Lightweight Federated Learning for Rice Leaf Disease Classification Using Non Independent and Identically Distributed Images
Abstract
:1. Introduction
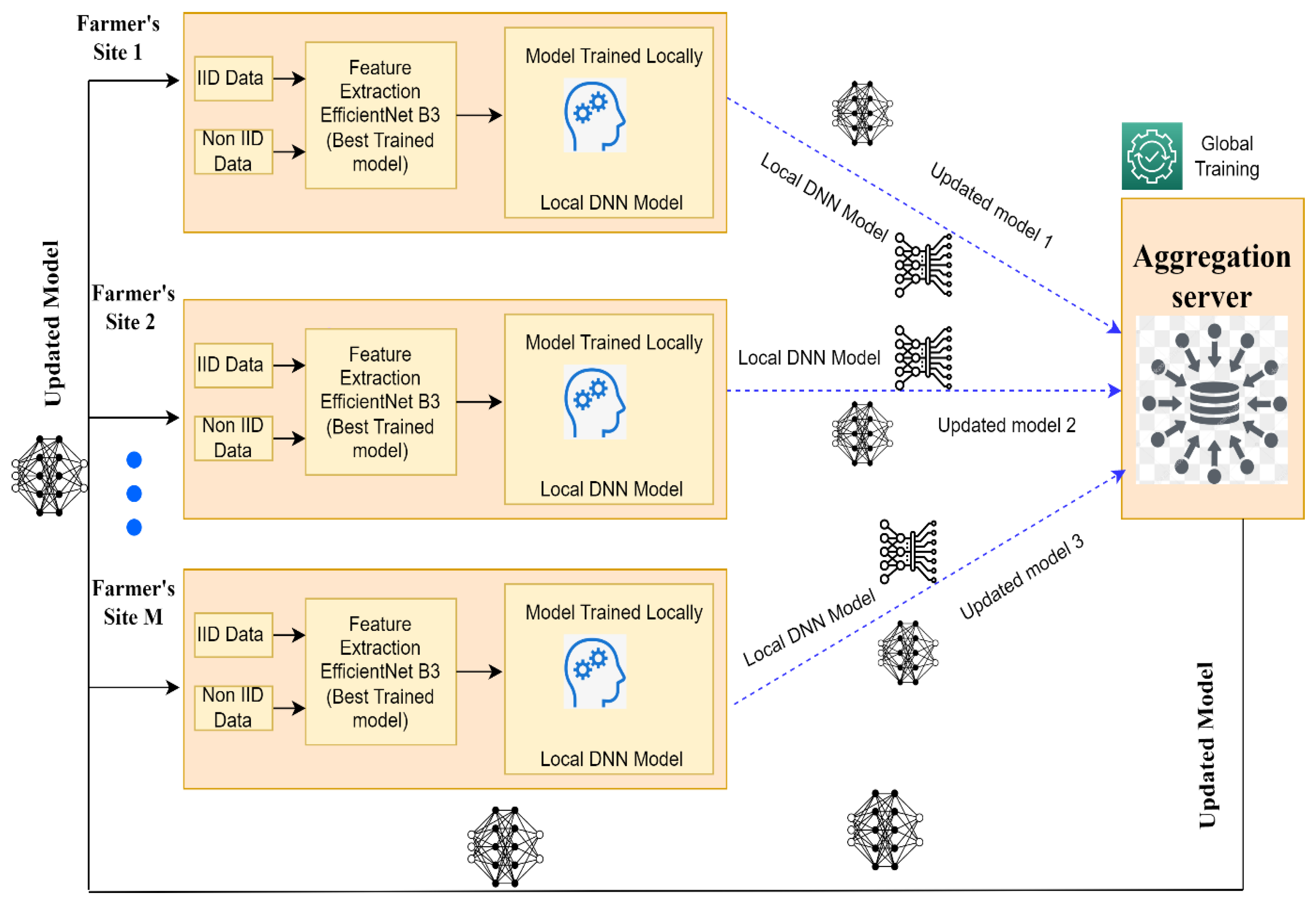
- i.
