Comparison of Bacterial Diversity in the Rhizosphere of Chromolaena odorata (L.) R.M. King and H.Rob. in Different Habitats
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sampling
2.2. Soil Microbe Determination and Analysis
2.3. Statistical Analysis
3. Results
3.1. Invasion Characteristics and Soil Physicochemical Properties of Chromolaena odorata in Three Habitats
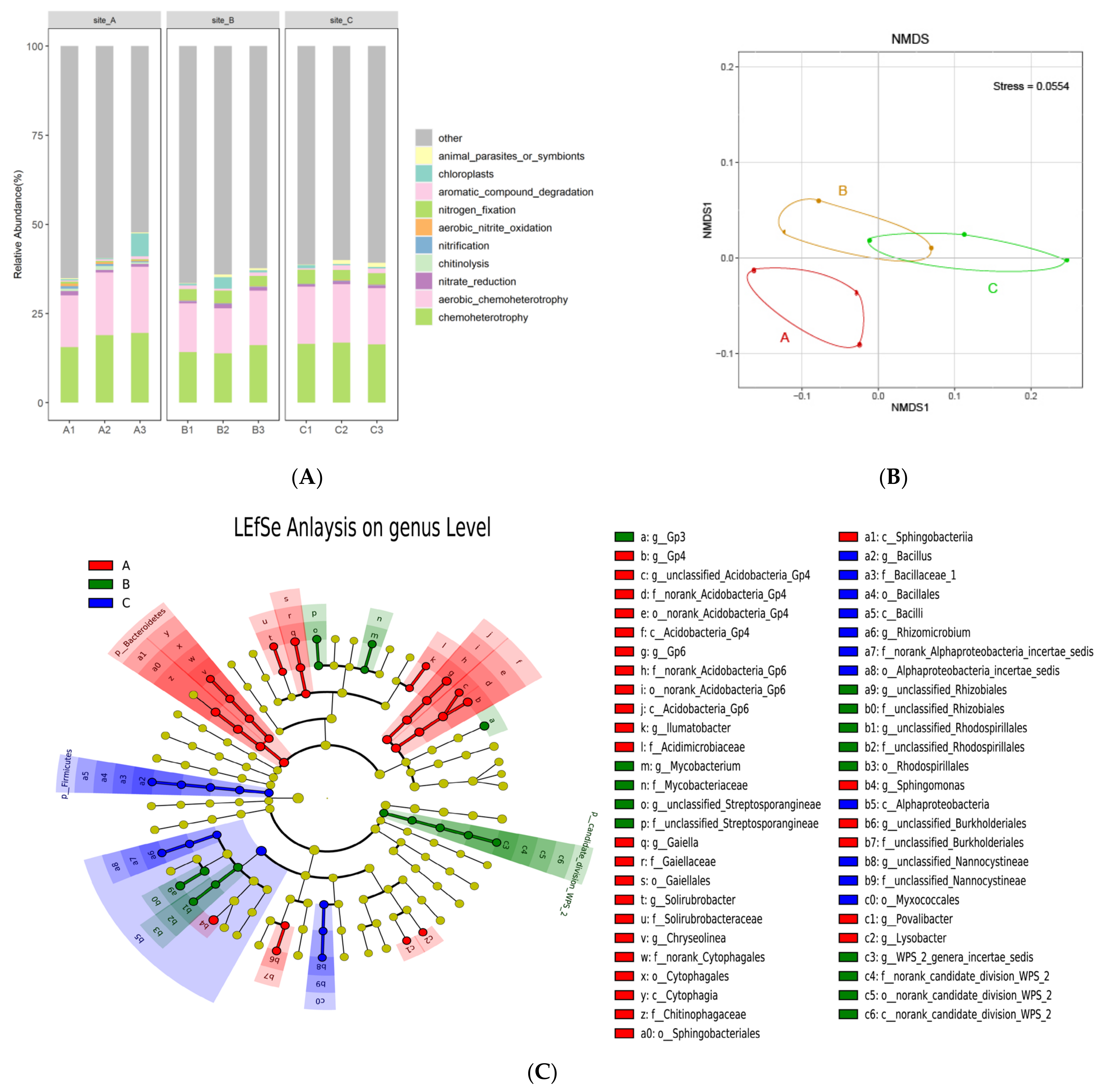
3.2. Community Composition of Rhizosphere Bacteria of Chromolaena odorata
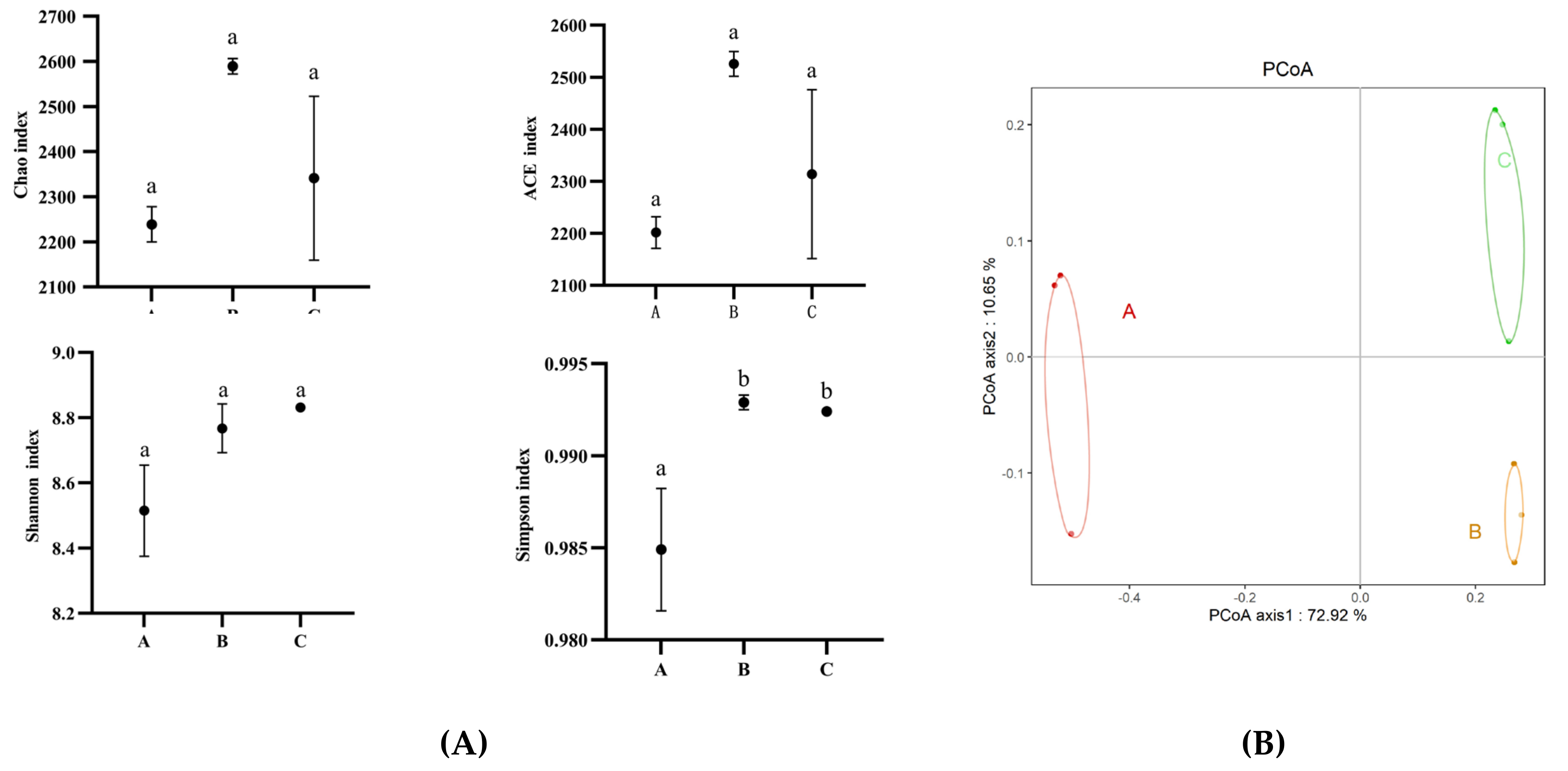
3.3. Diversity of Rhizosphere Bacterial Community of Chromolaena odorata
3.4. Functional Prediction of Rhizosphere Bacteria of Chromolaena odorata
3.5. Analysis of Influence Factors of Rhizosphere Bacterial Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bellard, C.; Cassey, P.; Blackburn, T.M. Alien species as a driver of recent extinctions. Biol. Lett. 2016, 12, 20150623. [Google Scholar] [CrossRef] [PubMed]
- Foxcroft, L.C.; Pysek, P.; Richardson, D.M.; Pergl, J.; Hulme, P.E. The Bottom Line: Impacts of Alien Plant Invasions in Protected Areas. In Plant Invasions in Protected Areas: Patterns, Problems and Challenges; Foxcroft, L.C., Pysek, P., Richardson, D.M., Genovesi, P., Eds.; Invading Nature-Springer Series in Invasion Ecology; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7, pp. 19–41. [Google Scholar]
- Milbau, A.; Stout, J.C.; Graae, B.J.; Nijs, I. A hierarchical framework for integrating invasibility experiments incorporating different factors and spatial scales. Biol. Invasions 2009, 11, 941–950. [Google Scholar] [CrossRef]
- Steinlein, T. Invasive Alien Plants and Their Effects on Native Microbial Soil Communities. In Progress in Botany 74; Luttge, U., Beyschlag, W., Francis, D., Cushman, J., Eds.; Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2013; Volume 74, pp. 293–319. [Google Scholar]
- Zhang, Z.; Liu, Y.; Brunel, C.; van Kleunen, M. Soil-microorganism-mediated invasional meltdown in plants. Nat. Ecol. Evol. 2020, 4, 1612–1621. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.E.; Wandrag, E.M.; Barrett, L.G.; Thrall, P.H.; Duncan, R.P. Soil biotic effects and competition; What are the mechanisms behind being a successful invader? Pedobiologia 2021, 87–88, 150749. [Google Scholar] [CrossRef]
- Bever, J.D. Soil community feedback and the coexistence of competitors: Conceptual frameworks and empirical tests. New Phytol. 2003, 157, 465–473. [Google Scholar] [CrossRef]
