Sustainable Remediation Using Hydrocarbonoclastic Bacteria for Diesel-Range Hydrocarbon Contamination in Soil: Experimental and In Silico Evaluation
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Samples
2.2. Soil Characterization Methods
2.3. Soil Contamination
2.4. Hydrocarbonoclastic Bacteria
2.5. Bacterial Identification
2.6. Biodegradation Experiment in Microcosms
2.7. Chromatographic Analysis
2.8. Homology Modeling
2.9. Molecular Docking
2.10. Molecular Dynamics Simulations
3. Results and Discussion
3.1. Soil Characterization
3.2. Microorganism Isolation and Characterization
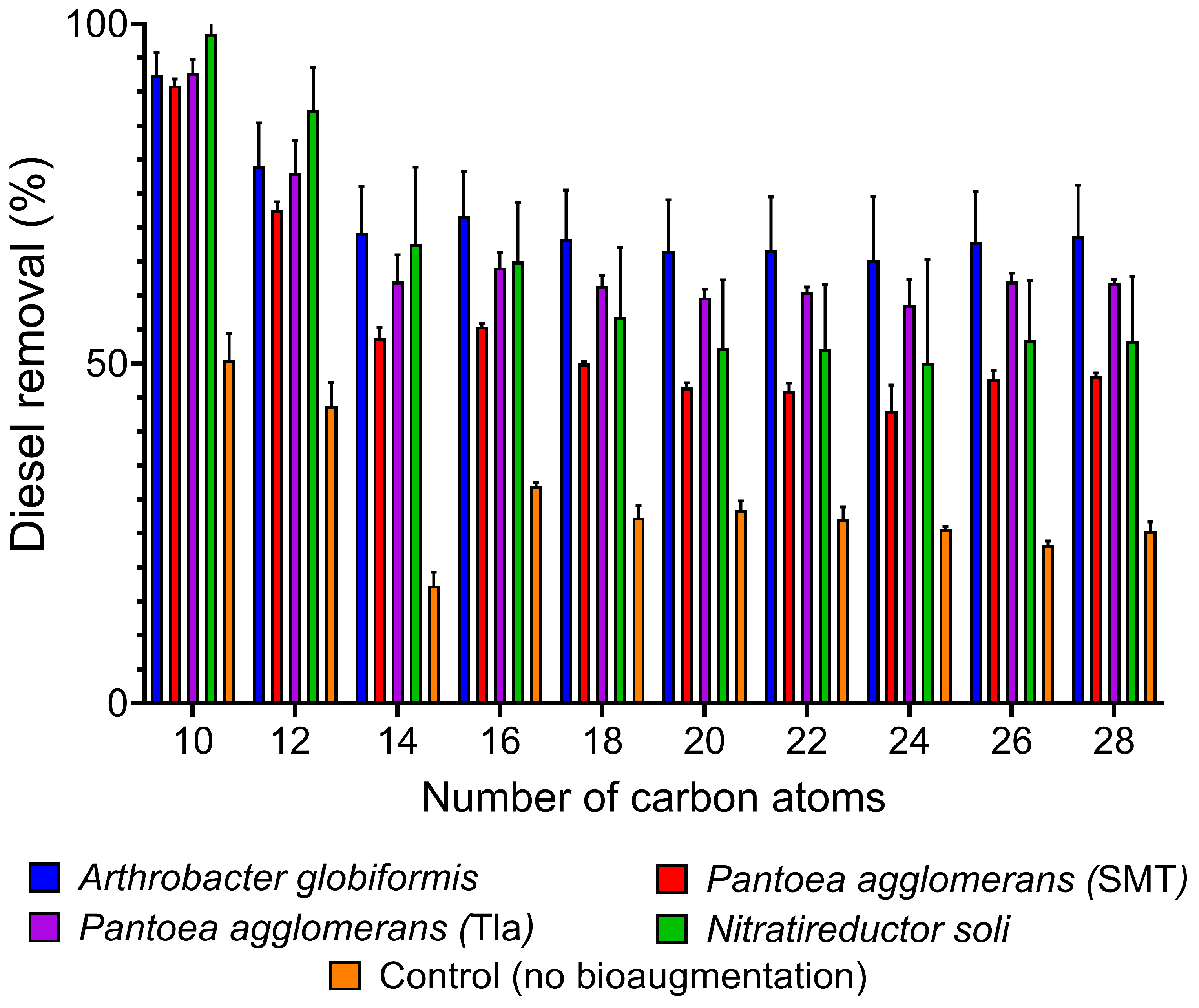
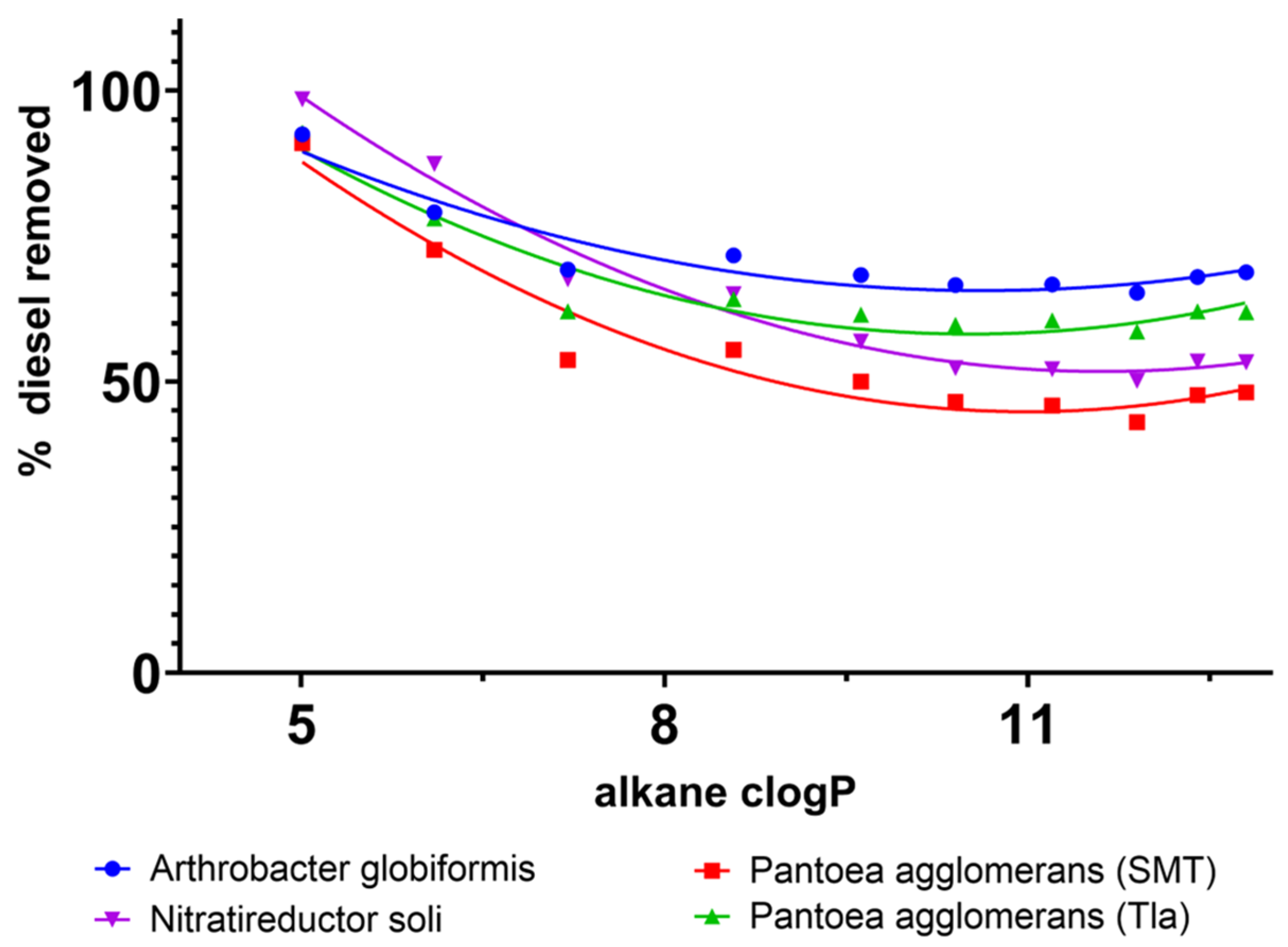
3.3. Biodegradation of Diesel-Range Hydrocarbons in Soil
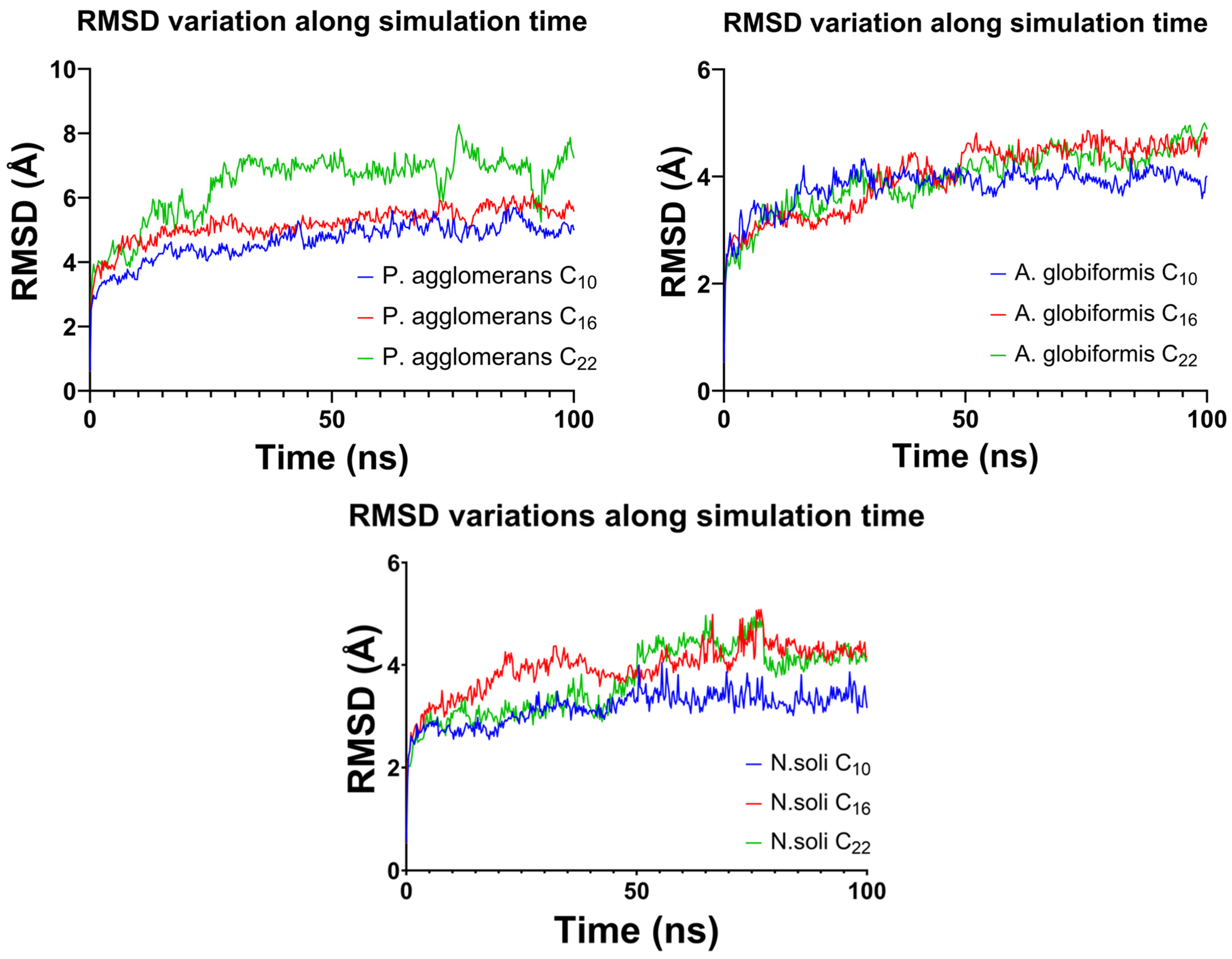
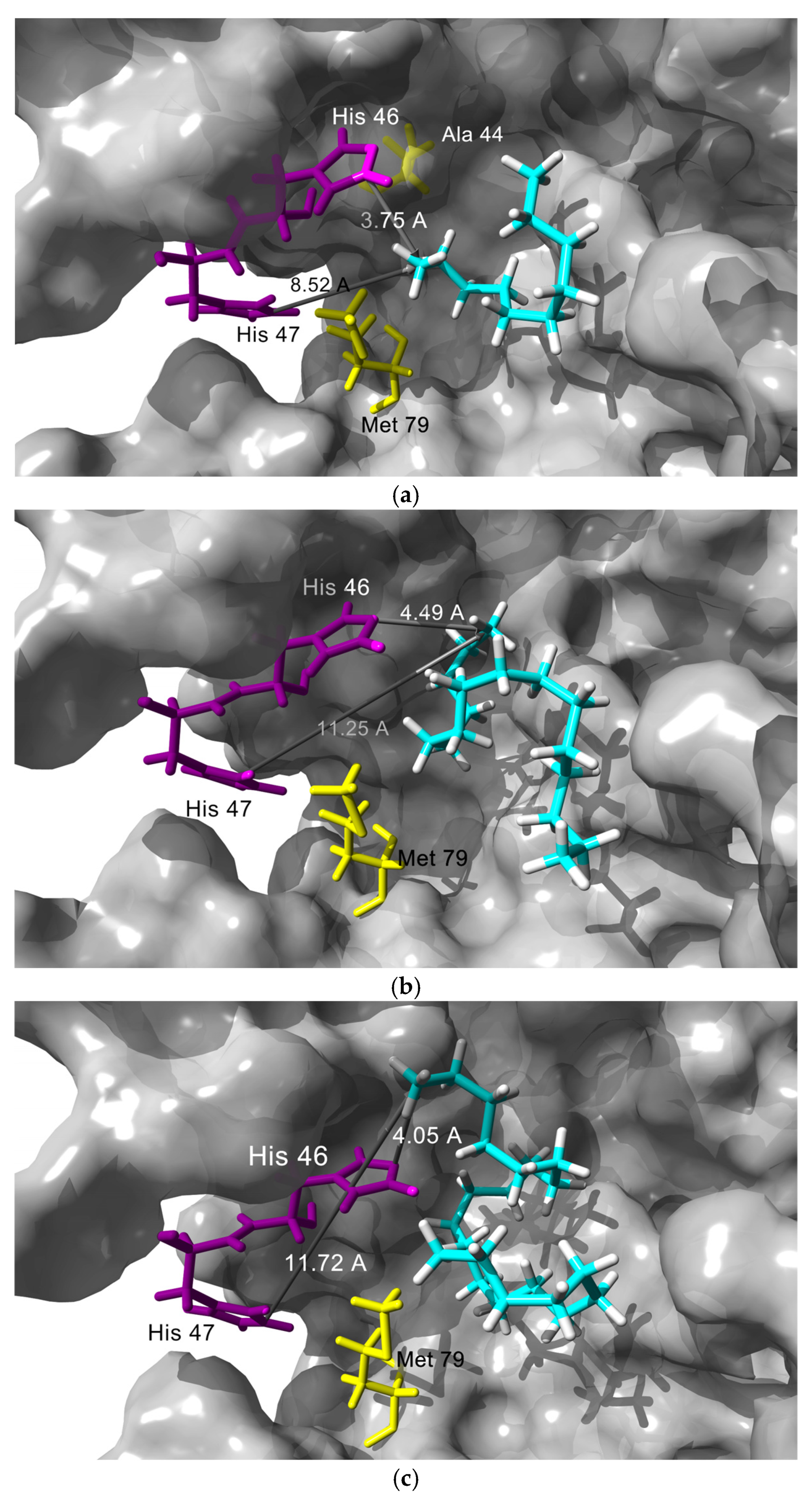
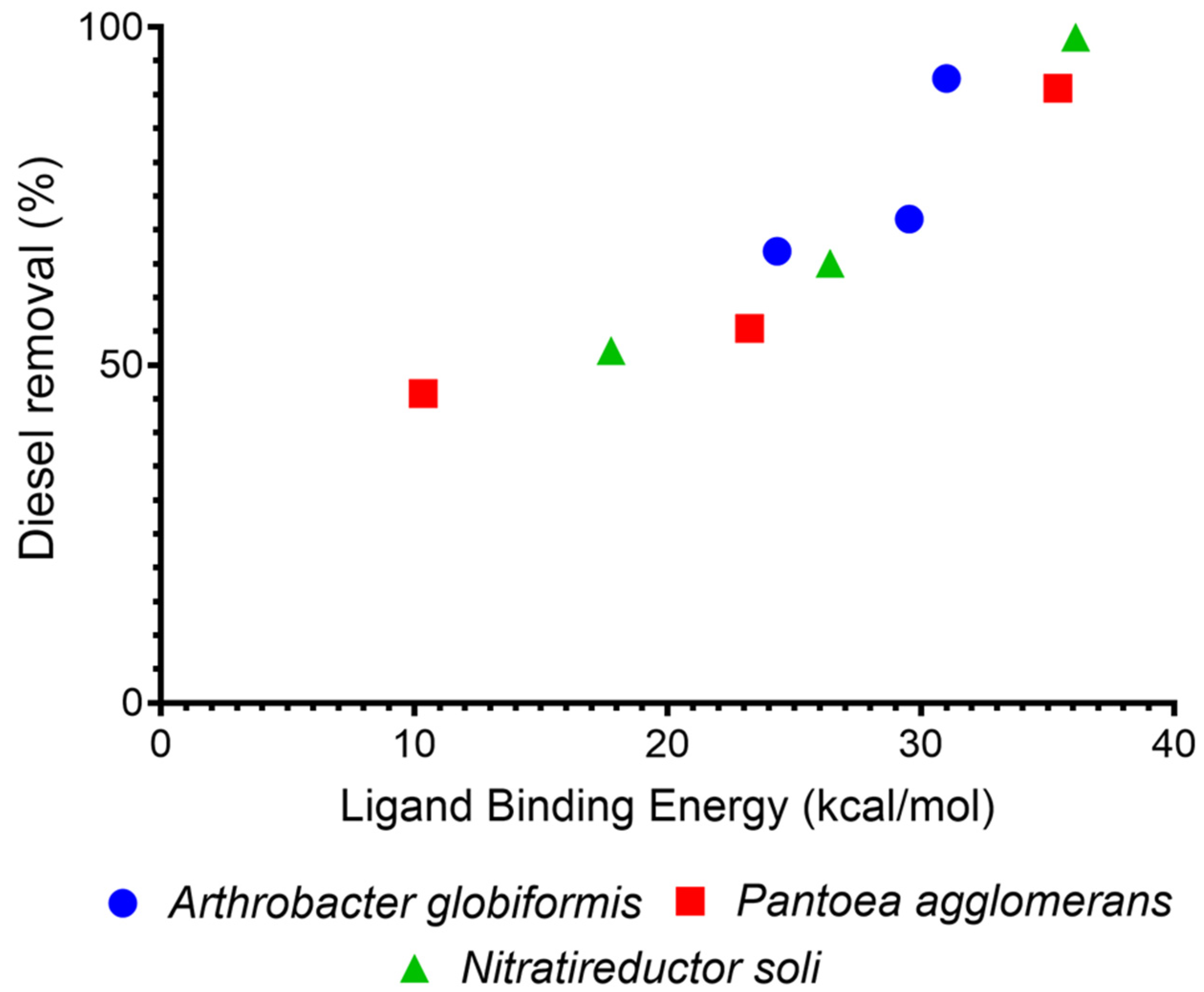
3.4. In Silico Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- British Petroleum. British Petroleum Full Report—Statistical Review of World Energy; British Petroleum: London, UK, 2021. [Google Scholar]
- Shahi, A.; Aydin, S.; Ince, B.; Ince, O. Evaluation of microbial population and functional genes during the bioremediation of petroleum-contaminated soil as an effective monitoring approach. Ecotoxicol. Environ. Saf. 2016, 125, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Khudur, L.S.; Shahsavari, E.; Miranda, A.F.; Morrison, P.D.; Nugegoda, D.; Ball, A.S. Evaluating the efficacy of bioremediating a diesel-contaminated soil using ecotoxicological and bacterial community indices. Environ. Sci. Pollut. Res. 2015, 22, 14809–14819. [Google Scholar] [CrossRef]
- Ikuesan, F.A. Evaluation of Crude Oil Biodegradation Potentials of Some Indigenous Soil Microorganisms. J. Sci. Res. Rep. 2017, 13, 1–9. [Google Scholar] [CrossRef]
- Sutormin, O.S.; Goncharov, A.S.; Kratasyuk, V.A.; Petrova, Y.Y.; Bajbulatov, R.Y.; Yartsov, A.E.; Shpedt, A.A. Effects of Oil Contamination on Range of Soil Types in Middle Taiga of Western Siberia. Sustainability 2024, 16, 11204. [Google Scholar] [CrossRef]
- Skrypnik, L.; Maslennikov, P.; Novikova, A.; Kozhikin, M. Effect of Crude Oil on Growth, Oxidative Stress and Response of Antioxidative System of Two Rye (Secale cereale L.) Varieties. Plants 2021, 10, 157. [Google Scholar] [CrossRef]
- Chorom, M.; Sharifi, H.S.; Motamedi, H. Bioremediation of a crude oil-polluted soil by application of fertilizers. J. Environ. Health Sci. Eng 2010, 7, 319–326. [Google Scholar]
- Rocha, I.M.V.; Silva, K.N.O.; Silva, D.R.; Martínez-Huitle, C.A.; Santos, E.V. Coupling electrokinetic remediation with phytoremediation for depolluting soil with petroleum and the use of electrochemical technologies for treating the effluent generated. Sep. Purif. Technol. 2019, 208, 194–200. [Google Scholar] [CrossRef]
- Eremeeva, A.M.; Kondrasheva, N.K.; Khasanov, A.F.; Oleynik, I.L. Environmentally Friendly Diesel Fuel Obtained from Vegetable Raw Materials and Hydrocarbon Crude. Energies 2023, 16, 2121. [Google Scholar] [CrossRef]
