Thai Local Chicken Breeds, Chee Fah and Fah Luang, Originated from Chinese Black-Boned Chicken with Introgression of Red Junglefowl and Domestic Chicken Breeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area, Environmental Data, and Spatial Habitat Suitability Model
2.2. Specimen Collection and DNA Extraction
2.3. Mitochondrial DNA D-Loop Sequencing, Quality Control, and Data Analysis
2.4. Microsatellite Genotyping and Data Analysis
3. Results
3.1. Land Suitability Map of Chee Fah and Fah Luang Chickens
3.2. Model Performance and Variable Importance of Habitat Suitability
3.3. Comparison of Environmental Factors between Local Chicken Farms in Mae Hong Son and Chiang Rai Provinces
3.4. Genetic Variability of Chee Fah and Fah Luang Chicken Populations Based on Mitochondrial DNA D-Loop Haplotypes
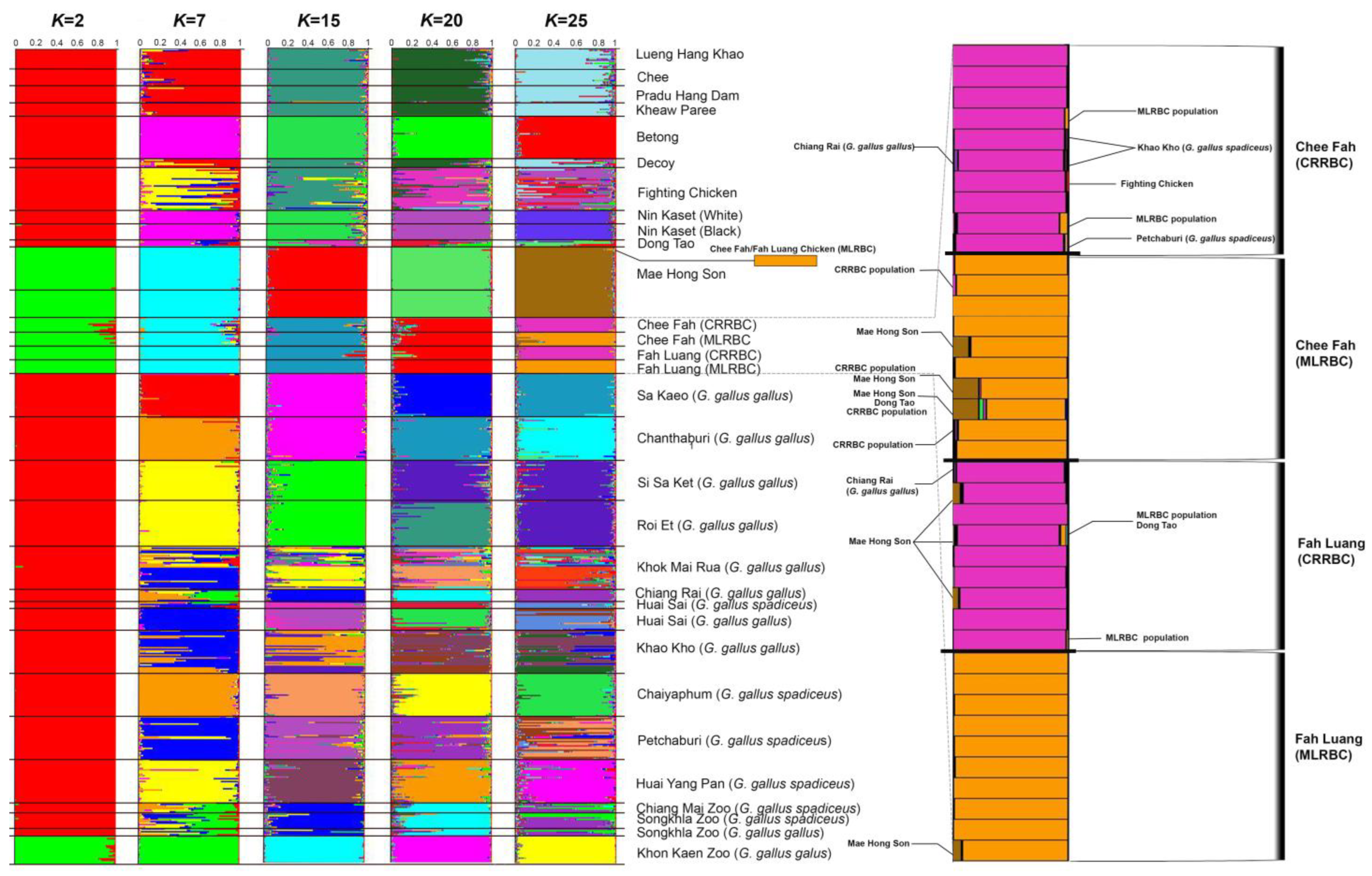
3.5. Genetic Variability of Chee Fah and Fah Luang Chicken Populations Based on Microsatellite Data
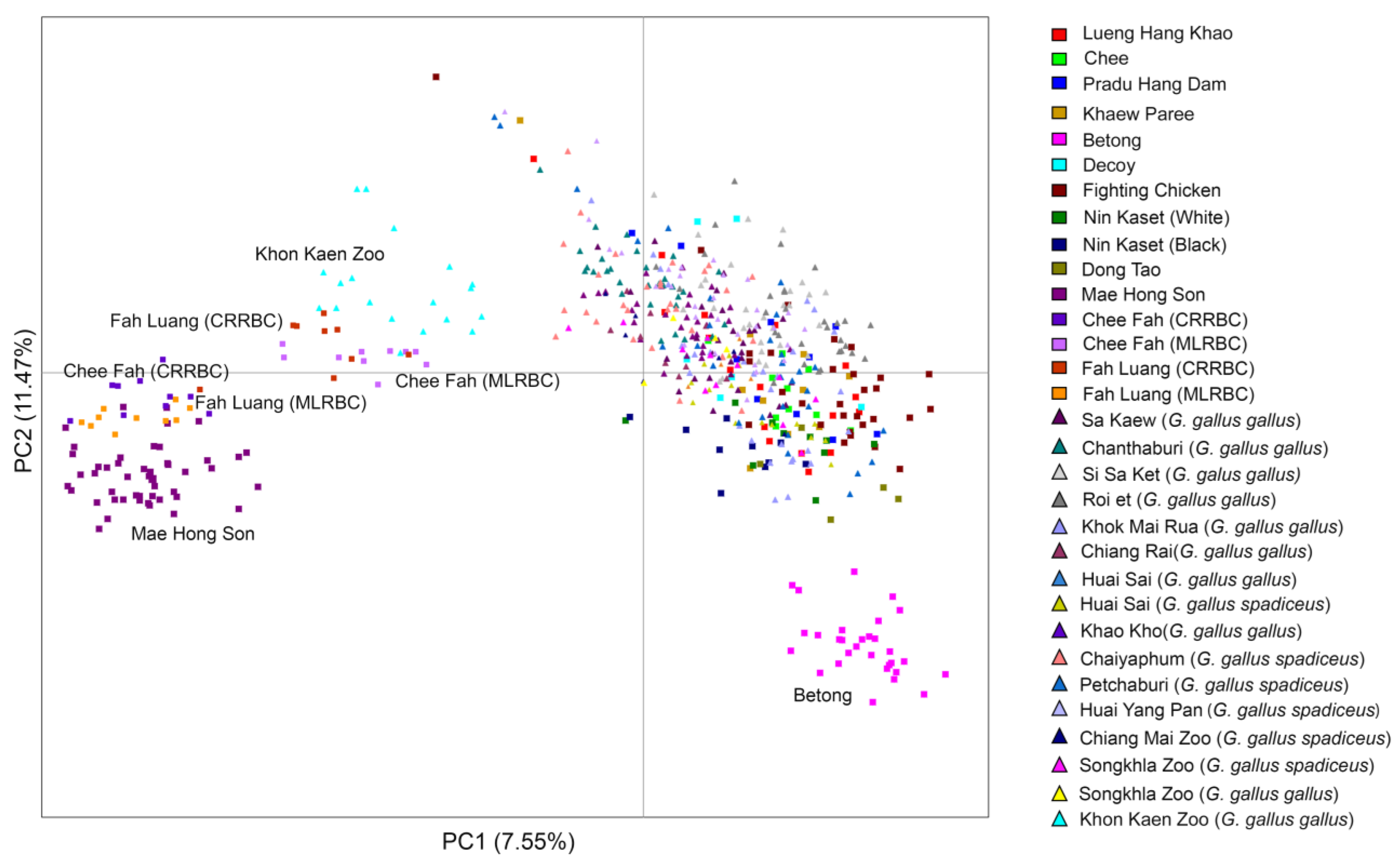
3.6. Genetic Differences among Chee Fah Chickens, Fah Luang Chickens, Red Junglefowl, and Other Thai Domestic Chicken Breeds
4. Discussion
4.1. Lineage of Chee Fah and Fah Luang Chickens Is the Same as Chinese Black-Boned Chicken Breeds
4.2. Introgression of Red Junglefowl and Thai Domestic Chicken Breeds into Chee Fah and Fah Luang Chickens
4.3. Chee Fah and Fah Luang Chicken Breeds from Two Localities Show Different Population Structures, Different Gene Pool Origins, and Potential Signs of Adaptation to High Elevation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noito, K.; Suwanasopee, T.; Koonawootrittriron, S. Evaluation of quality and nutrient contents of eggs in Nin Kaset black-meat chickens at 25 to 37 weeks of age. Khon Kaen Agr. J. 2019, 47, 369–374. [Google Scholar]