- Reinhart, K.O.; Callaway, R.M. Soil biota facilitate exotic Acer invasions in Europe and North America. Ecol. Appl. 2004, 14, 1737–1745. [Google Scholar] [CrossRef]
- Richardson, A.E.; Barea, J.-M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Yu, K.; Pieterse, C.M.J.; Bakker, P.A.H.M.; Berendsen, R.L. Beneficial microbes going underground of root immunity. Plant Cell Environ. 2019, 42, 2860–2870. [Google Scholar] [CrossRef]
- Marasco, R.; Rolli, E.; Ettoumi, B.; Vigani, G.; Mapelli, F.; Borin, S.; Abou-Hadid, A.F.; El-Behairy, U.A.; Sorlini, C.; Cherif, A.; et al. A Drought Resistance-Promoting Microbiome Is Selected by Root System under Desert Farming. PLoS ONE 2012, 7, e48479. [Google Scholar] [CrossRef]
- Van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant-soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Callaway, R.M.; Thelen, G.C.; Rodriguez, A.; Holben, W.E. Soil biota and exotic plant invasion. Nature 2004, 427, 731–733. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moenne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dye, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Hoque, M.N.; Hannan, A.; Imran, S.; Paul, N.C.; Mondal, M.F.; Sadhin, M.M.R.; Bristi, J.M.; Dola, F.S.; Abu Hanif, M.; Ye, W.; et al. Plant Growth-Promoting Rhizobacteria-Mediated Adaptive Responses of Plants Under Salinity Stress. J. Plant Growth Regul. 2022. [Google Scholar] [CrossRef]
- Yadegari, M.; Rahmani, H.A.; Noormohammadi, G.; Ayneband, A. Plant Growth Promoting Rhizobacteria Increase Growth, Yield and Nitrogen Fixation in Phaseolus vulgaris. J. Plant Nutr. 2010, 33, 1733–1743. [Google Scholar] [CrossRef]
- Cassan, F.; Vanderleyden, J.; Spaepen, S. Physiological and Agronomical Aspects of Phytohormone Production by Model Plant-Growth-Promoting Rhizobacteria (PGPR) Belonging to the Genus Azospirillum. J. Plant Growth Regul. 2014, 33, 440–459. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Chen, L.; Li, K.; Shang, J.; Wu, Y.; Chen, T.; Wanyan, Y.; Wang, E.; Tian, C.; Chen, W.; Chen, W.; et al. Plant growth-promoting bacteria improve maize growth through reshaping the rhizobacterial community in low-nitrogen and low-phosphorus soil. Biol. Fertil. Soils 2021, 57, 1075–1088. [Google Scholar] [CrossRef]
- Rout, M.E.; Callaway, R.M. Interactions between exotic invasive plants and soil microbes in the rhizosphere suggest that ‘everything is not everywhere’. Ann. Bot. 2012, 110, 213–222. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, Z.; Yang, G.; He, H.; Liu, H. Microbial activity and diversity in the rhizosphere soil of the invasive species Zizania latifolia in the wetland of Wuchang Lake, China. Mar. Freshw. Res. 2020, 71, 1702–1713. [Google Scholar] [CrossRef]
