM, and GB. It utilizes criteria such as splitter, random state, class weight, number of jobs (n_jobs), kernel, subsample, etc.
Table 1 shows the parameters used in HARPS from other algorithms.
To understand the detailed mathematics behind the HARPS framework, let us explore the key components individually. This includes an in-depth look at how the classifiers work, how ensemble methods like majority voting operate mathematically, and the performance metrics that evaluate the success of the hybrid algorithm.
The mathematical formula for the majority voting ensemble method can be expressed as follows:
where
represents the
n samples and
d features.
Each classifier learns its own decision function from the training data. It is assumed we have m classifiers, and each classifier predicts a label for the sample .
Mathematically, a classifier is a function defined as:
After training on the dataset, each classifier provides a prediction for a given sample. For
m classifiers, the set of predictions for a sample
would be:
Each classifier employs a different strategy inside HARPS:
Decision Trees partition the feature space recursively based on conditions (e.g., “feature ”).
Random forests combine multiple decision trees to reduce overfitting and improve accuracy by averaging or voting.
SVMs create a hyperplane in a high-dimensional space that separates different classes.
Gradient boosting iteratively improves weak learners by focusing on correcting the errors made by previous models.
The core of the HARPS framework lies in the ensemble approach. Instead of relying on a single classifier, we combine the predictions of multiple classifiers to make a final decision. The simplest form of ensemble learning is majority voting.
Majority Voting Mathematics
For a given sample
, the predictions from
m classifiers are:
Define as the prediction of the j-th classifier for sample , where .
We calculate the final prediction
by counting how many classifiers predicted each class and selecting the class with the most votes. Mathematically, this is expressed as:
where
is the Kronecker delta function, defined as:
The Kronecker delta function helps in counting how many classifiers predicted the class k. If a classifier predicts the label k, ; otherwise, it is 0. By summing over all classifiers, we count how many predicted each class, and the class with the most votes is selected.
Suppose we have three classifiers, and their predictions for a sample
are:
For each possible class
, the Kronecker delta function counts the number of votes:
Since class 1 received the most votes (2 votes), the final prediction for the sample
is:
Let be the input feature set, and let represent the set of class labels corresponding to Stable, Less Stable, and Not Stable proteins.
Let denote the prediction of the i-th base classifier, where corresponds to:
: DT
: RF
: SVM
: GB
Each base classifier returns a class label
. The final prediction of the HARPS ensemble,