- Qu, Y.; Zhao, H.; Han, N.; Zhou, G.; Song, G.; Gao, B.; Tian, S.; Zhang, J.; Zhang, R.; Meng, X.; et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 2013, 4, 2071. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Zhou, X.; Phuntsok, T.; Zhao, N.; Zhang, D.; Ning, C.; Li, D.; Zhao, H. Genomic analyses reveal genetic adaptations to tropical climates in chickens. IScience 2020, 23, 101644. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, A.F.; Zulkifli, I. Effects of high ambient temperature on blood parameters in red jungle fowl, village fowl and broiler chickens. J. Anim. Vet. Adv. 2010, 9, 1201–1207. [Google Scholar] [CrossRef]
- Pawar, S.S.; Basavaraj, S.; Dhansing, L.V.; Pandurang, K.N.; Sahebrao, K.A.; Vitthal, N.A.; Pandit, B.M.; Kumar, B.S. Assessing and mitigating the impact of heat stress in poultry. Adv. Anim. Vet. Sci. 2016, 4, 332–341. [Google Scholar] [CrossRef]
- Azoulay, Y.; Druyan, S.; Yadgary, L.; Hadad, Y.; Cahaner, A. The viability and performance under hot conditions of featherless broilers versus fully feathered broilers. Poult. Sci. 2011, 90, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Wolc, A.; Arango, J.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Dekkers, J.C.M. Genome wide association study for heat stress induced mortality in a white egg layer line. Poult. Sci. 2018, 98, 92–96. [Google Scholar] [CrossRef]
- Kang, S.; Kim, D.H.; Lee, S.; Lee, T.; Lee, K.W.; Chang, H.H.; Moon, B.; Ayasan, T.; Choi, Y.H. An acute, rather than progressive, increase in temperature-humidity index has severe effects on mortality in laying hens. Front. Vet. Sci. 2020, 7, 568093. [Google Scholar] [CrossRef]
- Boonkum, W.; Duangjinda, M.; Kananit, S.; Chankitisakul, V.; Kenchaiwong, W. Genetic effect and growth curve parameter estimation under heat stress in slow-growing Thai native chickens. Vet. Sci. 2021, 8, 297. [Google Scholar] [CrossRef]