- Maceiras, R.; Alfonsin, V.; Martinez, J.; Martinez Vara de Rey, C. Remediation of Diesel-Contaminated Soil by Ultrasonic Solvent Extraction. Int. J. Environ. Res. 2018, 12, 651–659. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, Q.; Yao, Y.; Chen, Z.; Zhou, J. Micro-bubbles enhanced removal of diesel oil from the contaminated soil in washing/flushing with surfactant and additives. J. Environ. Manage. 2021, 290, 112570. [Google Scholar] [CrossRef]
- Nemati, B.; Baneshi, M.M.; Akbari, H.; Dehghani, R.; Mostafaii, G. Phytoremediation of pollutants in oil-contaminated soils by Alhagi camelorum: Evaluation and modeling. Sci. Rep. 2024, 14, 5502. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, X.; Sun, K.; Lin, C.; Ma, J.; He, M.; Ouyang, W. Persulfate-based advanced oxidation processes (AOPs) for organic-contaminated soil remediation: A review. Chem. Eng. J. 2019, 372, 836–851. [Google Scholar] [CrossRef]
- Nguyen, T.-B.; Sherpa, K.; Bui, X.-T.; Nguyen, V.-T.; Vo, T.-D.-H.; Ho, H.-T.-T.; Chen, C.-W.; Dong, C.-D. Biochar for soil remediation: A comprehensive review of current research on pollutant removal. Environ. Pollut. 2023, 337, 122571. [Google Scholar] [CrossRef]
- Liu, Y.; Biswas, B.; Hassan, M.; Naidu, R. Green Adsorbents for Environmental Remediation: Synthesis Methods, Ecotoxicity, and Reusability Prospects. Processes 2024, 12, 1195. [Google Scholar] [CrossRef]
- Michael-Igolima, U.; Abbey, S.J.; Ifelebuegu, A.O. A systematic review on the effectiveness of remediation methods for oil contaminated soils. Environ. Adv. 2022, 9, 100319. [Google Scholar] [CrossRef]
- Adams, G.O.; Fufeyin, P.T.; Samson, E.O.; Ehinomen, I. Bioremediation, Biostimulation and Bioaugmention:A Review. Int. J. Environ. Bioremediat. Biodegrad. 2015, 3, 28–39. [Google Scholar]
- Rusănescu, C.O.; Istrate, I.A.; Rusănescu, A.M.; Constantin, G.A. Bioremediation of Soil Contamination with Polycyclic Aromatic Hydrocarbons—A Review. Land 2025, 14, 10. [Google Scholar] [CrossRef]
- Safdari, M.S.; Kariminia, H.R.; Rahmati, M.; Fazlollahi, F.; Polasko, A.; Mahendra, S.; Wilding, W.V.; Fletcher, T.H. Development of bioreactors for comparative study of natural attenuation, biostimulation, and bioaugmentation of petroleum-hydrocarbon contaminated soil. J. Hazard. Mater. 2018, 342, 270–278. [Google Scholar] [CrossRef]
- Wu, M.; Dick, W.A.; Li, W.; Wang, X.; Yang, Q.; Wang, T.; Xu, L.; Zhang, M.; Chen, L. Bioaugmentation and biostimulation of hydrocarbon degradation and the microbial community in a petroleum-contaminated soil. Int. Biodeterior. Biodegradat. 2016, 107, 158–164. [Google Scholar] [CrossRef]