- Every client trains the model locally at their respective local site using their own data set (images of rice leaf disease), then uploads the locally trained model to the main server;
- ii.
- The central server integrates local models, updates, trains a global model, then provides access to the updated model for all clients;
- iii.
- Every client uses the parameters received by the globally trained model from the central server to both inform their own decisions and take part in the next cycle of model updates.
- i.
- Implementing a lightweight federated learning approach for rice leaf disease classification;
- ii.
- Implementing an improved distributed training model using federated learning with IID and non-IID datasets;
- iii.
- The proposed procedure has demonstrated effectiveness compared to present techniques with data privacy for rice leaf diseases classification.
2. Related Study
3. Materials and Methods
3.1. Federated Learning
| Algorithm 1 (FedAvg: Federated Learning) |
| M- Clients from 1 to n F- fractions of clients used per round B- Mini batch size (local) E- Epoch number (local) |
|
Server() //At Global Server Initialize global weights: ῳ0 for t = 1,2,3… do T ← max (F.M,1) Rt←(random sets of T clients) for client m Є Rt do parallel ClientUpdate(m, ῳt) ῳt+1 ← ∑Mm=1 ῳmt+1 end end |
|
Client () // At Client site ClientUpdate (m, ῳ): // on client site ẞ ← Split data into B Batches for each t from 1 to E do for batch b Є ẞ do Update client with weights ῳ end end return ῳ to server |
3.2. Data Collection and Pre-Processing
3.3. Feature Extraction
3.4. IID and Non-IID Data
3.5. Proposed Federated Learning Framework for Rice Leaf Disease Images Classification
3.6. Model Validation
4. Results
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wani, J.A.; Sharma, S.; Muzamil, M.; Ahmed, S.; Sharma, S.; Singh, S. Machine Learning and Deep Learning Based Computational Techniques in Automatic Agricultural Diseases Detection: Methodologies, Applications, and Challenges. In Archives of Computational Methods in Engineering; Springer: Dordrecht, The Netherlands, 2022; Volume 29, pp. 641–677. [Google Scholar]
- Kodama, T.; Hata, Y. Development of Classification System of Rice Disease Using Artificial Intelligence. In Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Miyazaki, Japan, 7–10 October 2018; pp. 3699–3702. [Google Scholar] [CrossRef]
- Kivrak, K.G.; Eker, T.; Sari, H.; Sari, D.; Akan, K.; Aydinoglu, B.; Catal, M.; Toker, C. Integration of Extra-Large-Seeded and Double-Podded Traits in Chickpea (Cicer arietinum L.). Agronomy 2020, 10, 901. [Google Scholar] [CrossRef]
- Rice Knowledge Bank. Available online: http://www.knowledgebank.irri.org/ericeproduction/Importance_of_Rice.htm (accessed on 29 July 2023).
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Image Processing Techniques for Diagnosing Rice Plant Disease: A Survey. Procedia Comput. Sci. 2020, 167, 516–530. [Google Scholar] [CrossRef]