- Soleimani, A.F.; Zulkifli, I.; Omar, A.R.; Raha, A.R. Physiological responses of 3 chicken breeds to acute heat stress. Poult. Sci. 2011, 90, 1435–1440. [Google Scholar] [CrossRef]
- Gu, J.; Liang, Q.; Liu, C.; Li, S. Genomic analyses reveal adaptation to hot arid and harsh environments in native chickens of China. Front. Genet. 2020, 11, 582355. [Google Scholar] [CrossRef] [PubMed]
- Nanaei, A.H.; Kharrati-Koopaee, H.; Esmailizadeh, A. Genetic diversity and signatures of selection for heat tolerance and immune response in Iranian native chickens. BMC Genom. 2022, 23, 224. [Google Scholar] [CrossRef]
- Duangjinda, M.; Tunim, S.; Duangdaen, C.; Boonkum, W. Hsp70 genotypes and heat tolerance of commercial and native chickens reared in hot and humid conditions. Braz. J. Poult. Sci. 2017, 19, 7–18. [Google Scholar] [CrossRef]
- Nawab, A.; Ibtisham, F.; Li, G.; Kieser, B.; Wu, J.; Liu, W.; Zhao, Y.; Nawab, Y.; Li, K.; Xiao, M.; et al. Heat stress in poultry production: Mitigation strategies to overcome the future challenges facing the global poultry industry. J. Therm. Biol. 2018, 78, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Tarachai, P. Poultry breed and breeding. In Poultry Production; Maejo University: Chiang Mai, Thailand, 2017; Available online: http://www.as2.mju.ac.th/E-Book/t_prapakorn/%E0%B8%AA%E0%B8%A8241/ (accessed on 20 December 2022).
- Buranawit, K.; Chailungka, C.; Wongsunsri, C.; Laenoi, W. Phenotypic characterization of Thai native black-bone chickens indigenous to northern Thailand. Thai J. Vet. Med. 2016, 46, 547–554. [Google Scholar]
- Jaturasitha, S.; Srikanchai, T.; Kreuzer, M.; Wicke, M. Differences in carcass and meat characteristics between chicken indigenous to northern Thailand (Black-boned and Thai native) and imported extensive breeds (Bresse and Rhode Island red). Poult. Sci. 2008, 87, 160–169. [Google Scholar] [CrossRef]