, is obtained by majority voting across all classifiers:
where
is the indicator function defined as:
This formulation ensures that the class with the highest number of votes from the base classifiers is selected as the final output of the HARPS model.
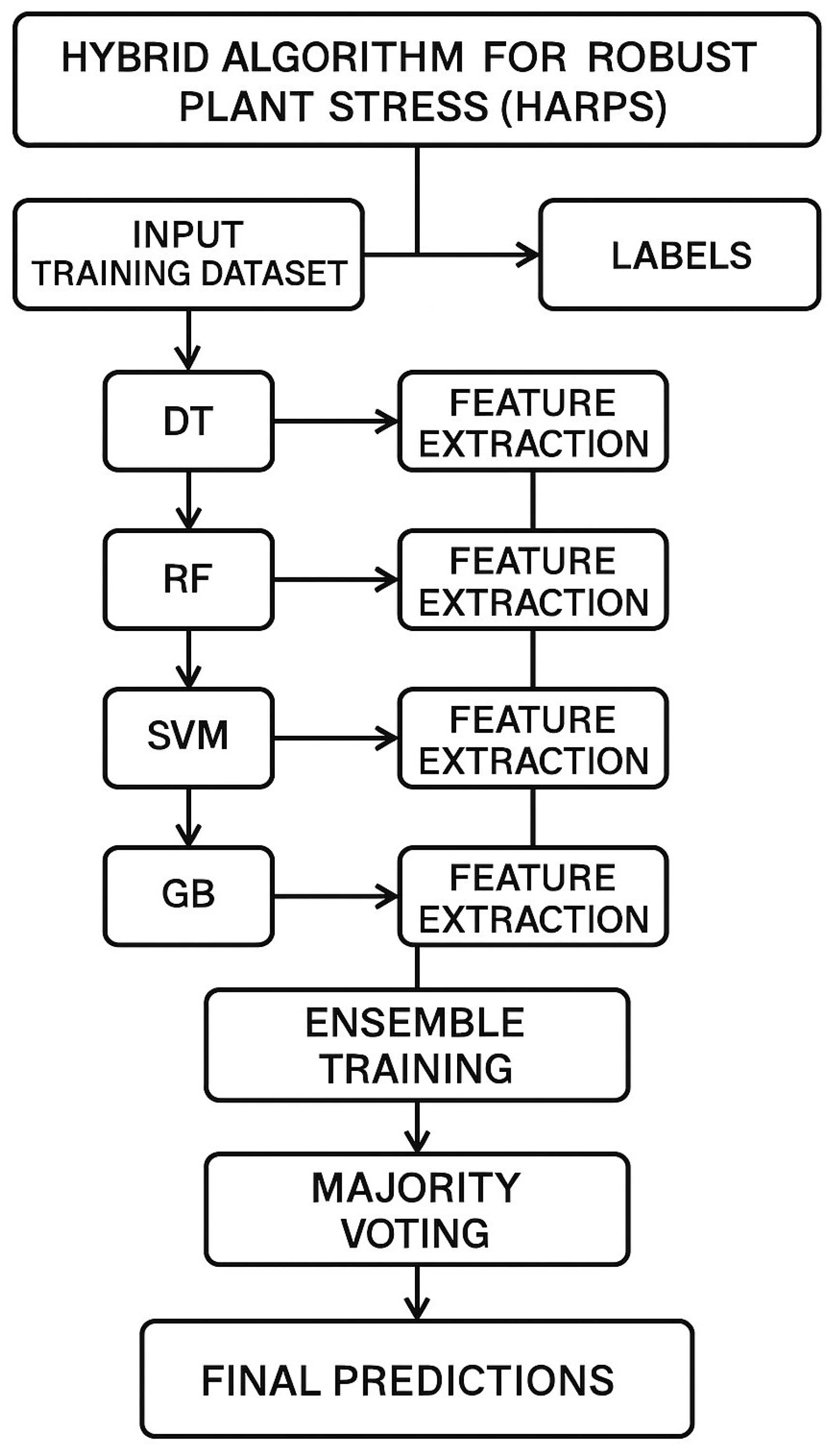
HARPS operates in three stages: feature extraction, ensemble construction, and majority voting. Initially, base classifiers are trained independently, and meaningful features are extracted from each model. These selected features are then used to retrain each base learner, ensuring a robust and diverse representation of the data. Finally, HARPS performs majority voting across all classifiers for each test sample to generate the final prediction. This ensemble-based hybrid strategy helps HARPS as shown in Algorithm 1 improve generalization, reduce overfitting, and enhance stress detection accuracy across varied plant phenotypes.
Based on a hybrid approach combining several methods and parameters,
Figure 1 presents the proposed framework of the HARPS approach. The initial input is a list of protein IDs, which belong to the organism’s response to heat stress. Using the ‘random_state’ option to modify the model’s random operations and algorithm setup is one technique to ensure repeatable results.
The model uses a ‘criterion’, such as entropy or Gini, to rate the quality of data partitions; tree-based models typically use these kinds of parameters [
25]. The term “n-estimators” [
26] denotes the combination of numerous estimators, suggesting the use of an ensemble approach whereby several models are trained and then merged to improve prediction accuracy. The term “bootstrap” [
27] implies that the model constructs individual estimators from resampled subsets of the data, hence enhancing the model’s robustness.
| Algorithm 1 Hybrid Algorithm for Robust Plant Stress (HARPS) |
Input: - Feature matrix X, label vector Y - Base classifiers: DT, RF, SVM, GB - Test split ratio r, random seed s Data Preparation: - Split into training set and testing set using ratio r and seed s Model Training: - Train DT, RF, SVM, and GB on Prediction: - Predict labels on using all four classifiers - Collect predictions: Majority Voting: for each sample i in the test set do Initialize array for all classes k for each base classifier j do Predict class for sample i Update count: end for Compute final predicted class: end for Evaluation: - Compute evaluation metrics: Accuracy, Precision, Recall, and F1-score using and final_pred Output: - Final predicted labels for all test samples - Performance metrics for the hybrid model
|
Later in the workflow, we examine a “regularization parameter” [
28], which refers to techniques to prevent overfitting by penalizing large coefficients, and a “kernel” [
29], which proposes data manipulation to facilitate separation and is often observed in SVM, and “learning rate” [
30], which suggests an iterative optimization technique, possibly with a gradient boosting framework.