- Nwankwegu, A.S.; Onwosi, C.O. Bioremediation of gasoline contaminated agricultural soil by bioaugmentation. Environ. Technol. Innov. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Brzeszcz, J.; Kapusta, P.; Steliga, T.; Turkiewicz, A. Hydrocarbon Removal by Two Differently Developed Microbial Inoculants and Comparing Their Actions with Biostimulation Treatment. Molecules 2020, 25, 661. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dash, H.R. Microbial Bioremediation: A Potential Tool for Restoration of Contaminated Areas. In Microbial Biodegradation and Bioremediation; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 2–21. ISBN 9780128004821. [Google Scholar]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Lima, S.D.; Oliveira, A.F.; Golin, R.; Lopes, V.C.P.; Caixeta, D.S.; Lima, Z.M.; Morais, E.B. Isolation and characterization of hydrocarbon-degrading bacteria from gas station leaking-contaminated groundwater in the Southern Amazon, Brazil. Brazilian J. Biol. 2020, 80, 354–361. [Google Scholar] [CrossRef]
- Nie, Y.; Chi, C.-Q.; Fang, H.; Liang, J.-L.; Lu, S.-L.; Lai, G.-L.; Tang, Y.-Q.; Wu, X.-L. Diverse alkane hydroxylase genes in microorganisms and environments. Sci. Rep. 2014, 4, 4968. [Google Scholar] [CrossRef]
- Groves, J.T.; Feng, L.; Austin, R.N. Structure and Function of Alkane Monooxygenase (AlkB). Acc. Chem. Res. 2023, 56, 3665–3675. [Google Scholar] [CrossRef]
- Sandoval-Pauker, C.; Yin, S.; Castillo, A.; Ocuane, N.; Puerto-Diaz, D.; Villagrán, D. Computational Chemistry as Applied in Environmental Research: Opportunities and Challenges. ACS ES&T Eng. 2024, 4, 66–95. [Google Scholar]
- Wang, R.; Chen, H.; Guo, S.; He, Z.; Ren, N.; Ho, S.-H. Machine Learning toward Realizing End-to-End Biochar Design for Environmental Remediation. ACS ES&T Eng. 2024, 4, 2332–2345. [Google Scholar]
- Mohebbi, E.; Minnelli, C.; Pavoni, E.; Sisti, L.; Laudadio, E.; Stipa, P. Layered Double Hydroxides as Systems for Capturing Small-Molecule Air Pollutants: A Density Functional Theory Study. Molecules 2024, 29, 4996. [Google Scholar] [CrossRef]
- Yadav, A.; Vuković, L.; Narayan, M. An Atomic and Molecular Insight into How PFOA Reduces α-Helicity, Compromises Substrate Binding, and Creates Binding Pockets in a Model Globular Protein. J. Am. Chem. Soc. 2024, 146, 12766–12777. [Google Scholar] [CrossRef]
- Aloui, S.; Raboudi, F.; Ghazouani, T.; Salghi, R.; Hamdaoui, M.H.; Fattouch, S. Use of molecular and in silico bioinformatic tools to investigate pesticide binding to insect (Lepidoptera) phenoloxidases (PO): Insights to toxicological aspects. J. Environ. Sci. Health B. 2014, 49, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Librando, V.; Pappalardo, M. In silico bioremediation of polycyclic aromatic hydrocarbon: A frontier in environmental chemistry. J. Mol. Graph. Model. 2013, 44, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tratnyek, P.G.; Bylaska, E.J.; Weber, E.J. In silico environmental chemical science: Properties and processes from statistical and computational modelling. Environ. Sci. Process. Impacts 2017, 19, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Chowdhary, P.; Raj, A. In silico bioremediation strategies for removal of environmental pollutants released from paper mills using bacterial ligninolytic enzymes. In Microorganisms for Sustainable Environment and Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 249–285. [Google Scholar]