- Lengkidworraphiphat, P.; Wongpoomchai, R.; Taya, S.; Jaturasitha, S. Effect of genotypes on macronutrients and antioxidant capacity of chicken breast meat. Asian-Australas. J. Anim. Sci. 2020, 33, 1817–1823. [Google Scholar] [CrossRef]
- Prapattong, P. Dynmics of Being Yunnanese–Chinese in North Thailand: The Integrations into Thai-State. Doctoral Dissertation, Graduate School, Mae Fah Luang University,, Chiang Rai, Thailand, 2010. [Google Scholar]
- Buranawit, K.; Laenoi, W. Genetic parameters for production traits in F1 reciprocal crossbred Chee Fah and Fah Luang chickens. Anim. Prod. Sci. 2021, 62, 114–120. [Google Scholar] [CrossRef]
- Choprakarn, C.; Wongpichet, K. Village chicken production systems in Thailand. In Proceedings of the The International Poultry Conference, Bangkok, Thailand, 5–7 November 2007. [Google Scholar]
- Intarachote, U.; Namkhun, S.; Leotaragul, A. Selection and improvement regional native chickens (Fahluang chickens) for raising in northern highland of Thailand. 1. Productive performance and genetic parameters of Fahluang chickens at generation 1. In Proceedings of the 41th Kasetsart University Annual Conference; Kasetsart University: Bangkok, Thailand, 2003; pp. 434–444, (Article in Thai with an English Abstract). [Google Scholar]
- Morathop, S.; Leotaragul, A.; Limwatthana, C. Selection and Improvement Regional Native Chickens (Chee Fah chicken) for Raising in the Northern Highland of Thailand; The Royal Project Foundation: Chiang Mai, Thailand, 2005. [Google Scholar]
- Harintharanon, T. Food Security. Bureau of Livestock Standards and Certification, Department of Livestock Development. Available online: https://certify.dld.go.th/certify/images/research/2563/630923/Food%20security.pdf (accessed on 15 January 2023).