- Narmadha, R.P.; Arulvadivu, G. Detection and Measurement of Paddy Leaf Disease Symptoms Using Image Processing. In Proceedings of the 2017 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 5–7 January 2017; pp. 5–8. [Google Scholar] [CrossRef]
- Kaur, A.; Guleria, K.; Trivedi, N. A Deep Learning Based Model for Rice Leaf Disease Detection. In Proceedings of the 10th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 13–14 October 2022; IEEE: Noida, India, 2022; pp. 1–5. [Google Scholar]
- Lu, Y.; Yi, S.; Zeng, N.; Liu, Y.; Zhang, Y. Identification of Rice Diseases Using Deep Convolutional Neural Networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Vimalajeewa, D.; Kulatunga, C.; Berry, D.P.; Balasubramaniam, S. A Service-Based Joint Model Used for Distributed Learning: Application for Smart Agriculture. IEEE Trans. Emerg. Top. Comput. 2022, 10, 838–854. [Google Scholar] [CrossRef]
- Chhabra, R.; Singh, S.; Khullar, V. Privacy Enabled Driver Behavior Analysis in Heterogeneous IoV Using Federated Learning. Eng. Appl. Artif. Intell. 2023, 120, 105881. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Tardy, M.; González Ballester, M.A.; Mateus, D.; Piella, G. Memory-Aware Curriculum Federated Learning for Breast Cancer Classification. Comput. Methods Programs Biomed. 2023, 229, 107318. [Google Scholar] [CrossRef] [PubMed]
- Truong, H.T.; Ta, B.P.; Le, Q.A.; Nguyen, D.M.; Le, C.T.; Nguyen, H.X.; Do, H.T.; Nguyen, H.T.; Tran, K.P. Light-Weight Federated Learning-Based Anomaly Detection for Time-Series Data in Industrial Control Systems. Comput. Ind. 2022, 140, 103692. [Google Scholar] [CrossRef]
- Zhang, H.; Bosch, J.; Olsson, H.H. Federated Learning Systems: Architecture Alternatives. In Proceedings of the 2020 27th Asia-Pacific Software Engineering Conference (APSEC), Singapore, 1–4 December 2020; pp. 385–394. [Google Scholar] [CrossRef]
- Islam, M.; Reza, M.T.; Kaosar, M.; Parvez, M.Z. Effectiveness of Federated Learning and CNN Ensemble Architectures for Identifying Brain Tumors Using MRI Images. Neural Process. Lett. 2023, 55, 3779–3809. [Google Scholar] [CrossRef]
- Durrant, A.; Markovic, M.; Matthews, D.; May, D.; Enright, J.; Leontidis, G. The Role of Cross-Silo Federated Learning in Facilitating Data Sharing in the Agri-Food Sector. Comput. Electron. Agric. 2022, 193, 106648. [Google Scholar] [CrossRef]
- Khullar, V.; Singh, H.P. F-FNC: Privacy Concerned Efficient Federated Approach for Fake News Classification. Inf. Sci. 2023, 639, 119017. [Google Scholar] [CrossRef]
- Aggarwal, M.; Khullar, V.; Goyal, N. Contemporary and Futuristic Intelligent Technologies for Rice Leaf Disease Detection. In Proceedings of the 2022 10th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 13–14 October 2022; IEEE: Noida, India, 2022; pp. 12–17. [Google Scholar]
- Ramesh, T.R.; Lilhore, U.K.; Poongodi, M.; Simaiya, S.; Kaur, A.; Hamdi, M. Predictive Analysis of Heart Diseases With Machine Learning Approaches. Malays. J. Comput. Sci. 2022, 1, 132–148. [Google Scholar] [CrossRef]
- Das, S.K.; Bebortta, S. Heralding the Future of Federated Learning Framework: Architecture, Tools and Future Directions. In Proceedings of the 2021 11th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Noida, India, 28–29 January 2021; pp. 698–703. [Google Scholar] [CrossRef]