The process ultimately results in a “Hybrid” model that uses these many approaches to forecast or analyze “HEAT”, which may be a particular output or metric associated with the reaction to heat stress in different scenarios ( to ). This hybrid model probably aims to offer an in-depth examination of the effects of heat stress, which could be helpful in computational biology and bioinformatics, among other areas.
5. Results and Discussions
In this analysis, the dataset is split so that 80% is used to train the model, and 20% is used to test it. The model learns from the 80% training data and then makes predictions on the 20% test data. This helps us see how well the model can work with new, unseen data. With a 20% test ratio, the model’s accuracy is 72.01%. This balance between training and testing data helps ensure the model is both well-trained and properly tested. A comprehensive evaluation is provided by precision, recall, and F1 scores, which measure the accuracy of positive predictions, recollection, and the ability to record all positive instances, respectively. F1 score strikes a balance between precision and recall by considering the harmonic sequence. Underline how crucial these measures are to comprehending a model’s performance across a range of classes and identifying false positives as well as false negatives. Therefore, emphasizing recall, precision, and F1 Score guarantees a more accurate and nuanced evaluation in a variety of categorization problems.
The results obtained show that HARPS facilitates moving to more intelligent, ecologically friendly agriculture while simultaneously increasing accuracy and computing efficiency. Therefore, the proposed model can assist in minimizing avoidable chemical treatments by reducing the misclassification of stress conditions, therefore promoting agroecological sustainability.
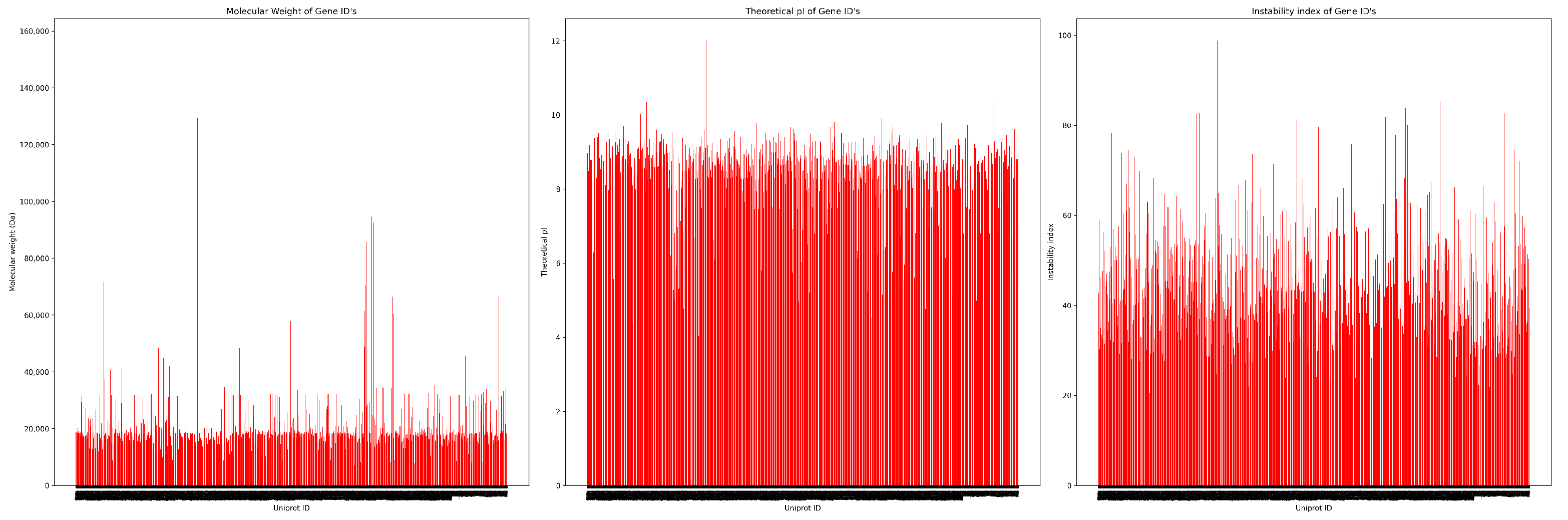
Figure 3 covers all three premises in different peaks, where theoretical pI shows stable results, but the unstable index gives a large gap among each other.