- Malomane, D.K.; Weigend, S.; Schmitt, A.O.; Weigend, A.; Reimer, C.; Simianer, H. Genetic diversity in global chicken breeds in relation to their genetic distances to wild populations. Genet. Sel. Evol. 2021, 53, 36. [Google Scholar] [CrossRef]
- Eda, M.; Shoocongdej, R.; Auetrakulvit, P.; Kachajiwa, J. The history of chicken and other bird exploitation in Thailand: Preliminary analysis of bird remains from four archaeological sites. Int. J. Osteoarchaeol. 2019, 29, 231–237. [Google Scholar] [CrossRef]
- Peters, J.; Lebrasseur, O.; Irving-Pease, E.K.; Paxinos, P.D.; Best, J.; Smallman, R.; Callou, C.; Gardeisen, A.; Trixl, S.; Frantz, L.; et al. The biocultural origins and dispersal of domestic chickens. Proc. Natl. Acad. Sci. USA 2022, 119, e2121978119. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Nunome, M.; Suwanasopee, T.; Duengkae, P.; Chaiwatana, S.; Chamchumroon, W.; Suzuki, T.; Koonawootrittriron, S.; Matsuda, Y.; Srikulnath, K. Origin and evolutionary history of domestic chickens inferred from a large population study of Thai red junglefowl and indigenous chickens. Sci. Rep. 2021, 11, 2035. [Google Scholar] [CrossRef] [PubMed]
- Singchat, W.; Chaiyes, A.; Wongloet, W.; Ariyaraphong, N.; Jaisamut, K.; Panthum, T.; Ahmad, S.F.; Chaleekarn, W.; Suksavate, W.; Inpota, M.; et al. Red junglefowl resource management guide: Bioresource reintroduction for sustainable food security in Thailand. Sustainability 2022, 14, 7895. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Supikamolseni, A.; Ngaoburanawit, N.; Sumontha, M.; Chanhome, L.; Suntrarachun, S.; Peyachoknagul, S.; Srikulnath, K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 2015, 14, 13981–13997. [Google Scholar] [CrossRef]
- Nishibori, M.; Hayashi, T.; Tsudzuki, M.; Yamamoto, Y.; Yasue, H. Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species. Anim. Genet. 2001, 32, 380–385. [Google Scholar] [CrossRef]
- Miao, Y.W.; Peng, M.S.; Wu, G.S.; Ouyang, Y.N.; Yang, Z.Y.; Yu, N.; Liang, J.P.; Pianchou, G.; Beja-Pereira, A.; Mitra, B.; et al. Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity 2013, 110, 277–282. [Google Scholar] [CrossRef]
- Tajima, A.; Pan, I.H.; Fucharoen, G.; Fucharoen, S.; Matsuo, M.; Tokunaga, K.; Juji, T.; Hayami, M.; Omoto, K.; Horai, S. Three major lineages of Asian Y chromosomes: Implications for the peopling of east and southeast Asia. Hum. Genet 2002, 110, 80–88. [Google Scholar] [CrossRef]
- Bentley, R.A.; Pietrusewsky, M.; Douglas, M.T.; Atkinson, T.C. Matrilocality during the prehistoric transition to agriculture in Thailand? Antiquity 2005, 79, 865–881. [Google Scholar] [CrossRef]
- Zhang, Y.; Colli, L.; Barker, J.S.F. Asian water buffalo: Domestication, history and genetics. Anim. Genet 2020, 51, 177–191. [Google Scholar] [CrossRef]
- Godinez, C.J.P.; Layos, J.K.N.; Yamamoto, Y.; Kunieda, T.; Duangjinda, M.; Liao, L.M.; Huang, X.H.; Nishibori, M. Unveiling new perspective of phylogeography, genetic diversity, and population dynamics of Southeast Asian and Pacific chickens. Sci. Rep. 2022, 12, 14609. [Google Scholar] [CrossRef] [PubMed]
- Walker, A. Matrilinial spirits, descend and territorial power in Northern Thailand. Aust. J. Anthropol. 2006, 17, 196–215. [Google Scholar] [CrossRef]