- Singh, P.K.; Imam, J.; Shukla, P. In Silico Approach in Bioremediation. In Microbial Biodegradation and Bioremediation; Elsevier: Amsterdam, The Netherlands, 2014; pp. 422–433. [Google Scholar]
- Vidal-Limon, A.; Aguilar-Toalá, J.E.; Liceaga, A.M. Integration of Molecular Docking Analysis and Molecular Dynamics Simulations for Studying Food Proteins and Bioactive Peptides. J. Agric. Food Chem. 2022, 70, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.E.; Fernandez, J.P.; Hadad, C.M.; MacKay, A.A. Molecular Docking as a Tool to Examine Organic Cation Sorption to Organic Matter. Environ. Sci. Technol. 2022, 56, 951–961. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, S.-S.; Xiao, Z.-C.; Min, Y.; Tian, T.; Zheng, Y.-M.; Zhao, Q.-B.; Yuan, Z.-H.; Yu, H.-Q. Re-evaluation and modification of dehydrogenase activity tests in assessing microbial activity for wastewater treatment plant operation. Water Res. 2023, 246, 120737. [Google Scholar] [CrossRef]
- Fernández, L.; Rojas, N.; Roldán, T.; Ramírez, M.; Zegarra, H.; Hernández, R.; Reyes, R.; Flores, D.; Arce, J. Manual de Técnicas de Análisis de Suelos Aplicadas a la Remediación de Sitios Contaminados; Instituto Mexicano del Petróleo, Secretaria de Medio Ambiente y Recursos Naturales, Instituto Nacional de Ecología: Mexico City, Mexico, 2006. [Google Scholar]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- US Environmental Protection Agency. Application, Performance, and Costs of Biotreatment Technologies for Contaminated Soils; US Environmental Protection Agency: Washington, DC, USA, 2002. [Google Scholar]
- US Environmental Protection Agency. Method 3546. Microwave Extraction; US Environmental Protection Agency: Washington, DC, USA, 2007. [Google Scholar]
- US Environmental Protection Agency. Method 8015C. Nonhalogenated organics by Gas Chromatography; US Environmental Protection Agency: Washington, DC, USA, 2007. [Google Scholar]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- McGreig, J.E.; Uri, H.; Antczak, M.; Sternberg, M.J.E.; Michaelis, M.; Wass, M.N. 3DLigandSite: Structure-based prediction of protein–ligand binding sites. Nucleic Acids Res. 2022, 50, W13–W20. [Google Scholar] [CrossRef]
- Jiménez, J.; Doerr, S.; Martínez-Rosell, G.; Rose, A.S.; De Fabritiis, G. DeepSite: Protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics 2017, 33, 3036–3042. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Vivar-Sierra, A.; Araiza-Macías, M.J.; Hernández-Contreras, J.P.; Vergara-Castañeda, A.; Ramírez-Vélez, G.; Pinto-Almazán, R.; Salazar, J.R.; Loza-Mejía, M.A. In Silico Study of Polyunsaturated Fatty Acids as Potential SARS-CoV-2 Spike Protein Closed Conformation Stabilizers: Epidemiological and Computational Approaches. Molecules 2021, 26, 711. [Google Scholar] [CrossRef] [PubMed]