It presents the distribution of three protein features, molecular weight, theoretical pI, and instability index across various UniProt Gene IDs. The molecular weight plot (left) shows a broad range, with most proteins clustering below 30,000 Da and a few significant peaks exceeding 150,000 Da. These high values likely represent large or multi-domain proteins.
Theoretical pI values appear more uniformly distributed, mostly ranging between 4 and 10. This stable pattern suggests that the majority of proteins maintain charge balance under physiological conditions. Proteins with pI ≥ 7.5 are especially relevant, as they are more resistant to denaturation during heat stress.
In contrast, the instability index displays a wide spread, ranging from very stable proteins (below 20) to highly unstable ones (above 80). This variation highlights the index’s importance in identifying heat-tolerant proteins. Overall, the consistent pI values and highly variable instability indices support their combined use in the HARPS model for accurate protein stability classification.
Among the individual classifiers, Gradient Boosting (96.1%) and SVM (91.4%) achieved the highest accuracies. However, the HARPS ensemble model surpassed both due to its ability to integrate the strengths of each base learner:
GB offers robust learning through sequential correction of errors.
SVM provides effective separation in high-dimensional feature spaces.
RF and DT add diversity and reduce overfitting through randomness and tree averaging.
By applying a majority voting strategy, HARPS reduces the impact of weak learners and enhances overall prediction stability, especially in multi-class classification settings.
The selected features contribute significantly to biological interpretability and classification performance under heat stress:
Theoretical pI ≥ 7.5 indicates basic proteins, which tend to remain more stable and functional under stress conditions due to improved solubility and charge-based interactions.
The instability index ≤ 38 identifies proteins with strong structural stability, minimizing the likelihood of heat-induced degradation. The removal of this feature led to significant performance drops in all models, especially GB and HARPS.
Molecular weight informs protein folding complexity; lower molecular weights often correspond to better heat tolerance and rapid cellular response.
In
Table 5 showing traditional classifiers (DT, RF, SVM, GB), we compared HARPS with state-of-the-art gradient boosting frameworks, including XGBoost and LightGBM. While XGBoost achieved a competitive accuracy of 95.7%, and LightGBM reached 94.8%, HARPS slightly outperformed both with an accuracy of 96.6%. This performance advantage is attributed to HARPS’s ensemble design, which integrates diverse learning paradigms, decision trees, random forests, support vector machines, and boosting through a majority voting mechanism.
HARPS also demonstrated greater stability across different test ratios and reduced susceptibility to overfitting, particularly when compared to individual models like DT and SVM. Despite the high accuracy of models such as SVM, HARPS maintained superior performance while requiring substantially lower computational time, completing inference in only 3 s compared to SVM’s 140 s.
The strength of HARPS lies in its ability to capture complementary decision patterns from different algorithmic families. Tree-based learners handle discrete splits and hierarchical patterns well, while margin-based SVMs offer robust separation in feature space. Boosting learners iteratively refine predictions. This synergy results in a more robust, generalizable, and efficient classifier for protein stability prediction under heat stress.
The proposed HARPS model outperformed traditional (DT, RF, SVM, GB) and state-of-the-art classifiers (XGBoost, LightGBM) in both accuracy and evaluation metrics, achieving the highest accuracy (96.6%), as well as the top ROC score (0.970) and AUC score (0.972). These results indicate HARPS’s superior ability to distinguish between classes with high confidence. Compared to strong models like XGBoost (AUC: 0.960) and SVM (AUC: 0.935), HARPS demonstrated more consistent class separability and predictive robustness. Additionally, HARPS required significantly less runtime (3 s), highlighting its efficiency alongside its high performance, making it a practical and powerful tool for protein stability prediction under stress conditions.
The classification accuracies using DT, RF, SVM, and GB are using a dataset of 8525 observations; the test ratio was systematically changed from 0.1 to 0.4 to assess the model. Consequently, to improve the model’s accuracy, it is important to consider extra performance indicators and investigate hyperparameter-changing strategies. The dataset consists of 8525 data points that were divided into two subsets: 6868 (80%) for training and 1657 (20%) for testing.
The DT algorithm achieved an accuracy of 72.1%, the RF algorithm improved slightly with an accuracy of 73.1%. When testing the SVM algorithm with a 20% test ratio, the accuracy is 91.40%. In contrast, the Gradient Boosting (GB) model shows a more noticeable improvement, with accuracy rising to 96.1% at the same 20% test ratio, indicating a more consistent performance with increasing test data.