- Aggarwal, M.; Khullar, V.; Goyal, N.; Singh, A.; Tolba, A.; Thompson, E.B.; Kumar, S. Pre-Trained Deep Neural Network-Based Features Selection Supported Machine Learning for Rice Leaf Disease Classification. Agriculture 2023, 13, 936. [Google Scholar] [CrossRef]
- Jiang, F.; Lu, Y.; Chen, Y.; Cai, D.; Li, G. Image Recognition of Four Rice Leaf Diseases Based on Deep Learning and Support Vector Machine. Comput. Electron. Agric. 2020, 179, 105824. [Google Scholar] [CrossRef]
- Krishnamoorthy, N.; Narasimha Prasad, L.V.; Pavan Kumar, C.S.; Subedi, B.; Abraha, H.B.; Sathishkumar, V.E. Rice Leaf Diseases Prediction Using Deep Neural Networks with Transfer Learning. Environ. Res. 2021, 198, 111275. [Google Scholar] [CrossRef]
- Rallapalli, S.M.; Saleem Durai, M.A. A Contemporary Approach for Disease Identification in Rice Leaf. Int. J. Syst. Assur. Eng. Manag. 2021, 1–11. [Google Scholar] [CrossRef]
- Prajapati, H.B.; Shah, J.P.; Dabhi, V.K. Detection and Classification of Rice Plant Diseases. Intell. Decis. Technol. 2017, 11, 357–373. [Google Scholar] [CrossRef]
- Kumar, P.R.; Kiran, R.; Singh, U.P.; Rathore, Y.; Janghel, R.R. Rice Leaf Disease Detection Using Mobile Net and Inception V.3. In Proceedings of the 2022 IEEE 11th International Conference on Communication Systems and Network Technologies (CSNT), Indore, India, 23–24 April 2022; IEEE: Indore, India, 2022; pp. 282–286. [Google Scholar]
- Azim, M.A.; Islam, M.K.; Rahman, M.M.; Jahan, F. An Effective Feature Extraction Method for Rice Leaf Disease Classification. Telkomnika (Telecommun. Comput. Electron. Control) 2021, 19, 463–470. [Google Scholar] [CrossRef]
- Pallathadka, H.; Ravipati, P.; Sekhar Sajja, G.; Phasinam, K.; Kassanuk, T.; Sanchez, D.T.; Prabhu, P. Application of Machine Learning Techniques in Rice Leaf Disease Detection. Mater. Today Proc. 2022, 51, 2277–2280. [Google Scholar] [CrossRef]
- Bhartiya, V.P.; Ram, J.R.; Yogesh, R.K. Rice Leaf Disease Prediction Using Machine Learning. In Proceedings of the 2022 Second International Conference on Power, Control and Computing Technologies (ICPC2T), Raipur, India, 1–3 March 2022; pp. 1–5. [Google Scholar]
- Khullar, V.; Singh, H.P. Privacy Protected Internet of Unmanned Aerial Vehicles for Disastrous Site Identification. Concurr. Comput. Pract. Exp. 2022, 34, e7040. [Google Scholar] [CrossRef]
- Lo, S.K.; Lu, Q.; Zhu, L.; Paik, H.Y.; Xu, X.; Wang, C. Architectural Patterns for the Design of Federated Learning Systems. J. Syst. Softw. 2022, 191, 111357. [Google Scholar] [CrossRef]
- Sheller, M.J.; Reina, G.A.; Edwards, B.; Martin, J.; Bakas, S. Multi-Institutional Deep Learning Modeling without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; Volume 11383, ISBN 9783030117221. [Google Scholar]
- Naeem, A.; Anees, T.; Naqvi, R.A.; Loh, W.K. A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis. J. Pers. Med. 2022, 12, 275. [Google Scholar] [CrossRef] [PubMed]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Deep Feature Based Rice Leaf Disease Identification Using Support Vector Machine. Comput. Electron. Agric. 2020, 175, 105527. [Google Scholar] [CrossRef]