- Yaemkong, S.; Rattanapradit, P.; Ngoc, T.N.; Charoensook, R.; Chirarat, N.; Soipethand, U.; Yaemkong, S. Diversity of traditional knowledge and local wisdom of indigenous chickens farmers in Bang Krathum, Nakhon Thai, Mueang and Chat Trakan districts Phitsanulok province. J. Appl. Anim. Res. 2017, 10, 39–46. [Google Scholar]
- Huang, X.; Weng, Z.; He, Y.; Miao, Y.; Luo, W.; Zhang, X.; Zhong, F.; Du, B. Mitochondrial DNA diversity and demographic history of Black-boned chickens in China. Mitochondrial DNA B Resour. 2021, 6, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Dorshorst, B.; Molin, A.M.; Rubin, C.J.; Johansson, A.M.; Stromstedt, L.; Pham, M.H.; Chen, C.F.; Hallbook, F.; Ashwell, C.; Andersson, L. A complex genomic rearrangement involving the endothelin 3 locus causes dermal hyperpigmentation in the chicken. PLoS Genet. 2011, 7, e1002412. [Google Scholar] [CrossRef]
- Shinomiya, A.; Kayashima, Y.; Kinoshita, K.; Mizutani, M.; Namikawa, T.; Matsuda, Y.; Akiyama, T. Gene duplication of endothelin 3 is closely correlated with the hyperpigmentation of the internal organs (Fibromelanosis) in silky chickens. Genetics 2012, 190, 627–638. [Google Scholar] [CrossRef]
- Lawal, R.A.; Martin, S.H.; Vanmechelen, K.; Vereijken, A.; Silva, P.; Al-Atiyat, R.M.; Aljumaah, R.S.; Mwacharo, J.M.; Wu, D.D.; Zhang, Y.P.; et al. The wild species genome ancestry of domestic chickens. BMC Biol. 2020, 18, 13. [Google Scholar] [CrossRef]
- Montgomery, M.E.W.; Nurthen, L.M.; Roderick, K.; Gilligan, D.M.; Briscoe, D.A.; Frankham, R. Relationships between population size and loss of genetic diversity: Comparisons of experimental results with theoretical predictions. Conserv. Genet. 2000, 1, 33–43. [Google Scholar] [CrossRef]
- Shi, S.; Shao, D.; Yang, L.; Liang, Q.; Han, W.; Xue, Q.; Qu, L.; Leng, L.; Li, Y.; Zhao, X.; et al. Whole genome analyses reveal novel genes associated with chicken adaptation to tropical and frigid environments. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- Harmon, L.J.; Braude, S. Conservation of small populations: Effective population sizes, inbreeding, and the 50/500 rule. In An Introduction to Methods and Models in Ecology, Evolution, and Conservation Biology; Braude, S., loe, S., Eds.; Princeton University Press: Princeton, NJ, USA, 2010; pp. 125–138. [Google Scholar] [CrossRef]
- Wolc, A.; Zhao, H.H.; Arango, J.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Preisinger, R.; Stricker, C.; Habier, D.; Fernando, R.L.; et al. Response and inbreeding from a genomic selection experiment in layer chickens. Genet. Sel. Evol. 2015, 47, 59. [Google Scholar] [CrossRef]
- Elferink, M.G.; Megens, H.J.; Vereijken, A.; Hu, X.; Crooijmans, R.P.M.A.; Groenen, M.A. Signatures of selection in the genomes of commercial and non-commercial chicken breeds. PLoS ONE 2012, 7, e32720. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.; Pettersson, M.E.; Honaker, C.F.; Siegel, P.B.; Carlborg, Ö. Standing genetic variation as a major contributor to adaptation in the Virginia chicken lines selection experiment. Genome Biol. 2015, 16, 219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, C.X.; Chamba, Y.; Ling, Y. Blood characteristics for high altitude adaptation in Tibetan chickens. Poult. Sci. 2007, 86, 1384–1389. [Google Scholar] [CrossRef]
- Wang, M.S.; Li, Y.; Peng, M.S.; Zhong, L.; Wang, Z.J.; Li, Q.Y.; Tu, X.L.; Dong, Y.; Zhu, C.L.; Wang, L.; et al. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 2015, 32, 1880–1889. [Google Scholar] [CrossRef]