At a 20% test ratio, the proposed algorithm HARPS delivers excellent performance, reaching an accuracy of 96.69%. This is a significant result, especially when compared to other algorithms, as HARPS achieves this accuracy in just 8 s. The algorithm performs very efficiently in terms of time complexity, making it a great choice when balancing both speed and accuracy. In comparison to other algorithms, HARPS’s ability to deliver strong results quickly sets it apart, particularly for applications requiring fast, reliable predictions.
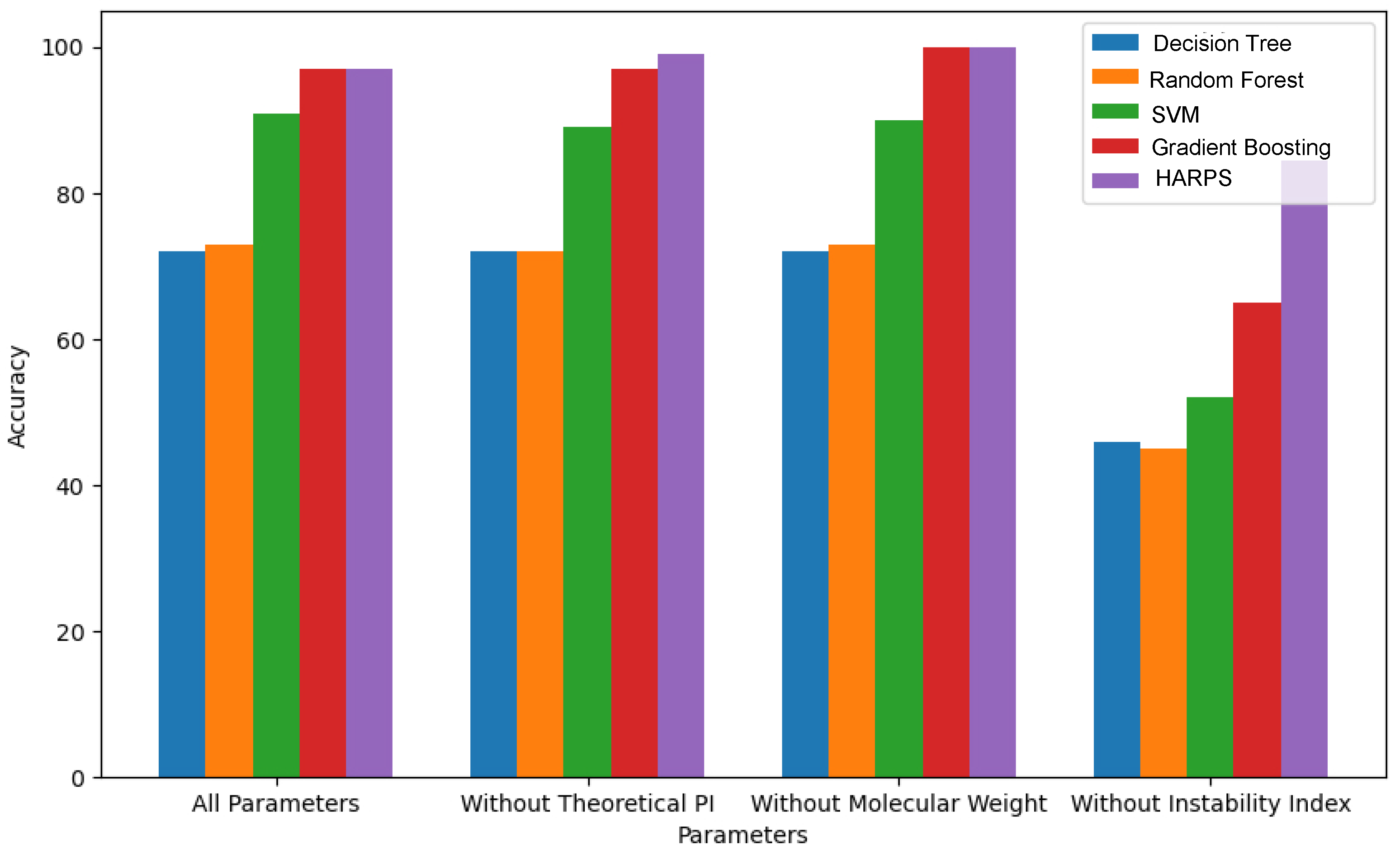
As part of an ablation study, the results shown in
Table 6 provide an overview of how accurate each model is in different situations and how particular items affect the overall performance. Interestingly, the results indicate that the instability index has a significant effect on the overall accuracy.