- Goodall, J.M.; Erasmus, D.J. Review of the status and integrated control of the invasive alien weed, Chromolaena odorata, in South Africa. Agric. Ecosyst. Environ. 1996, 56, 151–164. [Google Scholar] [CrossRef]
- Liao, Z.-Y.; Scheepens, J.F.; Li, Q.-M.; Wang, W.-B.; Feng, Y.-L.; Zheng, Y.-L. Founder effects, post-introduction evolution and phenotypic plasticity contribute to invasion success of a genetically impoverished invader. Oecologia 2020, 192, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.-Y.; Scheepens, J.F.; Li, W.-T.; Wang, R.-F.; Zheng, Y.-L.; Feng, Y.-L. Biomass reallocation and increased plasticity might contribute to successful invasion of Chromolaena odorata. Flora 2019, 256, 79–84. [Google Scholar] [CrossRef]
- Zheng, Y.-L.; Feng, Y.-L.; Zhang, L.-K.; Callaway, R.M.; Valiente-Banuet, A.; Luo, D.-Q.; Liao, Z.-Y.; Lei, Y.-B.; Barclay, G.F.; Silva-Pereyra, C. Integrating novel chemical weapons and evolutionarily increased competitive ability in success of a tropical invader. New Phytol. 2015, 205, 1350–1359. [Google Scholar] [CrossRef]
- Zhao, J.; Cao, L. Effects of invasive plants on soil microbial properties and soil enzyme activities of poplar forest in Hung-tse Lake Wetland. Ecol. Environ. Sci. 2018, 27, 2039–2046. [Google Scholar] [CrossRef]
- Li, W.; Song, Q.; Huang, R.; Zhao, J.; LI, Y.; Zhou, Q. Current status and conservation of Franois’ langurs (Trachypithecus francoisi) in Encheng National Nature Reserve in Guangxi, China. Acta Theriol. Sin. 2019, 39, 623–629. [Google Scholar] [CrossRef]
- Li, Q.; Wan, F.; Zhao, M. Distinct soil microbial communities under Ageratina adenophora invasions. Plant Biol. 2022, 24, 430–439. [Google Scholar] [CrossRef]
- Xu, Q.-Y.; Wang, D.; Quan, G.-M.; Xiang, H.-M.; Zhang, J.-E. Invasive Chromolaena odorata species specifically affects growth of its co-occurring weeds. Ann. N. Y. Acad. Sci. 2020, 1470, 57–66. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Nunan, N. The microbial habitat in soil: Scale, heterogeneity and functional consequences. J. Plant Nutr. Soil Sci. 2017, 180, 425–429. [Google Scholar] [CrossRef]
- Chen, Q.-L.; Ding, J.; Zhu, Y.-G.; He, J.-Z.; Hu, H.-W. Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environ. Int. 2020, 140, 105766. [Google Scholar] [CrossRef]
- Zhang, P.; Li, B.; Wu, J.; Hu, S. Invasive plants differentially affect soil biota through litter and rhizosphere pathways: A meta-analysis. Ecol. Lett. 2019, 22, 200–210. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. Jove-J. Vis. Exp. 2022, 183, e61715. [Google Scholar] [CrossRef]