- Passos, W.A.; Jesus, M.; Mata, F.; Menezes, M.S.; dos Santos, P.O.L.; Santos, B.L.P.; Santana, H.E.P.; Ruzene, D.S.; Silva, D.P. Bioremediation Potential of Sunflower-Derived Biosurfactants: A Bibliometric Description. Sustainability 2025, 17, 330. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14sb: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Haghollahi, A.; Fazaelipoor, M.H.; Schaffie, M. The effect of soil type on the bioremediation of petroleum contaminated soils. J. Environ. Manage. 2016, 180, 197–201. [Google Scholar] [CrossRef]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. The Role of Microorganisms in Bioremediation—A Review. Open J. Environ. Biol. 2017, 2, 038–046. [Google Scholar] [CrossRef]
- Ayilara, M.S.; Babalola, O.O. Bioremediation of environmental wastes: The role of microorganisms. Front. Agron. 2023, 5, 1183691. [Google Scholar] [CrossRef]
- Chaillan, F.; Chaîneau, C.H.; Point, V.; Saliot, A.; Oudot, J. Factors inhibiting bioremediation of soil contaminated with weathered oils and drill cuttings. Environ. Pollut. 2006, 144, 255–265. [Google Scholar] [CrossRef]
- Chaîneau, C.H.; Rougeux, G.; Yéprémian, C.; Oudot, J. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 2005, 37, 1490–1497. [Google Scholar] [CrossRef]
- Schjønning, P.; Thomsen, I.K.; Petersen, S.O.; Kristensen, K.; Christensen, B.T. Relating soil microbial activity to water content and tillage-induced differences in soil structure. Geoderma 2011, 163, 256–264. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: A review. Chemosphere 2017, 172, 52–71. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.H.; Jung, W.C.; Park, M.; Kim, M.S.; Lee, S.J.; Park, H. Changes in Soil Health with Remediation of Petroleum Hydrocarbon Contaminated Soils Using Two Different Remediation Technologies. Sustainability 2020, 12, 10078. [Google Scholar] [CrossRef]
- Girigiri, B.; Ariole, C.N.; Stanley, H.O. Bioremediation of Crude Oil Polluted Soil Using Biofertilizer from Nitrogen-fixing and Phosphate-solubilizing Bacteria. Am. J. Nanosci. 2019, 5, 27–38. [Google Scholar] [CrossRef]
- Marinescu, M.; Lacatusu, A.; Gament, E.; Plopeanu, G.; Carabulea, V. Bioremediation Potential of Native Hydrocarbons Degrading Bacteria in Crude Oil Polluted Soil. Bull. Univ. Agric. Sci. Vet. Med. Cluj Napoca. Agric. 2017, 74, 19. [Google Scholar] [CrossRef]
- Wang, X.; Lina, S.; Hui, W.; Hao, W.; Su, C.; Zheng, X. Surfactant-enhanced bioremediation of DDTs and PAHs in contaminated farmland soil. Environ. Technol. 2018, 39, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- Soltanighias, T.; Singh, A.E.A.; Satpute, S.K.; Banpurkar, A.G.; Koolivand, A.; Rahi, P. Assessment of biosurfactant-producing bacteria from oil contaminated soils and their hydrocarbon degradation potential. Environ. Sustain. 2019, 2, 285–296. [Google Scholar] [CrossRef]
- Bahobail, A.; El-Rab, S.M.F.G.; Amin, G.A.; Bahobail, A.; El-Rab, S.M.F.G.; Amin, G.A. Locally Isolated Bacterial Strains with Multiple Degradation Potential Capabilities on Petroleum Hydrocarbon Pollutants. Adv. Microbiol. 2016, 6, 852–866. [Google Scholar] [CrossRef]