The HARPS algorithm was developed by combining the characteristics of the DT, RF, SVM, and GB algorithms. These algorithms have individual accuracy percentages of 72.1%, 73.1%, 91.4%, and 96.1%, respectively. Once the theoretical pI, molecular weight, and instability index features were removed, the accuracies for DT, changed to 72.1%, 72.1%, and 45.6%, RF changed to 72.1%, 72.7%, and 45.4%, SVM changed to 89.5%, 89.5%, and 52.3%, and GB changed to 97.1%,100%, and 64.7%, respectively. The HARPS algorithm’s accuracy result was 96.6%; however, after the previously mentioned features were removed, it varied to 99.1%, 1.0%, and 86.5%. The HARPS hybrid algorithm demonstrated superior performance compared to the individual algorithms when their accuracies were compared.
The hybrid ML model (HARPS) combines multiple classifiers, including DT, RF, SVM, and GB, leveraging their individual strengths to achieve a high level of precision. By aggregating multiple perspectives and reducing errors, the model uses majority voting to make final predictions, ensuring greater accuracy. Majority voting is an ensemble learning technique that combines the predictions of individual models to produce an outcome. Each classifier in the ensemble casts a vote for a class label or numerical value, and the final prediction is determined by the majority vote. This approach minimizes the errors of individual models and enhances the overall adaptability and robustness of the forecast. The diverse set of algorithms greatly improves the effectiveness of the model, making it more reliable.
The results of ablation analyses for different ML techniques, such as DT, RF, SVM, GB and HARPS, are shown in
Figure 4. The accuracies achieved by DT and RF algorithms are 72.1% and 73.1%, respectively. However, their accuracies dropped to 45.6% and 45.4% when the instability index was ignored. With SVM, the accuracy remains at 91.4%. Gradient boosting outperformed all other techniques with an accuracy of 96.14%. The accuracy dropped significantly to 64.7% once the instability index was removed. HARPS outperforms all other techniques with an accuracy of 96.6%. The accuracy dropped significantly to 86.5% once the instability index was removed. The overall performance of the HARPS algorithm was better without the instability index. It dropped to 86.5% as compared to other algorithms but is still better compared to other models.
To obtain the desired results from the proposed hybrid approach, sometimes we need extra system resources. Processing times for all these existing and novel approaches are distinct, specifically on a system equipped with an 11th-generation Intel Core i7 processor and 8 GB of RAM. All evaluations were performed using 8 GB RAM and an 11th Gen Intel® Core™ i7 CPU (2.80 GHz), sourced from HP Inc., Palo Alto, CA, USA. DT took approximately 15 s to reach 72.1% accuracy, RF took 20 s to reach 73.1% accuracy, SVM utilized 55 s to reach 91.4% accuracy, GB took 18 s to reach 96.1% accuracy, and the HARPS algorithm utilized just 8 s to achieve an impressive accuracy of approximately 96.6%. The trade-offs between processing speed and accuracy are highlighted in this study. Even while SVM showed excellent accuracy, it consumes too much time than other models. On the other hand, HARPS demonstrated its data processing efficiency by achieving comparable or superior accuracy levels in a much shorter amount of time. This implies that HARPS might be a good option in situations when time and accuracy is critical.