- Dara, S.; Tumma, P. Feature Extraction by Using Deep Learning: A Survey. In Proceedings of the 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 29–31 March 2018; IEEE: Coimbatore, India, 2018; pp. 1795–1801. [Google Scholar]
- Ghosal, S.; Sarkar, K. Rice Leaf Diseases Classification Using CNN with Transfer Learning. In Proceedings of the 2020 IEEE Calcutta Conference (CALCON), Kolkata, India, 28–29 February 2020; pp. 230–236. [Google Scholar]
- Zhu, H.; Xu, J.; Liu, S.; Jin, Y. Federated Learning on Non-IID Data: A Survey. Neurocomputing 2021, 465, 371–390. [Google Scholar] [CrossRef]
- Dun, C.; Hipolito, M.; Jermaine, C.; Dimitriadis, D.; Kyrillidis, A. Efficient and Light-Weight Federated Learning via Asynchronous Distributed Dropout. In Proceedings of the 26th International Conference on Artificial Intelligence and Statistics, Valencia, Spain, 25–27 April 2023; Volume 206, pp. 1–31. [Google Scholar]
- Sharma, R.; Singh, A.; Kavita; Jhanjhi, N.Z.; Masud, M.; Jaha, E.S.; Verma, S. Plant Disease Diagnosis and Image Classification Using Deep Learning. Comput. Mater. Contin. 2022, 71, 2125–2140. [Google Scholar] [CrossRef]
- Haruna, Y.; Qin, S.; Mbyamm Kiki, M.J. An Improved Approach to Detection of Rice Leaf Disease with GAN-Based Data Augmentation Pipeline. Appl. Sci. 2023, 13, 1346. [Google Scholar] [CrossRef]
- Sudhesh, K.M.; Sowmya, V.; Sainamole Kurian, P.; Sikha, O.K. AI Based Rice Leaf Disease Identification Enhanced by Dynamic Mode Decomposition. Eng. Appl. Artif. Intell. 2023, 120, 105836. [Google Scholar] [CrossRef]
- Rani, S.; Ahmed, S.H.; Rastogi, R. Dynamic Clustering Approach Based on Wireless Sensor Networks Genetic Algorithm for IoT Applications. Wirel. Netw. 2020, 26, 2307–2316. [Google Scholar] [CrossRef]









| Reference | Area of Study | Algorithm | ML/DL | FL | Data Privacy | Evaluation Parameters |
|---|---|---|---|---|---|---|
| [21] | Rice leaf diseases | DCNN, SVM | √ | X | Not implemented | Accuracy = 96.8% |
| [22] | Rice leaf diseases | CNN InceptionNetV2 | √ | X | Not implemented | Accuracy = 95% |
| [23] | Rice leaf diseases | CNN, AlexNet M-Net | √ | X | Not implemented | Accuracy = 71.9% |
| [25] | Rice leaf diseases | MobileNet InceptionNetV2 | √ | X | Not implemented | Validation Accuracies 70.31%, 76.56% |
| [26] | Rice leaf diseases | XGBoost, SVM | √ | X | Not implemented | Accuracies 86.5%, 81% |
| [27] | Rice leaf diseases | CNN, SVM, Naïve Bayes, PCA | √ | X | Not implemented | better |
| [24] | Rice leaf diseases | KMeans, SVM | √ | X | Not implemented | Accuracy = 73.33% |
| [28] | Rice leaf diseases | Quadratic SVM | √ | X | Not implemented | Accuracy = 81.8% |
| [10] | Driver Behaviour | Bi-LSTM, CNN-Bi- LSTM, LSTM, CNN-LSTM | √ | √ | Implemented | Validation Accuracy = 89% |
| [29] | Disaster prediction | VGG16, DenseNet, ResNet, InceptionRes-NetV2 | √ | √ | Implemented | Validation Accuracy = 74% |