- Nihorimbere, V.; Ongena, M.; Smargiassi, M.; Thonart, P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. Et Environ. 2011, 15, 327–337. [Google Scholar]
- Cheng, D.; Tian, Z.; Feng, L.; Xu, L.; Wang, H. Diversity analysis of the rhizospheric and endophytic bacterial communities of Senecio vulgaris L. (Asteraceae) in an invasive range. Peerj 2019, 6, e6162. [Google Scholar] [CrossRef]
- Calvo, P.; Zebelo, S.; McNear, D.; Kloepper, J.; Fadamiro, H. Plant growth-promoting rhizobacteria induce changes in Arabidopsis thaliana gene expression of nitrate and ammonium uptake genes. J. Plant Interact. 2019, 14, 224–231. [Google Scholar] [CrossRef]
- Thiruvengadam, R.; Gandhi, K.; Vaithiyanathan, S.; Sankarasubramanian, H.; Loganathan, K.; Lingan, R.; Rajagopalan, V.R.; Muthurajan, R.; Iyadurai, J.E.; Kuppusami, P. Complete Genome Sequence Analysis of Bacillus subtilis Bbv57, a Promising Biocontrol Agent against Phytopathogens. Int. J. Mol. Sci. 2022, 23, 9732. [Google Scholar] [CrossRef]
- Sun, Y.-Y.; Zhang, Q.-X.; Zhao, Y.-P.; Diao, Y.-H.; Gui, F.-R.; Yang, G.-Q. Beneficial rhizobacterium provides positive plant-soil feedback effects to Ageratina adenophora. J. Integr. Agric. 2021, 20, 1327–1335. [Google Scholar] [CrossRef]
- Tilak, K.; Ranganayaki, N.; Pal, K.K.; De, R.; Saxena, A.K.; Nautiyal, C.S.; Mittal, S.; Tripathi, A.K.; Johri, B.N. Diversity of plant growth and soil health supporting bacteria. Curr. Sci. 2005, 89, 136–150. [Google Scholar]
- Wu, S.; Xue, S.; Iqbal, Y.; Xing, H.; Jie, Y. Seasonal Nutrient Cycling and Enrichment of Nutrient-Related Soil Microbes Aid in the Adaptation of Ramie (Boehmeria nivea L.) to Nutrient-Deficient Conditions. Front. Plant Sci. 2021, 12, 644904. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Ka, J.-O.; Cho, J.-C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. Fems Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.J.; Geppert, A.M.; Wanner, G.; Foesel, B.U.; Wuest, P.K.; Overmann, J. The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp nov., isolated from subtropical savannah soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 2971–2979. [Google Scholar] [CrossRef]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can We Use Functional Annotation of Prokaryotic Taxa (FAPROTAX) to Assign the Ecological Functions of Soil Bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