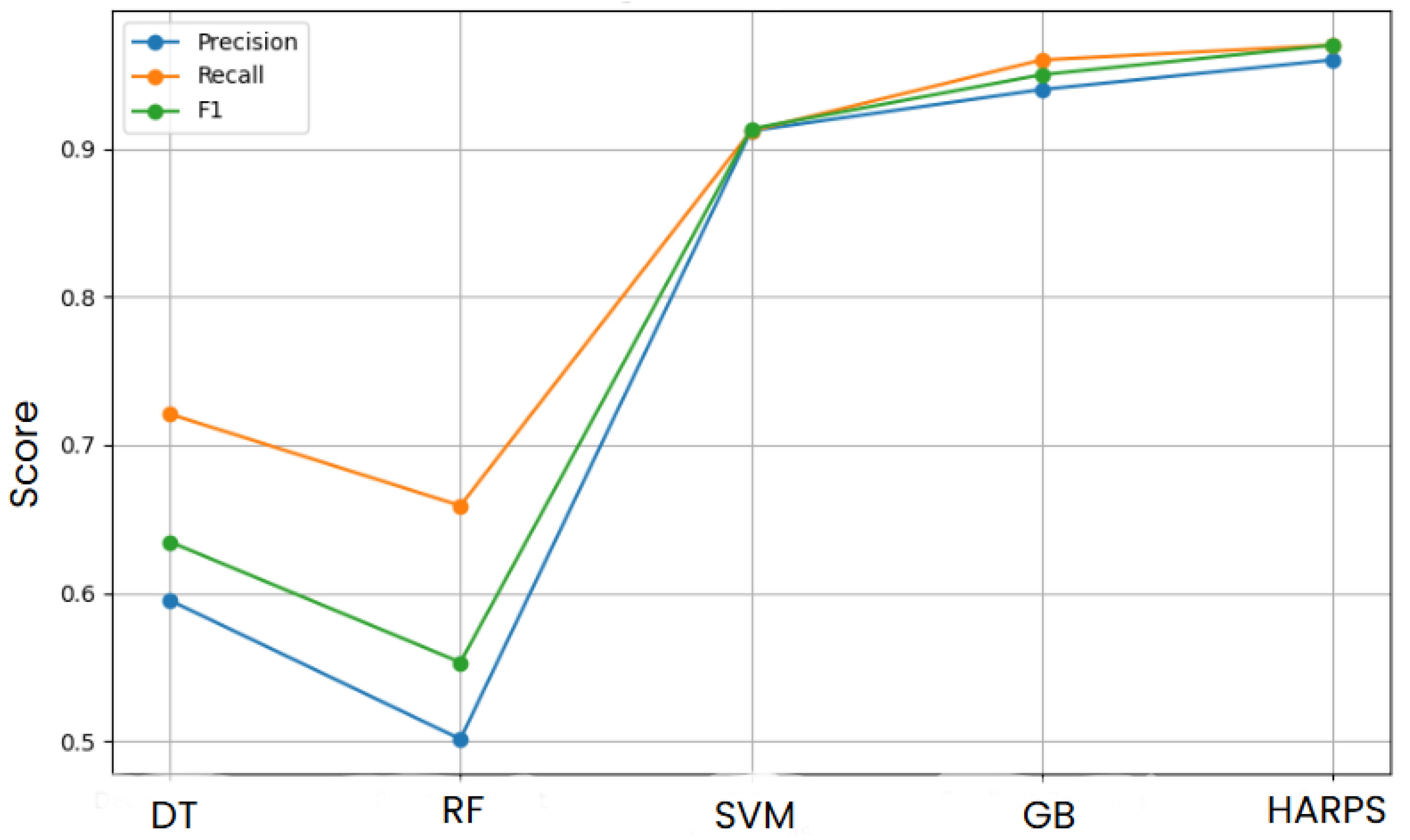
Based on three important metrics, the precision, recall and the F1 score in
Figure 5 offer a perceptive look at how different ML algorithms perform compared to each other on a dataset.
The DT algorithm, the simplest of the algorithms tested, clearly performs poorly on this particular dataset, with the lowest scores across all criteria. Decision trees may overfit training data, leading to poor generalization on unseen data, which could be the source of this.
The SVM algorithm’s performance is comparable to that of the RF, indicating that its strategy of identifying the best hyperplane for classification works effectively with this dataset. SVMs succeed at managing datasets with a large number of features, and their effectiveness in this high-dimensional space may be attributed to their capacity to operate in this environment.
GB improves performance, refining slightly in this evaluation compared to SVM and RF. It concentrates on hard circumstances that previous models misclassified, potentially increasing accuracy, by creating trees sequentially and having each tree attempt to correct the errors of the previous one.
Finally, the HARPS algorithm exhibits a combination of the attributes of the previous four algorithms and seems to be a personalized hybrid model, based on its leading outcomes. This implies that HARPS might be using the robustness of RF, the iterative improvement method of GB, and the dimensionality-handling capacity of SVMs. This could capture more data patterns and correlations, leading to better precision, recall, and F1 scores. The hybrid approach in question likely aims to take advantage of the unique benefits of each base algorithm, producing a very flexible and reliable model that performs well in various dataset dimensions. This performance could be especially helpful in real-world situations when it is necessary to strike a balance between various types of errors.