- Yuan, J.; Li, S.; Sheng, Z.; Zhang, M.; Liu, X.; Yuan, Z.; Yang, N.; Chen, J. Genome-wide run of homozygosity analysis reveals candidate genomic regions associated with environmental adaptations of Tibetan native chickens. BMC Genom. 2022, 23, 91. [Google Scholar] [CrossRef] [PubMed]
- Morehouse, S. The arc/info geographic information system. Comput. Geosci. 1992, 18, 435–441. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef]
- Potapov, P.; Hansen, M.C.; Kommareddy, I.; Kommareddy, A.; Turubanova, S.; Pickens, A.; Adusei, B.; Tyukavina, A.; Ying, Q. Landsat analysis ready data for global land cover and land cover change mapping. Remote Sens. 2020, 12, 246. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudı´k, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Pearson, R.G.; Raxworthy, C.J.; Nakamura, M.; Peterson, A.T. Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. J. Biogeogr. 2006, 34, 102–117. [Google Scholar] [CrossRef]
- Wisz, M.S.; Hijmans, R.J.; Li, J.; Peterson, A.T.; Graham, C.H.; Guisan, A. Effects of sample size on the performance of species distribution models. Divers. Distrib. 2008, 14, 763–773. [Google Scholar] [CrossRef]
- Baldwin, R.A. Use of maximum entropy modeling in wildlife research. Entropy 2009, 11, 854–866. [Google Scholar] [CrossRef]
- Araujo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Marmion, M.; Parviainen, M.; Luoto, M.; Heikkinen, R.K.; Thuiller, W. Evaluation of consensus methods in predictive species distribution modelling. Divers. Distrib. 2009, 15, 59–69. [Google Scholar] [CrossRef]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Wang, L.; Hu, L.; Gong, P. Comparison of classification algorithms and training sample sizes in urban land classification with Landsat thematic mapper imagery. Remote Sens. 2014, 6, 964–983. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1 km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium (IPDPS 2002), Fort Lauderdale, FL, USA, 15–19 April 2002. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic. Acids. Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Letunik, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E. A Monte Carlo method for combined segregation and linkage analysis. Am. J. Hum. Genet. 1992, 51, 1111–1126. [Google Scholar]
- Raymond, M.; Rousset, F. An exact test for population differentiation. Evolution 1995, 49, 1280–1283. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Welch, B.L. The generalization of ‘STUDENT’S’problem when several different population varlances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Park, S.D.E. The Excel Microsatellite Toolkit (version 3.1); Animal Genomics Laboratory, University College Dublin: Dublin, Ireland, 2001. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [CrossRef]
- Wang, J. COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 2011, 11, 141–145. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A program for detecting recent effective population size reductions from allele data frequencies. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]

| Breed | Population | N | Number of Haplotypes (H) | Theta (Per Site) from S | Average Number of Nucleotide Differences (k) | Overall Haplotype (h) | Nucleotide Diversities (π) |