| [16] | Fake News | Bi-LSTM, CNN-Bi- LSTM, LSTM, CNN-LSTM | √ | √ | Implemented | Validation Accuracy = 90–92% |
| [9] | Analyse Milk Quality | CNN, LSQR, PLSR, NNPLS | √ | √ | Implemented | Mean Accuracy LSQR = 82% PLSR = 84% CNN = 87% NNPLS = 89% |
| Leaf Disease Name | Original Images | Training Data (80%) | Testing Data (20%) |
|---|---|---|---|
| Bacterial leafblight | 1584 | 1267 | 317 |
| Blast | 1440 | 1140 | 300 |
| Brown spot | 1600 | 1290 | 310 |
| Tungro | 1308 | 1048 | 260 |
| Total | 5932 | 4745 | 1187 |
| Pre-Trained Model | Input Shape | Size (MB) | Parameters | Depth | Feature Layer |
|---|---|---|---|---|---|
| DenseNet201 | (224-224-3) | 80 | 20.2M | 402 | 2D Global average pooling |
| EfficientNetB3 | (300-300-3) | 48 | 12.3M | 210 | Dropout |
| EfficientNetB4 | (380-380-3) | 75 | 19.5M | 258 | Dropout |
| EfficientNetB5 | (456-456-3) | 118 | 30.6M | 312 | Dropout |
| EfficientNetB6 | (528-528-3) | 166 | 43.3M | 360 | Dropout |
| InceptionResnetV3 | (229-229-3) | 215 | 55.9M | 449 | 2D Global average pooling |
| ResNet101 | (224-224-3) | 171 | 44.7M | 209 | 2D Global average pooling |
| ResNet101V2 | (224-224-3) | 171 | 44.7M | 205 | 2D Global average pooling |
| ResNet152 | (224-224-3) | 232 | 60.4M | 311 | 2D Global average pooling |
| ResNet152V2 | (224-224-3) | 232 | 60.4M | 307 | 2D Global average pooling |
| VGG16 | (224-224-3) | 528 | 138.4M | 16 | Dense |
| VGG19 | (224-224-3) | 549 | 143.7M | 19 | Dense |
| Xception | (229-229-3) | 88 | 22.9M | 81 | 2D Global average pooling |
| Training Accuracy | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epochs | DN201 | ENB3 | ENB4 | ENB5 | ENB6 | IRV2 | RN101 | RN101V2 | RN152 | RN152V2 | VGG16 | VGG19 | Xception |
| 25 | 0.89 | 0.98 | 0.85 | 0.88 | 0.86 | 0.94 | 0.86 | 0.82 | 0.86 | 0.75 | 0.87 | 0.85 | 0.95 |
| 50 | 0.95 | 0.99 | 0.9 | 0.94 | 0.93 | 0.96 | 0.92 | 0.88 | 0.92 | 0.83 | 0.93 | 0.91 | 0.97 |
| 75 | 0.96 | 0.99 | 0.93 | 0.96 | 0.95 | 0.97 | 0.93 | 0.91 | 0.94 | 0.86 | 0.95 | 0.94 | 0.98 |
| 99 | 0.97 | 0.99 | 0.95 | 0.97 | 0.96 | 0.98 | 0.93 | 0.93 | 0.96 | 0.88 | 0.96 | 0.95 | 0.98 |
| Validation Accuracy | |||||||||||||
| Epochs | DN201 | ENB3 | ENB4 | ENB5 | ENB6 | IRV2 | RN101 | RN101V2 | RN152 | RN152V2 | VGG16 | VGG19 | Xception |
| 25 | 0.91 | 0.98 | 0.87 | 0.91 | 0.84 | 0.92 | 0.87 | 0.81 | 0.85 | 0.79 | 0.88 | 0.86 | 0.92 |
| 50 | 0.94 | 0.98 | 0.91 | 0.94 | 0.95 | 0.95 | 0.91 | 0.88 | 0.91 | 0.83 | 0.94 | 0.91 | 0.96 |
| 75 | 0.96 | 0.99 | 0.87 | 0.91 | 0.96 | 0.96 | 0.94 | 0.91 | 0.93 | 0.84 | 0.96 | 0.91 | 0.97 |
| 99 | 0.97 | 0.99 | 0.93 | 0.97 | 0.96 | 0.96 | 0.91 | 0.93 | 0.95 | 0.87 | 0.96 | 0.96 | 0.97 |
| Federated Learning (IID) | |||||||
|---|---|---|---|---|---|---|---|
| Comm.Round | Epoch | Training Accuracy | Change (+/−) | Training Loss | Change (+/−) | Validation Accuracy | Validation Loss |
| 1 | 1 | 0.99 | 0.0 | 0.023 | −0.009 | 0.96 | 0.2 |
| 10 | 0.99 | 0.014 | |||||
| 2 | 1 | 0.98 | 0.01 | 0.031 | −0.02 | 0.98 | 0.1 |
| 10 | 0.99 | 0.011 | |||||
| 3 | 1 | 0.98 | 0.01 | 0.051 | −0.035 | 0.95 | 0.2 |