- Ni, G.; Zhao, P.; Huang, Q.; Zhu, L.; Hou, Y.; Yu, Y.; Ye, Y.; Ouyang, L. Mikania micrantha invasion enhances the carbon (C) transfer from plant to soil and mediates the soil C utilization through altering microbial community. Sci. Total Environ. 2020, 711, 135020. [Google Scholar] [CrossRef]
- Capek, P.; Choma, M.; Tahovska, K.; Kana, J.; Kopacek, J.; Santruckova, H. Coupling the resource stoichiometry and microbial biomass turnover to predict nutrient mineralization and immobilization in soil. Geoderma 2021, 385, 114884. [Google Scholar] [CrossRef]
- Zhu, X.; Li, Q.; Li, Y.; Han, H.; Ma, K. Eupatorium adenophorum invasion alters soil bacterial community and diversity. Biodivers. Sci. 2015, 23, 665–672. [Google Scholar] [CrossRef]
- Li, S.; Xie, D.; Ge, X.; Dong, W.; Luan, J. Altered diversity and functioning of soil and root-associated microbiomes by an invasive native plant. Plant Soil 2022, 473, 235–249. [Google Scholar] [CrossRef]
- Wu, B.; Wang, S.; Wei, M.; Zhou, J.; Jiang, K.; Du, D.; Wang, C. The invasive tree staghorn sumac affects soil N-2-fixing bacterial communities in north China. Plant Biol. 2019, 21, 951–960. [Google Scholar] [CrossRef]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.A.; Valverde, A. The Functional Potential of the Rhizospheric Microbiome of an Invasive Tree Species, Acacia dealbata. Microb. Ecol. 2019, 77, 191–200. [Google Scholar] [CrossRef]
- Lindstroem, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef]
- Yousuf, J.; Thajudeen, J.; Rahiman, M.; Krishnankutty, S.; Alikunj, A.P.; Abdulla, M.H.A. Nitrogen fixing potential of various heterotrophic Bacillus strains from a tropical estuary and adjacent coastal regions. J. Basic Microbiol. 2017, 57, 922–932. [Google Scholar] [CrossRef]
- Moseman, S.M.; Zhang, R.; Qian, P.Y.; Levin, L.A. Diversity and functional responses of nitrogen-fixing microbes to three wetland invasions. Biol. Invasions 2009, 11, 225–239. [Google Scholar] [CrossRef]
- Xu, C.-W.; Yang, M.-Z.; Chen, Y.-J.; Chen, L.-M.; Zhang, D.-Z.; Mei, L.; Shi, Y.-T.; Zhang, H.-B. Changes in non-symbiotic nitrogen-fixing bacteria inhabiting rhizosphere soils of an invasive plant Ageratina adenophora. Appl. Soil Ecol. 2012, 54, 32–38. [Google Scholar] [CrossRef]
- Quan, G.M.; Mao, D.J.; Zhang, J.E.; Xie, J.F.; Xu, H.Q.; An, M. Response of invasive Chromolaena odorata and two coexisting weeds to contrasting irradiance and nitrogen. Photosynthetica 2015, 53, 419–429. [Google Scholar] [CrossRef]