- Hosseini, S.; Sharifi, R.; Habibi, A. Simultaneous removal of aliphatic and aromatic crude oil hydrocarbons by Pantoea agglomerans isolated from petroleum-contaminated soil in the west of Iran. Arch. Microbiol. 2024, 206, 98. [Google Scholar] [CrossRef]
- Chen, Q.; Ni, H.-Y.; Zhuang, W.; Sun, Z.-G.; Yang, Z.-Z.; Wang, H.-M.; He, Q.; He, J. Nitratireductor soli sp. nov., isolated from phenol-contaminated soil. Antonie Van Leeuwenhoek 2015, 108, 1139–1146. [Google Scholar] [CrossRef]
- Pan, X.-C.; Geng, S.; Mei, R.; Wang, Y.-N.; Cai, H.; Liu, X.-Y.; Tang, Y.-Q.; Nie, Y.; Ye, S.-Y.; Wu, X.-L. Nitratireductor shengliensis sp. nov., Isolated from an Oil-Polluted Saline Soil. Curr. Microbiol. 2014, 69, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Marasco, R.; Michoud, G.; Sefrji, F.O.; Fusi, M.; Antony, C.P.; Seferji, K.A.; Barozzi, A.; Merlino, G.; Daffonchio, D. The identification of the new species Nitratireductor thuwali sp. nov. reveals the untapped diversity of hydrocarbon-degrading culturable bacteria from the arid mangrove sediments of the Red Sea. Front. Microbiol. 2023, 14, 1155381. [Google Scholar] [CrossRef] [PubMed]
- Oro, C.E.D.; Saorin Puton, B.M.; Venquiaruto, L.D.; Dallago, R.M.; Tres, M. V Effective Microbial Strategies to Remediate Contaminated Agricultural Soils and Conserve Functions. Agronomy 2024, 14, 2637. [Google Scholar] [CrossRef]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Sansegundo-Lobato, P.; Redondo-Nieto, M.; Suman, J.; Cajthaml, T.; Blanco-Romero, E.; Martin, M.; Uhlik, O.; Rivilla, R. Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader Rhodococcus sp. Way2 revealed by its complete genome sequence. Microb. Genomics 2020, 6, e000363. [Google Scholar] [CrossRef]
- Striebich, R.C.; Smart, C.E.; Gunasekera, T.S.; Mueller, S.S.; Strobel, E.M.; McNichols, B.W.; Ruiz, O.N. Characterization of the F-76 diesel and Jet-A aviation fuel hydrocarbon degradation profiles of Pseudomonas aeruginosa and Marinobacter hydrocarbonoclasticus. Int. Biodeterior. Biodegradat. 2014, 93, 33–43. [Google Scholar] [CrossRef]
- Lyu, L.; Li, J.; Chen, Y.; Mai, Z.; Wang, L.; Li, Q.; Zhang, S. Degradation potential of alkanes by diverse oil-degrading bacteria from deep-sea sediments of Haima cold seep areas, South China Sea. Front. Microbiol. 2022, 13, 920067. [Google Scholar] [CrossRef]
- Pandolfo, E.; Barra Caracciolo, A.; Rolando, L. Recent Advances in Bacterial Degradation of Hydrocarbons. Water 2023, 15, 375. [Google Scholar] [CrossRef]
- Vandera, E.; Koukkou, A.I. Bacterial Community Response to Hydrocarbon Contamination in Soils and Marine Sediments: A Critical Review of Case Studies. In Microbial Ecotoxicology; Cravo-Laureau, C., Cagnon, C., Lauga, B., Duran, R., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 185–226. ISBN 978-3-319-61795-4. [Google Scholar]
- Ma, Y.; Zheng, X.; Anderson, S.H.; Lu, J.; Feng, X. Diesel oil volatilization processes affected by selected porous media. Chemosphere 2014, 99, 192–198. [Google Scholar] [CrossRef]