|---|---|---|---|---|---|---|---|
| Chee Fah | MLRBC 1 | 10 | 9 | 0.004 | 3.689 | 0.978 ± 0.054 | 0.004 ± 0.00047 |
| CRRBC 2 | 10 | 8 | 0.009 | 6.644 | 0.933 ± 0.077 | 0.007 ± 0.00144 | |
| Fah Luang | MLRBC 1 | 10 | 10 | 0.011 | 7.600 | 1.000 ± 0.045 | 0.008 ± 0.00144 |
| CRRBC 2 | 9 | 8 | 0.005 | 5.611 | 0.972 ± 0.064 | 0.006 ± 0.00103 | |
| Overall | 39 | 18 | 0.00853 | 6.959 | 0.994 ± 0.019 | 0.007 ± 0.00083 | |
| Breed | Population | Na1 | AR2 | Nea3 | I4 | Ho5 | He6 | PIC7 | F8 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Chee Fah | MLRBC 9 | Mean | 3.750 | 3.676 | 2.535 | 1.007 | 0.682 | 0.549 | 0.498 | −0.208 |
| S.E. | 0.228 | 1.163 | 0.171 | 0.071 | 0.064 | 0.035 | 0.176 | 0.077 | ||
| CRRBC 10 | Mean | 5.393 | 5.224 | 3.525 | 1.369 | 0.441 | 0.680 | 0.635 | 0.350 | |
| S.E. | 0.346 | 1.712 | 0.214 | 0.071 | 0.058 | 0.025 | 0.144 | 0.081 | ||
| Total | Mean | 4.571 | 4.450 | 3.030 | 1.188 | 0.562 | 0.614 | 0.566 | 0.076 | |
| S.E. | 0.233 | 1.650 | 0.151 | 0.055 | 0.046 | 0.023 | 0.174 | 0.067 | ||
| Fah Luang | MLRBC 9 | Mean | 4.857 | 4.703 | 3.039 | 1.208 | 0.669 | 0.617 | 0.569 | −0.092 |
| S.E. | 0.320 | 1.62 | 0.241 | 0.073 | 0.058 | 0.029 | 0.155 | 0.078 | ||
| CRRBC 10 | Mean | 5.107 | 4.765 | 3.609 | 1.342 | 0.446 | 0.669 | 0.628 | −0.092 | |
| S.E. | 0.323 | 1.535 | 0.253 | 0.078 | 0.046 | 0.032 | 0.172 | 0.078 | ||
| Total | Mean | 4.982 | 4.734 | 3.324 | 1.275 | 0.558 | 0.643 | 0.598 | 0.115 | |
| S.E. | 0.226 | 1.564 | 0.177 | 0.054 | 0.040 | 0.022 | 0.165 | 0.056 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budi, T.; Singchat, W.; Tanglertpaibul, N.; Wongloet, W.; Chaiyes, A.; Ariyaraphong, N.; Thienpreecha, W.; Wannakan, W.; Mungmee, A.; Thong, T.; et al. Thai Local Chicken Breeds, Chee Fah and Fah Luang, Originated from Chinese Black-Boned Chicken with Introgression of Red Junglefowl and Domestic Chicken Breeds. Sustainability 2023, 15, 6878. https://doi.org/10.3390/su15086878
Budi T, Singchat W, Tanglertpaibul N, Wongloet W, Chaiyes A, Ariyaraphong N, Thienpreecha W, Wannakan W, Mungmee A, Thong T, et al. Thai Local Chicken Breeds, Chee Fah and Fah Luang, Originated from Chinese Black-Boned Chicken with Introgression of Red Junglefowl and Domestic Chicken Breeds. Sustainability. 2023; 15(8):6878. https://doi.org/10.3390/su15086878
Chicago/Turabian StyleBudi, Trifan, Worapong Singchat, Nivit Tanglertpaibul, Wongsathit Wongloet, Aingorn Chaiyes, Nattakan Ariyaraphong, Worawit Thienpreecha, Wannapa Wannakan, Autchariyapron Mungmee, Thanyapat Thong, and et al. 2023. "Thai Local Chicken Breeds, Chee Fah and Fah Luang, Originated from Chinese Black-Boned Chicken with Introgression of Red Junglefowl and Domestic Chicken Breeds" Sustainability 15, no. 8: 6878. https://doi.org/10.3390/su15086878
APA StyleBudi, T., Singchat, W., Tanglertpaibul, N., Wongloet, W., Chaiyes, A., Ariyaraphong, N., Thienpreecha, W., Wannakan, W., Mungmee, A., Thong, T., Wattanadilokchatkun, P., Panthum, T., Ahmad, S. F., Lisachov, A., Muangmai, N., Chuenka, R., Prapattong, P., Nunome, M., Chamchumroon, W., ... Srikulnath, K. (2023). Thai Local Chicken Breeds, Chee Fah and Fah Luang, Originated from Chinese Black-Boned Chicken with Introgression of Red Junglefowl and Domestic Chicken Breeds. Sustainability, 15(8), 6878. https://doi.org/10.3390/su15086878