- Feng, Y.-L.; Lei, Y.-B.; Wang, R.-F.; Callaway, R.M.; Valiente-Banuet, A.; Inderjit; Li, Y.-P.; Zheng, Y.-L. Evolutionary tradeoffs for nitrogen allocation to photosynthesis versus cell walls in an invasive plant. Proc. Natl. Acad. Sci. USA 2009, 106, 1853–1856. [Google Scholar] [CrossRef]
- Yu, H.; Le Roux, J.J.; Jiang, Z.; Sun, F.; Peng, C.; Li, W. Soil nitrogen dynamics and competition during plant invasion: Insights from Mikania micrantha invasions in China. New Phytol. 2021, 229, 3440–3452. [Google Scholar] [CrossRef]




| Sample | Longitude | Latitude | Altitude | Functional Zone | Other |
|---|---|---|---|---|---|
| A | 107.094 | 22.778 | 229 | Experimental | The abandonment time is unknown, without trees, and shrubs, with grazing activities. |
| B | 106.978 | 22.798 | 378.53 | Experimental | The trees are 8 years old, with a density of 0.342 trees/m2, understory shrubs with Mussaenda pubescens Dryand. |
| C | 106.979 | 22.793 | 368 | Experimental | The trees are 1 year old, with a density of 0.188 trees/m2, understory shrubs with Vernicia fordii (Hemsl.) Airy Shaw |
| Sample | pH | SOC (g∙kg−1) | NH4+-N (mg∙kg−1) | NO3−-N (mg∙kg−1) | TK (g∙kg−1) | Biomass (kg) | Density |
|---|---|---|---|---|---|---|---|
| A | 8.00 ± 0.04 a | 40.97 ± 7.75 a | 8.61 ± 1.43 b | 2.82 ± 1.57 b | 12.61 ± 0.49 a | 0.64 ± 0.09 a | 10.56 ± 2.99 b |
| B | 5.37 ± 0.14 b | 32.58 ± 6.26 a | 19.72 ± 4.52 a | 4.25 ± 0.95 b | 6.29 ± 0.41 b | 0.04 ± 0.02 b | 31.00 ± 7.81 b |
| C | 4.99 ± 0.09 c | 40.64 ± 0.64 a | 17.97 ± 1 ab | 13.65 ± 1.92 a | 6.26 ± 0.09 b | 0.19 ± 0.02 b | 97.33 ± 32.04 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, X.; Zhao, C.; Li, J.; Li, B.; Zhu, J. Comparison of Bacterial Diversity in the Rhizosphere of Chromolaena odorata (L.) R.M. King and H.Rob. in Different Habitats. Sustainability 2023, 15, 2315. https://doi.org/10.3390/su15032315
Ni X, Zhao C, Li J, Li B, Zhu J. Comparison of Bacterial Diversity in the Rhizosphere of Chromolaena odorata (L.) R.M. King and H.Rob. in Different Habitats. Sustainability. 2023; 15(3):2315. https://doi.org/10.3390/su15032315
Chicago/Turabian StyleNi, Xinying, Caiyun Zhao, Junsheng Li, Bai Li, and Jinfang Zhu. 2023. "Comparison of Bacterial Diversity in the Rhizosphere of Chromolaena odorata (L.) R.M. King and H.Rob. in Different Habitats" Sustainability 15, no. 3: 2315. https://doi.org/10.3390/su15032315
APA StyleNi, X., Zhao, C., Li, J., Li, B., & Zhu, J. (2023). Comparison of Bacterial Diversity in the Rhizosphere of Chromolaena odorata (L.) R.M. King and H.Rob. in Different Habitats. Sustainability, 15(3), 2315. https://doi.org/10.3390/su15032315