- Ausma, S.; Grant, C.E.; Colleen, R.F.-H.; Laurie, H.-M.; Terry, J.G.; Mortimer, W.P. Volatile Hydrocarbon Emissions from a Diesel Fuel-Contaminated Soil Bioremediation Facility. J. Air Waste Manage. Assoc. 2002, 52, 769–780. [Google Scholar] [CrossRef]
- Chen, Y.-A.; Grace Liu, P.-W.; Whang, L.-M.; Wu, Y.-J.; Cheng, S.-S. Effect of soil organic matter on petroleum hydrocarbon degradation in diesel/fuel oil-contaminated soil. J. Biosci. Bioeng. 2020, 129, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, V.S.; Padwa, R.S. Role of Additives and their Influence on Performance of Engine for Petroleum Diesel Fuel, Oxygenated-Diesel Blend: A Review. Int. J. Eng. Res. Technol. 2015, 4, 1–9. [Google Scholar]
- Rakowska, J. Remediation of diesel-contaminated soil enhanced with firefighting foam application. Sci. Rep. 2020, 10, 8824. [Google Scholar] [CrossRef] [PubMed]
- Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B.; Throne-Holst, M. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 2007, 76, 1209–1221. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, J.; Han, L.; Lee, J.; Williams, S.C.; Forsberg, A.; Xu, Y.; Austin, R.N.; Feng, L. Structure and mechanism of the alkane-oxidizing enzyme AlkB. Nat. Commun. 2023, 14, 2180. [Google Scholar] [CrossRef]
- Jurcik, A.; Bednar, D.; Byska, J.; Marques, S.M.; Furmanova, K.; Daniel, L.; Kokkonen, P.; Brezovsky, J.; Strnad, O.; Stourac, J.; et al. CAVER Analyst 2.0: Analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 2018, 34, 3586–3588. [Google Scholar] [CrossRef]
- Ramírez, D.; Caballero, J. Is It Reliable to Take the Molecular Docking Top Scoring Position as the Best Solution without Considering Available Structural Data? Molecules 2018, 23, 1038. [Google Scholar] [CrossRef]
- Tran-Nguyen, V.K.; Bret, G.; Rognan, D. True Accuracy of Fast Scoring Functions to Predict High-Throughput Screening Data from Docking Poses: The Simpler the Better. J. Chem. Inf. Model. 2021, 61, 2788–2797. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, H.; Yao, X.; Li, D.; Xu, L.; Li, Y.; Tian, S.; Hou, T. Comprehensive evaluation of ten docking programs on a diverse set of protein–ligand complexes: The prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016, 18, 12964–12975. [Google Scholar] [CrossRef]
- Kim, D.; Noh, M.H.; Park, M.; Kim, I.; Ahn, H.; Ye, D.-Y.; Jung, G.Y.; Kim, S. Enzyme activity engineering based on sequence co-evolution analysis. Metab. Eng. 2022, 74, 49–60. [Google Scholar] [CrossRef]
- Amin, A.; Naveed, M.; Sarwar, A.; Rasheed, S.; Saleem, H.G.M.; Latif, Z.; Bechthold, A. In vitro and in silico Studies Reveal Bacillus cereus AA-18 as a Potential Candidate for Bioremediation of Mercury-Contaminated Wastewater. Front. Microbiol. 2022, 13, 847806. [Google Scholar] [CrossRef] [PubMed]