| 10 | 0.99 | 0.016 | |||||
| 4 | 1 | 0.99 | 0.0 | 0.041 | −0.033 | 0.99 | 0.06 |
| 10 | 0.99 | 0.008 | |||||
| 5 | 1 | 0.98 | 0.01 | 0.039 | −0.034 | 0.99 | 0.08 |
| 10 | 0.99 | 0.005 | |||||
| 6 | 1 | 0.99 | 0.01 | 0.024 | −0.02 | 1 | 0.009 |
| 10 | 1 | 0.004 | |||||
| 7 | 1 | 0.98 | 0.0 | 0.113 | −0.094 | 0.98 | 0.01 |
| 10 | 0.98 | 0.019 | |||||
| 8 | 1 | 0.97 | 0.01 | 0.06 | −0.04 | 0.97 | 0.33 |
| 10 | 0.98 | 0.02 | |||||
| 9 | 1 | 1 | −0.01 | 0.009 | −0.001 | 0.97 | 0.19 |
| 10 | 0.99 | 0.005 | |||||
| 10 | 1 | 0.98 | 0.01 | 0.05 | −0.004 | 0.99 | 0.03 |
| 10 | 0.99 | 0.006 | |||||
| Federated Learning (Non-IID) | |||||||
|---|---|---|---|---|---|---|---|
| Comm.Round | Epoch | Training Accuracy | Change (+/−) | Training Loss | Change (+/−) | Validation Accuracy | Validation Loss |
| 1 | 1 | 0.95 | 0.02 | 0.11 | −0.03 | 0.94 | 0.13 |
| 10 | 0.97 | 0.08 | |||||
| 2 | 1 | 0.96 | −0.01 | 0.15 | −0.04 | 1 | 0.02 |
| 10 | 0.95 | 0.11 | |||||
| 3 | 1 | 0.93 | 0.07 | 0.19 | −0.13 | 1 | 0.01 |
| 10 | 0.99 | 0.06 | |||||
| 4 | 1 | 1 | −0.04 | 0.02 | 0.03 | 0.96 | 0.1 |
| 10 | 0.96 | 0.05 | |||||
| 5 | 1 | 0.95 | 0.02 | 0.12 | −0.04 | 0.97 | 0.15 |
| 10 | 0.97 | 0.08 | |||||
| 6 | 1 | 0.97 | 0.01 | 0.09 | −0.04 | 0.96 | 0.18 |
| 10 | 0.98 | 0.05 | |||||
| 7 | 1 | 0.97 | 0.01 | 0.08 | −0.02 | 0.92 | 0.33 |
| 10 | 0.98 | 0.06 | |||||
| 8 | 1 | 0.95 | 0.02 | 0.22 | 0.08 | 0.94 | 0.17 |
| 10 | 0.97 | 0.14 | |||||
| 9 | 1 | 0.95 | 0.01 | 0.16 | −0.05 | 0.95 | 0.12 |
| 10 | 0.96 | 0.11 | |||||
| 10 | 1 | 0.97 | 0.02 | 0.07 | −0.03 | 0.95 | 0.08 |
| 10 | 0.99 | 0.04 | |||||
| Data | Accuracy | Loss | Precision | Recall-Rate | |
|---|---|---|---|---|---|
| Training | IID | 98.22 | 0.13 | 98.22 | 98.22 |
| Training | Non-IID | 96.17 | 0.13 | 96.1 | 96.54 |
| Evaluation | IID | 99.79 | 0.01 | 99.79 | 99.79 |
| Evaluation | Non-IID | 97.48 | 0.09 | 97.74 | 97.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aggarwal, M.; Khullar, V.; Goyal, N.; Alammari, A.; Albahar, M.A.; Singh, A. Lightweight Federated Learning for Rice Leaf Disease Classification Using Non Independent and Identically Distributed Images. Sustainability 2023, 15, 12149. https://doi.org/10.3390/su151612149
Aggarwal M, Khullar V, Goyal N, Alammari A, Albahar MA, Singh A. Lightweight Federated Learning for Rice Leaf Disease Classification Using Non Independent and Identically Distributed Images. Sustainability. 2023; 15(16):12149. https://doi.org/10.3390/su151612149
Chicago/Turabian StyleAggarwal, Meenakshi, Vikas Khullar, Nitin Goyal, Abdullah Alammari, Marwan Ali Albahar, and Aman Singh. 2023. "Lightweight Federated Learning for Rice Leaf Disease Classification Using Non Independent and Identically Distributed Images" Sustainability 15, no. 16: 12149. https://doi.org/10.3390/su151612149
APA StyleAggarwal, M., Khullar, V., Goyal, N., Alammari, A., Albahar, M. A., & Singh, A. (2023). Lightweight Federated Learning for Rice Leaf Disease Classification Using Non Independent and Identically Distributed Images. Sustainability, 15(16), 12149. https://doi.org/10.3390/su151612149