- Tasleem, M.; Hussein, W.M.; El-Sayed, A.-A.A.A.; Alrehaily, A. An In Silico Bioremediation Study to Identify Essential Residues of Metallothionein Enhancing the Bioaccumulation of Heavy Metals in Pseudomonas aeruginosa. Microorganisms 2023, 11, 2262. [Google Scholar] [CrossRef] [PubMed]

| Organic Matter | 4.8% |
| Texture | Sand 70% Silt 27%, sandy loam clay 3% |
| Field capacity | 29.04% |
| pH | 5.9 |
| Humidity | 18.9% |
| Total nitrogen | 0.04% |
| Soluble phosphorus | 2.66 ppm |
| Hydrocarbonoclastic bacteria | 2.4 × 106 CFU/g d.s. |
| Hydrocarbonoclastic fungi | 3.76 × 104 CFU/g d.s. |
| Isolated Strain (Code) | Species Identification | Sequence Identity (%) | Site | Metabolic Characteristic: Hydrocarbonoclastic (H), Phosphorus-Solubilizing (P), or Nitrogen-Fixing (N) | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| H1 | Arthrobacter globiformis | 95.72 | 98.64 | Juchitepec | H |
| F1 | Pantoea agglomerans | 96.60 | 96.33 | Santa María | H + P |
| F2 | Pantoea agglomerans | 95.84 | 98.45 | Tlapala | H + P |
| N2 | Nitratireductor soli | 92.47 | 94.10 | Juchitepec | H + N |
| Alkane | A. globiformis | P. agglomerans | N. soli |
|---|---|---|---|
| C10 | −55.6 | −56.1 | −61.6 |
| C12 | −64.2 | −62.6 | −73.3 |
| C14 | −72.5 | −66.7 | −76.7 |
| C16 | −71.5 | −72.8 | −80.4 |
| C18 | −83.1 | −81.1 | −86.4 |
| C20 | −82.1 | −81.9 | −88.7 |
| C22 | −85.6 | −86.1 | −86.9 |
| C24 | −91.2 | −85.8 | −99.4 |
| C26 | −77.9 | −83.2 | −94.2 |
| C28 | −76.3 | −81.5 | −91.7 |
| Alkane | A. globiformis | P. agglomerans | N. soli |
|---|---|---|---|
| C10 | 31.00 | 35.40 | 36.10 |
| C16 | 29.54 | 23.24 | 26.41 |
| C22 | 25.37 | 10.36 | 17.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinosa-López, F.; Pelcastre-Guzmán, K.; Cerón-Nava, A.; Rivera-Noriega, A.; Loza-Mejía, M.A.; Islas-García, A. Sustainable Remediation Using Hydrocarbonoclastic Bacteria for Diesel-Range Hydrocarbon Contamination in Soil: Experimental and In Silico Evaluation. Sustainability 2025, 17, 5535. https://doi.org/10.3390/su17125535
Espinosa-López F, Pelcastre-Guzmán K, Cerón-Nava A, Rivera-Noriega A, Loza-Mejía MA, Islas-García A. Sustainable Remediation Using Hydrocarbonoclastic Bacteria for Diesel-Range Hydrocarbon Contamination in Soil: Experimental and In Silico Evaluation. Sustainability. 2025; 17(12):5535. https://doi.org/10.3390/su17125535
Chicago/Turabian StyleEspinosa-López, Fernanda, Karen Pelcastre-Guzmán, Anabelle Cerón-Nava, Alicia Rivera-Noriega, Marco A. Loza-Mejía, and Alejandro Islas-García. 2025. "Sustainable Remediation Using Hydrocarbonoclastic Bacteria for Diesel-Range Hydrocarbon Contamination in Soil: Experimental and In Silico Evaluation" Sustainability 17, no. 12: 5535. https://doi.org/10.3390/su17125535
APA StyleEspinosa-López, F., Pelcastre-Guzmán, K., Cerón-Nava, A., Rivera-Noriega, A., Loza-Mejía, M. A., & Islas-García, A. (2025). Sustainable Remediation Using Hydrocarbonoclastic Bacteria for Diesel-Range Hydrocarbon Contamination in Soil: Experimental and In Silico Evaluation. Sustainability, 17(12), 5535. https://doi.org/10.3390/su17125535










